Abstract
Background: β-ketolipoyl coenzyme A synthase (KCS) is an essential limiting catalyst involved in carbon chain elongation during fatty acid biosynthesis, characterized by strict substrate specificity. C18:1 (oleic acid) plays a vital role in cell membranes and is essential for nutrient storage and stress defense. There are indications of significant accumulation and rapid synthesis of C18:1 during the early growth stages of Ginkgo biloba L. episperm. The KCS gene family in G. biloba has yet to be analyzed, and the role of KCS in oleic acid synthesis remains unexplored. Methods: In this study, this issue was investigated using transcriptomic and metabolomic data, bioinformatics analysis to screen a key gene from the KCS gene family, and dual validation using yeast and Arabidopsis thaliana expression systems to probe its function. Results: A total of 11 members of the GbKCS gene family were identified, and the dynamics of these genes were analyzed during exocarp development in the G. biloba genome. Among them, the gene designated GbKCS7 showed a highly direct association with the content of C18:1. Heterologous expression of GbKCS7 in yeast increased C18:1N12 and C18:1 content by 3.18-fold and 2.07-fold, respectively. Overexpression of GbKCS7 in Arabidopsis showed that C18:1 was increased by 27.70% and 31.43% in GbKCS7-OE-1 and GbKCS7-OE-2 strains, correspondingly, in juxtaposition to the non-transgenic plants. In addition, the content of VLCFAs increased to varying degrees. Conclusions: These outcomes offer important insights for investigating the role of KCS genes in fatty acid synthesis to further improve G.biloba resistance.
1. Introduction
Land plants are susceptible to harsh conditions, such as water scarcity, salinity, and extreme temperatures, as well as pests and pathogens during growth and development. Fatty acids and their derivatives have attracted considerable attention due to their extensive involvement in the defense pathways of various plant species [1,2]. Among fatty acids, C16 and C18 fatty acids were not only core intermediates in lipid synthesis but also important antecedents engaged in the synthesis of very-long-chain fatty acids (VLCFAs), such as plant epidermal cuticle substances and waxes. Additionally, they were crucial for minimizing non-stomatal transpiration and conferring tolerance against environmental stresses, including drought, low temperatures, and UV exposure, alongside biotic challenges like pest and pathogen attacks [3,4,5,6].
A common unsaturated fatty acid in plants, C18:1 (oleic acid), served as an energy reserve component and precursor of bioactive molecules (mainly jasmonic acid and jasmonic acid methyl ester) and extracellular barriers (cutin and suberin), plying an important role in plant growth, development, and defense processes [7,8]. In Arabidopsis thaliana, C18:1 was extensively involved in defense signaling pathways mediated by salicylic acid (SA) and jasmonic acid (JA) [9]. Notably, loss-of-function mutations in SSI2, which encoded the predominant stearoyl-ACP desaturase (SACPD) isoform, caused a significant reduction in C18:1 level. These metabolic changes induced developmental defects, including stunted growth and spontaneous necrotic lesions in Arabidopsis, while concurrently suppressing jasmonate signaling and compromising disease resistance against fungal pathogens, including B. cinerea [10,11]. In Arabidopsis, C18:1 was found to stimulate phospholipase D (PLD) activity, initiating programmed cell death through H2O2 signaling while concurrently enhancing tolerance to abiotic stresses [12].
Plant fatty acid biosynthesis occurred through two distinct phases: (1) de novo synthesis of C16 and C18 fatty acids catalyzed by the fatty acid synthase (FAS) complex, followed by (2) elongation to VLCFAs (C > 18) mediated by the endoplasmic reticulum-localized fatty acid elongase (FAE) complex [13,14]. The fatty acid elongase (FAE) complex comprised four enzymatic components: β-ketoacyl-CoA synthase (KCS), β-ketoacyl-CoA reductase (KCR), β-hydroxyacyl-CoA dehydratase (HCD), and enoyl-CoA reductase (ECR). These enzymes sequentially mediated the four-step elongation cycle involving condensation, reduction, dehydration, and final reduction reactions during fatty acid chain extension [15,16]. Exhibiting precise substrate selectivity, KCS served as the pivotal rate-controlling enzyme during fatty acid chain elongation. The final fatty acid profile, including chain length and saturation pattern, was principally regulated by the enzyme’s catalytic properties and substrate binding affinity [17,18,19]. There was a fine division in the KCS gene family. Studies in Arabidopsis revealed that while AtKCS1 and AtKCS11 demonstrated catalytic competence toward both saturated and mono-unsaturated C16–C24 acyl-CoA substrates, AtKCS17 displayed preferential activity for saturated C16–C22 acyl-CoA species [20]. AtKCS1 expression in heterologous systems (yeast and Nicotiana benthamiana) elongated C16 fatty acids into C18–C22 derivatives [21]. Moreover, AtKCS2 and AtKCS20 mainly synthesized C22 and C24 and AtKCS5 and AtKCS6 mainly synthesized C24–C28, while FAE1/AtKCS18 preferentially catalyzed C20/C22 production [22,23].
An ancient relict gymnosperm plant, G.biloba L. (G. biloba) is renowned for its significant medicinal, nutritional, and ornamental value [24,25,26,27]. Known for its high resistance to various stresses, G. biloba demonstrates remarkable adaptability to diverse environments and has a long lifespan. G. biloba seeds comprise four parts: the fleshy outer seed coat (episperm), bony middle seed coat, membranous inner seed coat, and kernel. As it matures, the surface of the episperm becomes coated with a hydrophobic white waxy substance [28]. The fleshy sarcocarp has an important protective function for G. biloba seeds before the embryo is fully developed [29]. Therefore, it is generally believed that the episperm of G. biloba is an important protective tissue. Through the pre-laboratory analysis of metabolomic data from exocarp stages—EP1 (early stage of exocarp differentiation), EP2 (rapid growth stage of exocarp), and EP3 (mature stage of exocarp)—we observed that oleic acid was rapidly synthesized and accumulated during the early developmental stage of the exocarp, particularly in the C18:1 metabolite trend. β-ketoacyl-CoA synthase is crucial for fatty acid production; however, research on the KCS gene family remains lacking, and GbKCS functions remain unexplored in G. biloba. Our understanding of this enzyme’s role in oleic acid synthesis was also limited. To address these gaps, we characterized the GbKCS gene family and identified GbKCS7 through transcriptomic analysis as a key gene associated with C18:1 fatty acid production. Subsequent functional validation in yeast and Arabidopsis systems confirmed its ability to enhance oleic acid accumulation, demonstrating its biological significance in plant lipid metabolism.
2. Materials and Methods
2.1. Systematic Analysis of GbKCS Genes in G. biloba Genome
Protein homology comparisons were performed using the BLASTp program in TBtools software to identify the KCS gene in the genome downloaded from the G. biloba Genome Database (http://gigadb.org/dataset/100209, accessed on 6 December 2023) [30]. The NCBI-CDD (https://www.ncbi.nlm.nih.gov/cdd, accessed on 6 December 2023) and Pfam (http://pfam.xfam.org/, accessed on 6 December 2023) databases were used to search for conserved structural domains of the resulting KCS to ensure data accuracy. Prior transcriptome and metabolomics data of G. biloba episperm were used to screen KCS genes showing varied expression during G. biloba episperm development [31]. Then, the submitted KCS protein sequences were analyzed using PROTPARAM (https://web.expasy.org/protparam/, accessed on 17 December 2023) for the isoelectric point, instability coefficient, hydrophilic index, and molecular weight to analyze their physicochemical properties. The subcellular localization of GbKCS proteins was predicted using CELLO software (http://cello.life.nctu.edu.tw/, accessed on 17 December 2023).
2.2. Mutiple Sequence Alignment and Phylogenetic Analysis
We performed multiple sequence alignments between AtKCS protein sequences, obtained from the Arabidopsis Information Repository (TAIR) database (https://www.arabidopsis.org/, accessed on 21 December 2023), and GbKCS proteins using the Clustal Omega online program (https://www.ebi.ac.uk/Tools/msa/clustalo/, accessed on 21 December 2023). The alignment results were subsequently visualized and displayed using Jalview software (Jalview version 2.10.5). The apple (Malus × domestica Borkh.) MdKCS protein sequences were downloaded from the apple genome database (https://iris.angers.inra.fr/gddh13/the-apple-genome-downloads.html, accessed on 21 December 2023) [32]. To study the phylogenetic relationship of GbKCS protein, the evolutionary tree was constructed by MEGA 11.0 using 21 Arabidopsis AtKCS, 28 apple (Malus × domestica) MdKCS, and 11 GbKCS protein sequences. The parameter settings were as follows: phylogeny: maximum likelihood (ML) tree, test of phylogeny: bootstrap method, bootstrap replication: 1000. The phylogenetic results were beautified using the online website ITOL (https://itol.embl.de/, accessed on 21 December 2023).
2.3. Chromosomal Location, Conserved Motifs and Gene Structure Analysis
The structure of the GbKCS was determined using the gff3 annotation file from G. biloba transcriptome data, and its chromosomal location was mapped with the MapGene2 Chromosome (MG2C v2.1) (http://mg2c.iask.in/mg2c_v2.0/, accessed on 25 December 2023). The gene structure was visualized using the Gene Structure Display Server (GSDS 2.0) (http://gsds.gao-lab.org/, accessed on 25 December 2023), while the conserved motifs of protein sequences were predicted and analyzed using the MEME (https://meme-suite.org/meme/tools/meme, accessed on 25 December 2023) online program. Default settings were applied, and the resulting XML format file was submitted to TBtools software for the visualization of conserved motifs [30].
2.4. Prediction of Cis-Acting Elements and Expression Analyis of GbKCS Genes
The upstream 2000 bp sequences of all the GbKCS genes were extracted from the genomic DNA sequence and submitted to PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 5 January 2024) to predict the potential cis-acting elements in the promoter region, and the cis-acting elements were analyzed using the TBtools software [33]. Additionally, the number, function, and type of cis-acting elements in GbKCS genes were compiled. FPKM-based heatmaps by TBtools software displayed GbKCS expression dynamics in G. biloba episperm. Correlation analysis was performed between the content of C18:1 and the expression levels of GbKCS genes, joining analysis of transcriptome-metabolome data.
2.5. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR) Analysis
To confirm the confidence of the transcriptome data, we designed primers for all GbKCS family members, normalized the data between samples using GbGAPDH as a housekeeping gene, and detected these genes by qRT-PCR [34]. Primer sequences were presented in the Supplementary Material (Table S1). RNA was extracted and reverse-transcribed using the Plant RNA Kit (OMEGA, Norcross, GA, USA) and RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA), respectively. Quantitative RT-PCR was performed using the SYBR Green Premix Pro Taq HS qPCR Kit (High Rox Plus (Accurate Biology), Hunan, China) and the ABI Step One Plus real-time PCR system. Total RNA was extracted from the samples (G. biloba episperm at three developmental stages) and synthesized into first-strand cDNA, and the reaction system containing cDNA was used for qRT-PCR. The parameters were set as pre-denaturation at 95 °C for 30 s, denaturation at 95 °C for 5 s, and annealing extension at 60 °C for 30 s for a total of 40 cycles, and a melting curve was plotted. Internal replicates were included, and three biological replicates were performed for all samples. Gene expression quantification was performed via the 2−△△Ct analytical method.
2.6. Gene Cloning and Vector Construction
The coding sequence (CDS) of GbKCS7 was amplified from G. biloba episperm cDNA using gene-specific primer P1 and subsequently ligated into the pClone007 Versatile Simple Vector (Tsingke, Cat. 007VS) for downstream applications. The positive clones were sequenced to confirm the accuracy of the cloned fragments. The CDS sequence of the GbKCS7 gene was cloned into the pCAMBIA1300 vector using a specific primer (P2) containing BamH I and Xbal I restriction sites, fused to the GFP reporter under the control of the CaMV35S promoter. The recombinant vector was named 35S: GbKCS7-GFP. The coding sequence was directionally inserted into the pYES2 expression vector through BamH I and Xho I restriction sites engineered into primer P3, generating the recombinant construct designated pYES2-GbKCS7. Primer sequences are enumerated in the Supplementary Material (Table S1).
2.7. Subcellular Localization of GbKCS7 Gene
The Agrobacterium tumefaciens strain GV3101 containing the 35S: GbKCS7-GFP or 35S: GFP vector was infiltrated into N. benthamiana leaves. The GFP fluorescence signal was detected using a fluorescence microscope (Carl Zeiss, Oberkochen, Germany). The DsRed protein, connected with AtPIP2A (a cellular membrane aquaporin), was co-expressed with 35S: GbKCS7-GFP or 35S: GFP in N. benthamiana leaves [35].
2.8. Ectopic Expression of GbKCS7 Gene in Yeast
Plasmids pYES2-GbKCS7 and pYES2 were transformed into yeast (INVscI)-competent cells using Gene Pulser Xcell (BIO-RAD, Alfred Nobel Drive Hercules, CA, USA). Transformed yeast cells were cultured in a synthetic complete medium lacking uracil (SC/-Ura) containing 2% (w/v) glucose, followed by induction in SC/-Ura medium supplemented with 2% (w/v) galactose and 2% (w/v) raffinose for 24 h.
2.9. Generation of GbKCS7 Overexpression Lines in Arabidopsis
The Arabidopsis plants were transformed with the 35S: GbKCS7-GFP vector via floral dipping [36]. Homozygous T3 transgenic lines were identified over three consecutive generations through hygromycin (25 mg L−1) resistance screening on Murashige and Skoog (MS) solid medium. The genomic DNA of resistant seedlings was used as a template and verified by PCR using a specific primer (P16). The primer (P16) was designed from a 750 bp sequence between the CDS of the GbKCS7 and GFP sequence. Quantitative RT-PCR analysis of GbKCS7 expression in positive transgenic plants utilized AtACT2 as a control gene and primers P11 and P17. Primer sequences are listed in the Supplementary Material (Table S1).
2.10. Extraction and GC-MS-Based Analysis of Fatty Acid Composition
The methods for extracting and analyzing fatty acids were adapted from previous research with minor adjustments [21]. A suitable volume of yeast suspension (OD600 = 1.4) was taken, and fatty acids were extracted from the yeast. The appropriate amount of Arabidopsis leaves was weighed and the fatty acids were extracted from them. An internal standard of 5 μl nonacosanoic acid (10 mg/mL) was incorporated. The fatty acid profile was determined via gas chromatography–mass spectrometry (GC-MS) with the specified equipment: (1) a Trace 1300 gas chromatograph (Thermo Fisher) with TG-FAME column (550 m × 0.25 mm × 0.20 μm), (2) helium carrier gas at constant flow (0.63 mL/min), and (3) an ISQ 7000 mass detector (Thermo Fisher) with EI ionization source The content of fatty acids was relatively quantified according to the intensity of the peaks related to the content of nonacosanoic acid.
2.11. Statistical Analysis
Each measurement involved three separate biological replicates. Specific primers were engineered employing Primer 5.0 software. Data processing was carried out using Microsoft Excel 2021 (Redmond, WA, USA). The statistical analysis for the t-test was performed using SPSS 26.0 (IBM, New York, NY, USA), with a significance threshold set at p < 0.05. Data from three independent experiments were analyzed and presented as mean values ± standard deviation using GraphPad Prism version 9.0 for graphical representation. Adobe Illustrator(Adobe Inc., Mountain View, CA, USA) was utilized to enhance and refine the images.
3. Results
3.1. Variation of C16:1 and C18:1 Content in G. biloba Episperm at Diverse Ontogenetic Phases
The relative contents of C16:1 and C18:1 were analyzed based on metabolomic data of G. biloba episperm at different growth periods. Along with the development of episperm, the relative content of C16:1 gradually decreased from 5.2 × 106 to 1.6 × 106. The range of C18:1 content was between 2.6 × 108 and 3.2 × 108 and showed a downward trend at the Ep3 stage (Figure 1).
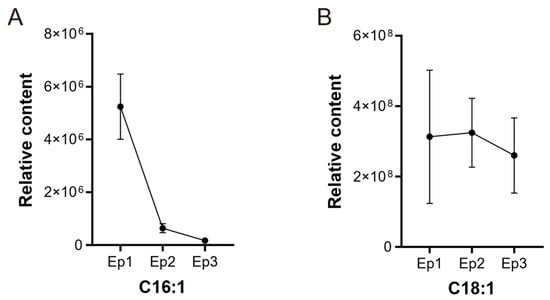
Figure 1.
The relative content of two fatty acids during distinct developmental phases of G. biloba episperm. (A) The relative content of C16:1. (B) The relative content of C18:1.
3.2. Identification of the GbKCS Genes and Their Location in G. biloba Chromosomes
In silico analysis revealed the presence of 11 KCS gene family members in the G. biloba genomic sequence, and they were evenly spread on 9 chromosomes (Figure 2). Chromosome 1 (Chr1) and Chr3 each possess two GbKCS genes, and the remaining 7 chromosomes each possess one GbKCS gene. The identified GbKCS genes were successively named from GbKCS1 to GbKCS11 according to their position on the chromosome.
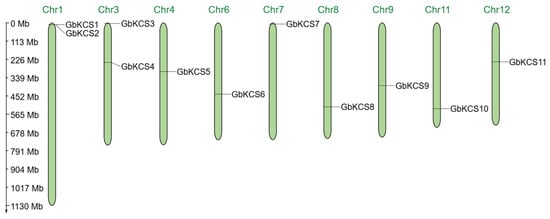
Figure 2.
Chromosomal localization of the GbKCS genes. The top margin of every green vertical bar contained its assigned chromosome number designation, which reflected the approximate chromosomal locations of individual GbKCS genes across the genome.
The CDS length of the 11 GbKCS genes ranges from 1377 bp (GbKCS1) to 1827 bp (GbKCS5). The GbKCS proteins exhibited variable physicochemical parameters (Table S2), with GbKCS5 (608 amino acids) and GbKCS1 (458 amino acids) showing the maximum and minimum lengths, correspondingly. The theoretical isoelectric point of these KCS proteins ranges from 7.82 (GbKCS9) to 9.36 (GbKCS1), all of which are greater than 7.0, indicating that they are basic proteins. The average grand value of hydropathicity (GRAVY) ranges from −1.150 (GbKCS6) to 0.019 (GbKCS2), suggesting that most GbKCSs are hydrophilic proteins.
3.3. Phylogenetic Analysis of GbKCS Genes and Their Homologues from Other Species
Evolutionary relationships between G. biloba and other species were inferred from comparative analysis of 11 GbKCS and 21 AtKCS proteins, including sequence alignment and domain conservation assessment. These analyses revealed that all identified proteins possessed two characteristic conserved domains: FAE1_CUT1_RppA and ACP_syn_III_C (Supplementary Figure S1). These domains exhibited remarkable sequence conservation across all examined KCS proteins, suggesting their critical functional importance in the enzymatic activity. In addition, the C-terminus of GbKCS proteins is highly conserved, whereas the N-terminus is more flexible.
A total of 60 KCS protein sequences derived from A. thaliana (AtKCS), M. domestica (MdKCS), and G. biloba (GbKCS) were used to assess the phylogenetic relationship (Figure 3). A total of 21 AtKCSs were segregated into four main groups (KCS1-like, FAE1-like, FDH-like, and CER6) [32,37]. They were further classified into eight subgroups, including α, β, γ, δ, ζ, ε, η, and θ, based on protein homology relationships and structural features [38,39]. The θ subgroup contains the most members of GbKCSs and MdKCSs. The α, γ, δ, ζ, and ε subgroups each contained 1 GbKCS protein. Additionally, the η subgroup contained 2 GbKCS proteins (GbKCS1 and GbKCS2), while no GbKCS and MdKCS are classified into the β subgroup.
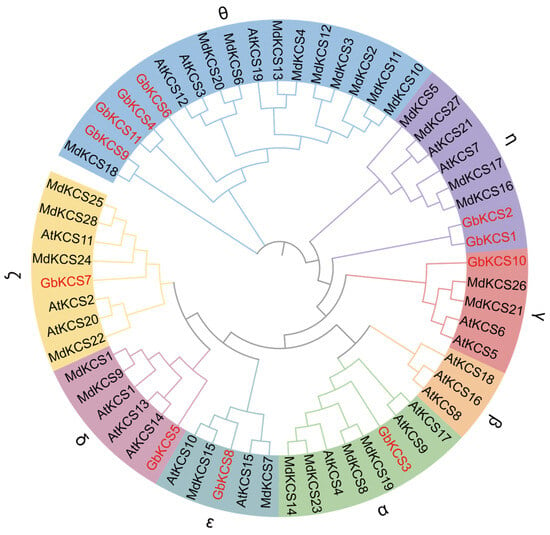
Figure 3.
Phylogenetic tree of the KCS gene family in A. thaliana, G. biloba, and M. domestica. The evolutionary tree was generated from full-length protein sequences employing maximum likelihood algorithms. Eight subgroups were assigned in disparate shades, while G. biloba KCS family members were highlighted in red for immediate identification.
3.4. Conserved Motifs and Gene Structure of the GbKCS Genes
Most GbKCS proteins have identical type and number of conserved motifs (Figure 4A). Four GbKCS proteins (GbKCS1, GbKCS4, GbKCS6, and GbKCS11) contain 12 conserved motifs, and GbKCS2 contains 13 conserved motifs, while the remaining 6 proteins contain 14 conserved motifs. All identified proteins consistently contained motifs 1 through 10 and motif 12 in their sequences. GbKCS proteins of the same subgroup exhibit similar patterns of conserved motifs. For example, GbKCS4, GbKCS6, and GbKCS11 have the same type and number of conserved motifs. While only GbKCS1 and GbKCS2 contain the motif 15.
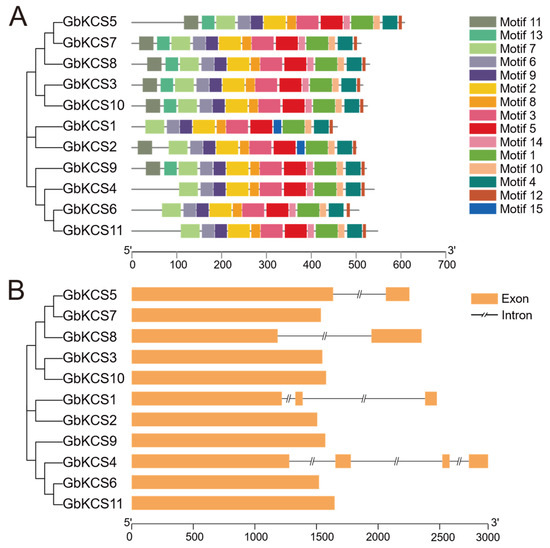
Figure 4.
The conserved motifs and gene structure analysis of GbKCS genes. (A) The conserved motifs of GbKCS proteins. (B) The gene structure of GbKCS. Different motifs are shown as boxes with different colors. The yellow bar represents exons (expressed regions).
Analysis of the GbKCS gene structure established that seven GbKCS genes were intronless. GbKCS5 and GbKCS8, which belong to the δ and ε subgroups, respectively, each contained one intron. GbKCS1, which is part of the η subgroup, contained two introns. In contrast, GbKCS4, which belongs to the θ subgroup, contained three introns and four exons (Figure 4B).
3.5. Cis-Acting Element Analysis of the GbKCS Gene Promoters
The cis-acting element prediction information of the 2000 bp promoter sequence upstream of GbKCS demonstrated that there were four types of elements in this segment: plant hormone signaling, stress adaptation, plant growth and development, and light response (Figure 5A). The MeJA response elements (CGTCA-motif and TGACG-motif) were found in 7 GbKCS genes (GbKCS1, GbKCS2, GbKCS4, GbKCS5, GbKCS7, GbKCS9, and GbKCS10) (Figure 5B). The gibberellin response elements P-box and GARE-motif were identified in 5 and 2 GbKCS genes, respectively. In addition, phytohormone response elements such as abscisic acid (ABRE), auxin (AuxRR-core and TGA-element), and salicylic acid (TCA-element) were found. These discoveries implied that most of the GbKCS genes were potentially under the control of a variety of phytohormones, the modulation of the expression of which methyl jasmonate (MeJA) and gibberellin may have been vital. Furthermore, stress response elements were detected in the promoter portions of the GbKCS genes. Including MBS, LTR, ARE, Wbox, and WUN-motif, these cis-elements were associated with defense and stress processes such as drought induction, low temperature response, anerobic induction, pathogen trauma response, and mechanical injury response. The promoter regions of GbKCS genes also contained cis-elements associated with plant growth, development (CAT-box, MSA-like, and circadian), and light response.
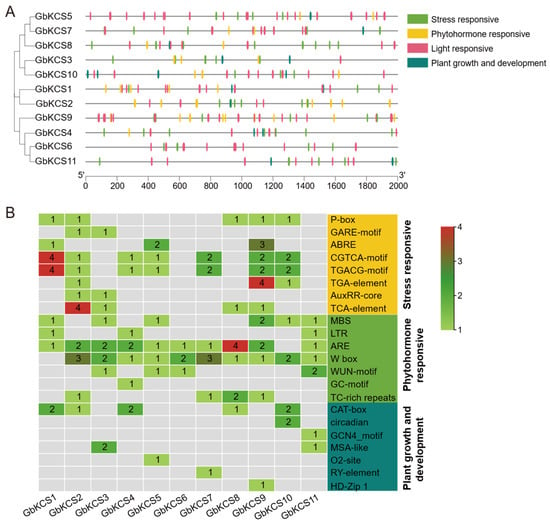
Figure 5.
The cis-acting elements analysis in GbKCS genes. (A) Location of the four classifications of cis-acting elements in the promoter region. (B) Name and number of plant growth and development, stress response, and phytohormone response cis-acting elements of GbKCS genes. Distinct color codes were assigned to categorize various classes of cis-regulatory elements.
3.6. Expression Pattern of the GbKCS Genes
In light of the clustering of expression patterns, the 11 GbKCS genes were categorized into upregulated and downregulated expression patterns (Figure 6A). During episperm development, all GbKCS genes were downregulated, except for GbKCS3. Notably, most GbKCS genes were highly expressed at the Ep1 stage, except for GbKCS7, which showed predominant expression at both the Ep1 (FPKM = 80.18) and Ep2 (FPKM = 76.36) stages. Additionally, qRT-PCR validation of GbKCS expression profiles demonstrated strong concordance with the FPKM values, verifying the reliability of our transcriptomic data (Supplemental Figure S2).
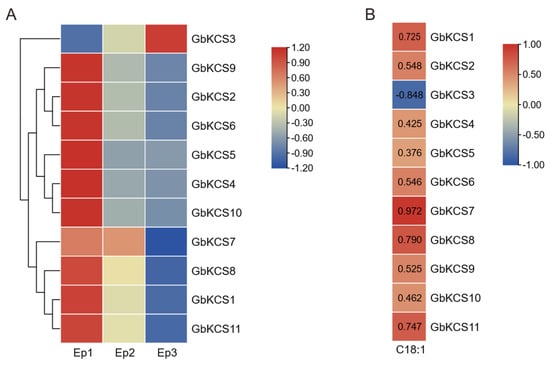
Figure 6.
Heatmap of GbKCS gene expression in G. biloba episperm and correlation analysis with C18:1 content. (A) Heatmap of the GbKCS genes’ expression in G. biloba episperm at different developmental stages. (B) Heatmap of Pearson correlation between C18:1 and GbKCS genes in G. biloba episperm, with red and blue color codes denoting high and low correlation values, individually.
Pearson correlation analysis was performed between C18:1 content and the expression levels of the GbKCS genes to explore the key GbKCS genes involved in C18:1 biosynthesis. The outcomes revealed that the C18:1 content in the episperm was positively associated with the expression of GbKCS, except for GbKCS3 (Figure 6B). Among these, the correlation coefficient between the C18:1 content and expression of GbKCS7 was 0.972, indicating a significant correlation between them. Thus, we hypothesized that GbKCS7 likely participates in C18:1 biosynthesis in G. biloba episperm.
3.7. Cloning and Subcellular Localization of GbKCS7 Gene
Sequence analysis predicted GbKCS7 to encode 511 amino acids, with its full-length CDS measuring 1536 base pairs (Figure S3). The outcomes of subcellular localization indicate that the GbKCS7-GFP was highly abundant in the plasma membrane (Figure 7). Thus, the protein encoded by the GbKCS7 gene functions in the plasma membrane.
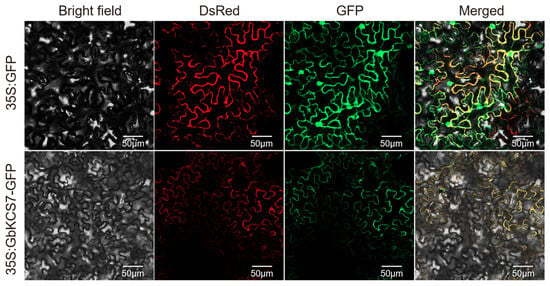
Figure 7.
Subcellular localization of the GbKCS7 gene. 35S: GFP and 35S: GbKCS7-GFP represent the control and GbKCS7 expression vectors, respectively. They were co-expressed with DsRed fused with plasma membrane marker protein in N. benthamiana leaf epidermal cells. Scale bar = 50 μm.
3.8. Heterologous Expression of GbKCS7 Gene and Analysis of Fatty Acid Composition in Yeast
The pYES2-GbKCS7 vector was transformed into yeast (INVscI) for ectopic expression, and the total fatty acids in transformed yeast cells were extracted for qualitative and quantitative analysis using GC-MS. The outcomes revealed that while the overexpression of GbKCS7 did not affect the types of fatty acids in yeast cells, it did alter the content of certain fatty acids (Figure 8 and Figure S4). Specifically, the C16:1 content was significantly reduced to 20.42% of that in the control group (CK) due to GbKCS7. Conversely, compared to CK, the levels of C18:1N12 and C18:1 rose by 3.18- and 2.07-fold, respectively. Moreover, the contents of C18:0, C20:1, and C22:1 also grew by 48%, 85%, and 34%, separately. These findings suggest that GbKCS7 primarily catalyzes the extension process from C16:1 to C18:1 and accelerates the subsequent biosynthesis of C18–C22 fatty acids.
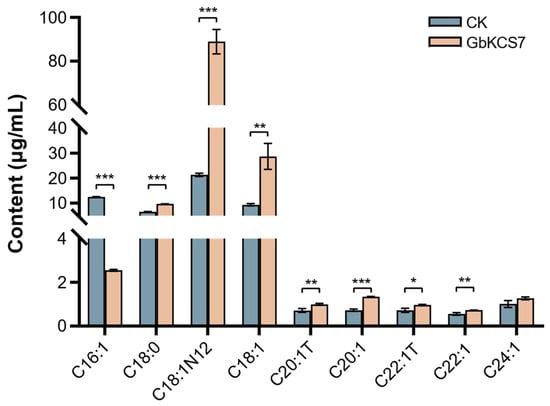
Figure 8.
Changes in the fatty acid composition of the yeast by the induction of GbKCS7. Data represent mean ± standard error (SE) of three biological replicates. The *, **, and *** show significance at p < 0.05, p < 0.01 and p < 0.001, respectively, among control and overexpression (GbKCS7) groups according to Student’s t-test.
3.9. Overexpression of GbKCS7 Gene and Analysis of Fatty Acid Composition in Arabidopsis
The 35S: GbKCS7-GFP vector was transformed into Arabidopsis, and aggregate 5 homozygous transgenic lineages were confirmed by PCR with specific primers (Figure S5). Following this, qRT-PCR was applied to measure the relative expression of GbKCS7 between different lines (Figure S6). GbKCS7-OE-1 and GbKCS7-OE-2, two lines with the highest expression level of GbKCS7, were picked for further examination. Morphological observation demonstrates that overexpression of the GbKCS7 gene exerted no significant effect on the growth of transgenic lines (Figure S6).
The results of the fatty acid composition we measured in the leaves of transgenic Arabidopsis and WT showed that C18:1 in the GbKCS7-OE-1 and GbKCS7-OE-2 lines increased by 27.70% and 31.43%, respectively, compared with that in WT (Figure 9). This result further validates our speculation that the GbKCS7 gene specially catalyzes the extension of C16:1 to produce C18:1. In addition, in the GbKCS7-OE-1 and GbKCS7-OE-2, C20:1 was increased by 12.96% and 11.87%, C20:0 was increased by 39.44% and 14.98%, and C22:0 was increased by 180.36% and 170.94%, respectively.
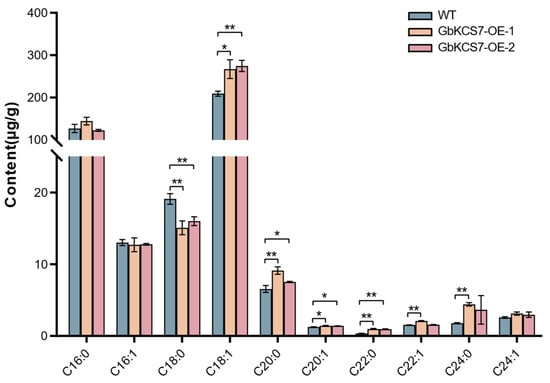
Figure 9.
Fatty acid composition of Arabidopsis plants. Data represent mean ± standard error (SE) of three biological replicates. The * and ** show significance at p < 0.05 and p < 0.01, respectively, among WT and GbKCS7-OE Arabidopsis according to Student’s t-test.
4. Discussion
In plants, the KCS genes are comprehensively associated with the growth and development processes. For instance, in cotton (Gossypium barbadense), the GbKCS genes are closely associated with fiber elongation [40,41]. The VvKCS genes have a potential role in grapevine (Vitis vinifera L.) pollen wall formation [42]. Beyond involvement in growth and development, KCS genes are widely involved in plant resistance in response to both biological and environmental stressors. For example, the heterologous expression of CsKCS6 in Arabidopsis promotes the accumulation of VLCFAs, which then improves the tolerance of transgenic plants to drought [43]. VvKCS11 (grapevine L.) and CqKCS2B.1 (Chenopodium quinoa) improve the resistance of transgenic Arabidopsis to salt stress [44,45].
In this investigation, 11 KCS genes identified from G. biloba genomic and transcriptomic data that were variably expressed in the episperm all had two completely conserved domains (FAE1_CUT1_RppA and ACP_syn_III_C), which were typical of the KCS gene family. The FAE1_CUT1_RppA structural domain encompassed the protein’s active site and regions involved in substrate binding [46]. Meanwhile, in the context of both plant and bacterial systems, the ACP_syn_III_C structural domain was crucial for beginning the fatty acid synthesis enzyme process chain, facilitating the combination interaction between acetyl-CoA and malonyl ACP [47]. GbKCS proteins were weakly basic, and our analysis of the amino acid sequences of AtKCS and GbKCS proteins revealed that the N terminus of KCS proteins was less conserved. This finding aligned with observations in sorghum (Sorghum bicolor) [48], leading us to hypothesize that this might be linked to variations in substrate-binding recognition, despite the presence of conservation. The multispecies phylogenetic tree revealed that KCS proteins were separated into eight subfamilies, aligning with the findings in Arabidopsis [38]. Conversely, the 11 GbKCS gene-encoded proteins were organized into seven subclades, with any GbKCS proteins found in the FAE-like (β) subclade, similar to those in peanut and passion fruit [13,17].
The identification and analysis of cis-acting elements within gene promoter regions were instrumental in predicting their potential functions and transcriptional regulation mechanisms. Previous investigations had indicated that GbKCS genes might be a key factor in plant hormone signal transduction and stress response [49]. In this investigation, we identified numerous cis-acting elements upstream of the GbKCS gene, including methyl jasmonate elements (CGTCA-motif, TGACG-motif), abscisic acid response element (ABRE), MYB binding site (MBS), and anaerobic response element (ARE). In sweet cherry (Prunus avium L.), exogenous MeJA significantly increased the expression levels of KCS1 and KCS6 and increased the waxy component in the epidermis of ripe fruit [50]. Cuticular wax is an important barrier that reduces fruit water loss and resists environmental stress and was essential for the proper development of fruits and seeds [29].
The distinct expression patterns of GbKCS genes in G. biloba episperm suggested that they played essential roles at various developmental stages. Specifically, the expression level of GbKCS3 gradually elevated as the exocarp matured, whereas that of GbKCS7 was highly expressed during Ep1 and Ep2 but was downregulated at Ep3. In citrus, Cs7g13310 and Cs7g28170 were highly expressed in the exocarp of fruits during early developmental stages, whereas CsKCS2 (Cs6g02360) and CsKCS11 (Cs4g17260) were prominently expressed during fruit ripening stages, indicating their potential roles in wax synthesis in both young and ripe fruits [19]. Based on these observations, we hypothesized that GbKCS7 was crucial for fatty acid synthesis and accumulation during the pre-expansion stage of exocarp development. Further expression studies in yeast and Arabidopsis systems were necessary to validate its functions.
Heterologous expression in yeast is a common method to study the substrate specificity and catalytic function of proteins encoded by KCS genes in plants [22]. Several studies have shown that KCS genes in multiple species, such as Arabidopsis and rape, can exhibit catalytic activity in wild-type or Δelo3 yeast [20,51,52]. Functional characterization of HaKCS1 through heterologous expression in Helianthus annuus in wild-type yeast increased the accumulation of VLCFAs, specifically C22:0, C24:0, and C26:0 [53]. Using the same method to discover the catalytic function of five ZmKCS genes in maize (Zea mays), ZmKCS2 produced C20 and C22 VLCFAs; ZmKCS4, ZmKCS11, and ZmKCS20 produced C24; and ZmKCS15 is involved in the synthesis of C26 [54]. In this study, the GbKCS7 gene was heterologously expressed in yeast, mainly manifested by the decrease in C16:1 and the increase in C18:1, C20:1, and C22:1. This result is similar to that of Batsale et al. [21], where AtKCS1 extended the C16 fatty acid in yeast to C18 up to C22. It follows that both GbKCS7 and AtKCS1 seem to exhibit the ability to produce C18:1.
In Arabidopsis, the overexpression of GbKCS7 resulted in a notable rise in the levels of C18:1 and C20:1, along with an increase in the content of saturated fatty acids such as C20:0 and C22:0. This proposes that GbKCS7 is participating in the synthesis of C18:1 and extension of VLCFAs. However, the increase in saturated fatty acid content is not reflected in the yeast expression system, which may be due to the lack of a specific cofactor or a certain KCS protein interacting with GbKCS7, leading to functional differences from those shown in yeast. Some proteins in plants are cofactors for FAE to complete the carbon chain elongation process, for instance, the synthesis of VLCFAs requires the participation of CER2-like proteins in Arabidopsis and rice [55,56,57]. Moreover, it was found that homo- and hetero-dimerization can be formed between KCS2, KCS6, and KCS9 proteins in Arabidopsis, indicating that FAE may contain a variety of KCS and that KCS can form heterodimers [58]. The research of Huang et al. showed that KCS3 was able to bind to KCS6 to form a heterodimer, resulting in a decrease in KCS6 activity [59]. In both yeast and Arabidopsis, the outcomes of observing overexpression of GbKCS substantiated the findings from the early-stage combined metabolomic and transcriptomic analyses. This evidence underscores the pivotal role of KCS7 in oleic acid accumulation and its significant impact on modifying VLCFA content.
Fatty acids C22 and C24 serve as crucial substrates in the synthesis of cuticle wax and aliphatic suberins, with the penetrability of the plant epidermis typically linked to the cuticular wax content. KCS2/DAISY and KCS20 in Arabidopsis extend C20 to C22, and thus participate in the production and regulation of cuticle wax and aliphatic suberins [60]. Overexpression of the CqKCS2B.1 gene from quinoa in Arabidopsis promoted the accumulation of C22 and C24 and increased the salt resistance of the plants [44]. MdKCS2 and CsKCS6 diminish the permeability of the Arabidopsis epidermis and enhance the tolerance of plants to abiotic stresses [43,61]. In this research, overexpression of GbKCS7 resulted in an increased C22 content in Arabidopsis, which may enhance the plant’s resilience to abiotic stress. Nevertheless, to better inform genetic breeding efforts aimed at developing highly resistant G. biloba varieties, it is essential to further analyze its biological function concerning the cuticular wax content in overexpressed Arabidopsis.
5. Conclusions
In G. biloba episperm, we identified 11 GbKCS genes differentially expressed in this study, and their physicochemical properties, phylogeny, cis-acting elements, and expression pattern were evaluated to furnish new understanding of the gene family. A gene named GbKCS7 was selected, and its function in fatty acid biosynthesis was further verified. Heterologous expression of GbKCS7 increased the content of C18:1N12 and C18:1 in yeast by 3.18- and 2.07-fold, respectively, and this gene increased the content of C18:1 and VLCFAs in Arabidopsis and reduced the permeability of the leaf epidermis. Our findings demonstrate that GbKCS7 plays an essential function in fatty acid synthesis regulation in G. biloba episperm and has a potential role in plant endurance to abiotic stress. This information provides a reference for genetic breeding with high resistance of G. biloba varieties.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes16070773/s1, Table S1. Primer sequences for PCR amplification and qRT-PCR, Table S2. Physiochemical properties of the GbKCS genes; Figure S1. Multi-sequence alignment and domain analysis of the AtKCS and GbKCS Proteins. Figure S2. Transcriptome FPKM expression data were verified by qRT-PCR. Figure S3. PCR electrophoresis diagram of GbKCS7 genes. Figure S4. Heatmap of fatty acid content clustering in yeast overexpressing. Figure S5. Arabidopsis genomic DNA was used as a template for PCR validation. Figure S6. Relative expression levels of GbKCS7 in overexpressed Arabidopsis plants. Figure S7. Phenotype of GbKCS7 overexpressing Arabidopsis.
Author Contributions
Conceptualization, X.Z., K.F., M.L. and Y.W.; methodology, X.Z., K.F., M.L. and Y.W.; software, X.Z.; validation, X.Z. and K.F.; formal analysis, X.Z.; investigation, Z.F., Z.Y., J.L., S.Z., X.M., Y.W. and F.W.; writing—original draft preparation, X.Z. and K.F.; writing—review and editing, X.Z., K.F., Z.F., M.L. and Y.W.; supervision, M.L. and Y.W.; project administration, Y.W.; funding acquisition, Y.W. All authors have read and agreed to the published version of the manuscript.
Funding
The National Natural Science Foundation of China (32171842); The National Natural Science Foundation of China (31570682); The Central South University of Forestry and Technology graduate Science and Technology Innovation Fund (2023CX02004); Innovation Project of Central South University of Forestry and Technology (90102-63223068).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The transcriptome data of G. biloba episperm has been deposited as a BioProject under accession PRJNA1026889.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Lim, G.H.; Singhal, R.; Kachroo, A.; Kachroo, P. Fatty Acid– and Lipid-Mediated Signaling in Plant Defense. Annu. Rev. Phytopathol. 2017, 55, 505–536. [Google Scholar] [CrossRef] [PubMed]
- Xiao, R.; Zou, Y.; Guo, X.; Li, H.; Lu, H. Fatty acid desaturases (FADs) modulate multiple lipid metabolism pathways to improve plant resistance. Mol. Biol. Rep. 2022, 49, 9997–10011. [Google Scholar] [CrossRef] [PubMed]
- García Coronado, H.; Tafolla Arellano, J.C.; Hernández Oñate, M.Á.; Burgara Estrella, A.J.; Robles Parra, J.M.; Tiznado Hernández, M.E. Molecular Biology, Composition and Physiological Functions of Cuticle Lipids in Fleshy Fruits. Plants 2022, 11, 1133. [Google Scholar] [CrossRef] [PubMed]
- Lewandowska, M.; Keyl, A.; Feussner, I. Wax biosynthesis in response to danger: Its regulation upon abiotic and biotic stress. New Phytol. 2020, 227, 698–713. [Google Scholar] [CrossRef]
- Philippe, G.; Sørensen, I.; Jiao, C.; Sun, X.; Fei, Z.; Domozych, D.S.; Rose, J.K.C. Cutin and suberin: Assembly and origins of specialized lipidic cell wall scaffolds. Curr. Opin. Plant Biol. 2020, 55, 11–20. [Google Scholar] [CrossRef]
- Zhukov, A.; Popov, V. Synthesis of C20–38 Fatty Acids in Plant Tissues. Int. J. Mol. Sci. 2022, 23, 4731. [Google Scholar] [CrossRef]
- He, M.; Qin, C.-X.; Wang, X.; Ding, N.-Z. Plant Unsaturated Fatty Acids: Biosynthesis and Regulation. Front. Plant Sci. 2020, 11, 390. [Google Scholar] [CrossRef]
- Walley, J.W.; Kliebenstein, D.J.; Bostock, R.M.; Dehesh, K. Fatty acids and early detection of pathogens. Curr. Opin. Plant Biol. 2013, 16, 520–526. [Google Scholar] [CrossRef]
- Kachroo, A.; Venugopal, S.C.; Lapchyk, L.; Falcone, D.; Hildebrand, D.; Kachroo, P. Oleic acid levels regulated by glycerolipid metabolism modulate defense gene expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 5152–5157. [Google Scholar] [CrossRef]
- Gao, Q.M.; Venugopal, S.; Navarre, D.; Kachroo, A. Low Oleic Acid-Derived Repression of Jasmonic Acid-Inducible Defense Responses Requires the WRKY50 and WRKY51 Proteins. Plant Physiol. 2011, 155, 464–476. [Google Scholar] [CrossRef]
- Kachroo, P.; Venugopal, S.C.; Navarre, D.A.; Lapchyk, L.; Kachroo, A. Role of Salicylic Acid and Fatty Acid Desaturation Pathways in ssi2-Mediated Signaling. Plant Physiol. 2005, 139, 1717–1735. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, C.; Qin, C.; Wood, T.; Olafsdottir, G.; Welti, R.; Wang, X. The Oleate-Stimulated Phospholipase D, PLDδ, and Phosphatidic Acid Decrease H2O2-Induced Cell Death in Arabidopsis. Plant Cell 2003, 15, 2285–2295. [Google Scholar] [CrossRef] [PubMed]
- Rizwan, H.M.; Shaozhong, F.; Li, X.; Bilal Arshad, M.; Yousef, A.F.; Chenglong, Y.; Shi, M.; Jaber, M.Y.M.; Anwar, M.; Hu, S.-Y.; et al. Genome-Wide Identification and Expression Profiling of KCS Gene Family in Passion Fruit (Passiflora edulis) Under Fusarium kyushuense and Drought Stress Conditions. Front. Plant Sci. 2022, 13, 872263. [Google Scholar] [CrossRef] [PubMed]
- Xue, D.; Zhang, X.; Lu, X.; Chen, G.; Chen, Z.-H. Molecular and Evolutionary Mechanisms of Cuticular Wax for Plant Drought Tolerance. Front. Plant Sci. 2017, 8, 621. [Google Scholar] [CrossRef]
- Kogure, K.; Watanabe, A.; Ito, Y. Interaction of ONION2 ketoacyl CoA synthase with ketoacyl CoA reductase of rice. Mol. Biol. Rep. 2022, 49, 1643–1647. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, C.; Li, X.; Liu, Z.-Y.; Liu, X.; Wang, C.-L. Comprehensive analysis of KCS gene family in pear reveals the involvement of PbrKCSs in cuticular wax and suberin synthesis and pear fruit skin formation. Plant Mol. Biol. 2023, 112, 341–356. [Google Scholar] [CrossRef]
- Huai, D.; Xue, X.; Li, Y.; Wang, P.; Li, J.; Yan, L.; Chen, Y.; Wang, X.; Liu, N.; Kang, Y.; et al. Genome-Wide Identification of Peanut KCS Genes Reveals That AhKCS1 and AhKCS28 Are Involved in Regulating VLCFA Contents in Seeds. Front. Plant Sci. 2020, 11, 406. [Google Scholar] [CrossRef]
- Millar, A.A.; Kunst, L. Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J. 1997, 12, 121–131. [Google Scholar] [CrossRef]
- Yang, H.; Mei, W.; Wan, H.; Xu, R.; Cheng, Y. Comprehensive analysis of KCS gene family in Citrinae reveals the involvement of CsKCS2 and CsKCS11 in fruit cuticular wax synthesis at ripening. Plant Sci. 2021, 310, 110972. [Google Scholar] [CrossRef]
- Blacklock, B.J.; Jaworski, J.G. Substrate specificity of Arabidopsis 3-ketoacyl-CoA synthases. Biochem. Biophys. Res. Commun. 2006, 346, 583–590. [Google Scholar] [CrossRef]
- Batsale, M.; Alonso, M.; Pascal, S.; Thoraval, D.; Haslam, R.P.; Beaudoin, F.; Domergue, F.; Joubès, J. Tackling functional redundancy of Arabidopsis fatty acid elongase complexes. Front. Plant Sci. 2023, 14, 1107333. [Google Scholar] [CrossRef] [PubMed]
- Batsale, M.; Bahammou, D.; Fouillen, L.; Mongrand, S.; Joubès, J.; Domergue, F. Biosynthesis and Functions of Very-Long-Chain Fatty Acids in the Responses of Plants to Abiotic and Biotic Stresses. Cells 2021, 10, 1284. [Google Scholar] [CrossRef] [PubMed]
- Tresch, S.; Heilmann, M.; Christiansen, N.; Looser, R.; Grossmann, K. Inhibition of saturated very-long-chain fatty acid biosynthesis by mefluidide and perfluidone, selective inhibitors of 3-ketoacyl-CoA synthases. Phytochemistry 2012, 76, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Biernacka, P.; Adamska, I.; Felisiak, K. The Potential of Ginkgo biloba as a Source of Biologically Active Compounds—A Review of the Recent Literature and Patents. Molecules 2023, 28, 3993. [Google Scholar] [CrossRef]
- Liu, W.; Zou, M.; Wang, Y.; Cao, F.; Su, E. Ginkgo Seed Proteins: Characteristics, Functional Properties and Bioactivities. Plant Foods Hum. Nutr. 2021, 76, 281–291. [Google Scholar] [CrossRef]
- Wang, L.; Cui, J.; Jin, B.; Zhao, J.; Xu, H.; Lu, Z.; Li, W.; Li, X.; Li, L.; Liang, E.; et al. Multifeature analyses of vascular cambial cells reveal longevity mechanisms in old Ginkgo biloba trees. Proc. Natl. Acad. Sci. USA 2020, 117, 2201–2210. [Google Scholar] [CrossRef]
- Yang, J.; Feng, Z.; Liu, W.; Wang, Y.; Wang, G.; Yu, W.; Yang, G.; Yang, T.; Wang, Y.; Li, M. Exogenous hormone on episperm development and ginkgolic acid accumulation in Ginkgo biloba L. Ind. Crops Prod. 2021, 160, 113–140. [Google Scholar] [CrossRef]
- Li, F.; Liu, G.; Zhao, L.; Gao, X.; Shen, Z.; Cao, F.; Guo, Q. Morphological Characteristics, Ultrastructure, and Chemical Constituents of the Endotesta in Ginkgo (Ginkgo biloba L.). Plants 2023, 12, 3560. [Google Scholar] [CrossRef]
- Barry, C.S. Factors Influencing the Ripening and Quality of Fleshy Fruits. In Annual Plant Reviews Online; Blackwell Publishing Ltd.: Hoboken, NJ, USA, 2018; pp. 296–325. [Google Scholar]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Feng, Z.; Fan, K.; Yao, Z.; Wang, H.; Wu, X.; Tang, L.; Wang, Q.; Wang, Y.; Wang, Y.; Li, M. Integrative metabolome and transcriptome analysis reveals GbKCS and GbMYB involved in the biosynthesis of ginkgolic acids. Ind. Crops Prod. 2024, 220, 119225. [Google Scholar] [CrossRef]
- Lian, X.Y.; Wang, X.; Gao, H.N.; Jiang, H.; Mao, K.; You, C.X.; Li, Y.Y.; Hao, Y.J. Genome wide analysis and functional identification of MdKCS genes in apple. Plant Physiol. Biochem. 2020, 151, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Du, Z.; Huo, X.; Zhou, J.; Chen, Y.; Zhang, J.; Pan, A.; Wang, X.; Wang, F.; Zhang, J. Genome-wide analysis of PRR gene family uncovers their roles in circadian rhythmic changes and response to drought stress in Gossypium hirsutum L. PeerJ 2020, 8, e9936. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Han, X.; Qu, Y.; Zhang, Y.; Rong, H.; Wu, K.; Xu, L. Genome-Wide Identification of the Ginkgo (Ginkgo biloba L.) LBD Transcription Factor Gene and Characterization of Its Expression. Int. J. Mol. Sci. 2022, 23, 5474. [Google Scholar] [CrossRef] [PubMed]
- Nelson, B.K.; Cai, X.; Nebenführ, A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007, 51, 1126–1136. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Agrobacterium. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Costaglioli, P.; Joubès, J.; Garcia, C.; Stef, M.; Arveiler, B.; Lessire, R.; Garbay, B. Profiling candidate genes involved in wax biosynthesis in Arabidopsis thaliana by microarray analysis. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2005, 1734, 247–258. [Google Scholar] [CrossRef]
- Joubès, J.; Raffaele, S.; Bourdenx, B.; Garcia, C.; Laroche-Traineau, J.; Moreau, P.; Domergue, F.; Lessire, R. The VLCFA elongase gene family in Arabidopsis thaliana: Phylogenetic analysis, 3D modelling and expression profiling. Plant Mol. Biol. 2008, 67, 547–566. [Google Scholar] [CrossRef]
- Mo, L.; Yao, X.; Tang, H.; Li, Y.; Jiao, Y.; He, Y.; Jiang, Y.; Tian, S.; Lu, L. Genome-Wide Investigation and Functional Analysis Reveal That CsKCS3 and CsKCS18 Are Required for Tea Cuticle Wax Formation. Foods 2023, 12, 2011. [Google Scholar] [CrossRef]
- Rui, C.; Chen, X.; Xu, N.; Wang, J.; Zhang, H.; Li, S.; Huang, H.; Fan, Y.; Zhang, Y.; Lu, X.; et al. Identification and Structure Analysis of KCS Family Genes Suggest Their Reponding to Regulate Fiber Development in Long-Staple Cotton Under Salt-Alkaline Stress. Front. Genet. 2022, 13, 812449. [Google Scholar] [CrossRef]
- Xiao, G.H.; Wang, K.; Huang, G.; Zhu, Y.X. Genome-scale analysis of the cotton KCS gene family revealed a binary mode of action for gibberellin A regulated fiber growth. J. Integr. Plant Biol. 2016, 58, 577–589. [Google Scholar] [CrossRef]
- Zheng, H.; Liang, Y.; Hong, B.; Xu, Y.; Ren, M.; Wang, Y.; Huang, L.; Yang, L.; Tao, J. Genome-Scale Analysis of the Grapevine KCS Genes Reveals Its Potential Role in Male Sterility. Int. J. Mol. Sci. 2023, 24, 6510. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Wu, Q.; Yang, L.; Hu, W.; Liu, D.; Liu, Y. Ectopic Expression of CsKCS6 From Navel Orange Promotes the Production of Very-Long-Chain Fatty Acids (VLCFAs) and Increases the Abiotic Stress Tolerance of Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 564656. [Google Scholar] [CrossRef] [PubMed]
- Tariq, F.; Zhao, S.; Ahmad, N.; Wang, P.; Shao, Q.; Ma, C.; Yang, X. Overexpression of β-Ketoacyl CoA Synthase 2B.1 from Chenopodium quinoa Promotes Suberin Monomers’ Production and Salt Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 13204. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Yang, X.; Dong, S.; Ge, Y.; Zhang, X.; Zhao, X.; Han, N. Overexpression of β-Ketoacyl-CoA Synthase From Vitis vinifera L. Improves Salt Tolerance in Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 564385. [Google Scholar] [CrossRef]
- Funa, N.; Ohnishi, Y.; Ebizuka, Y.; Horinouchi, S. Alteration of reaction and substrate specificity of a bacterial type III polyketide synthase by site-directed mutagenesis. Biochem. J. 2002, 367, 781–789. [Google Scholar] [CrossRef]
- Abbadi, A.; Brummel, M.; Schütt, B.S.; Slabaugh, M.B.; Schuch, R.; Spener, F. Reaction mechanism of recombinant 3-oxoacyl-(acyl-carrier-protein) synthase III from Cuphea wrightii embryo, a fatty acid synthase type II condensing enzyme. Biochem. J. 1999, 345, 153–160. [Google Scholar] [CrossRef]
- Zhang, A.; Xu, J.; Xu, X.; Wu, J.; Li, P.; Wang, B.; Fang, H. Genome-wide identification and characterization of the KCS gene family in sorghum (Sorghum bicolor (L.) Moench). PeerJ 2022, 10, e14156. [Google Scholar] [CrossRef]
- Porto, M.S.; Pinheiro, M.P.N.; Batista, V.G.L.; dos Santos, R.C.; de Albuquerque Melo Filho, P.; de Lima, L.M. Plant Promoters: An Approach of Structure and Function. Mol. Biotechnol. 2014, 56, 38–49. [Google Scholar] [CrossRef]
- Balbontín, C.; Gutiérrez, C.; Schreiber, L.; Zeisler-Diehl, V.V.; Marín, J.C.; Urrutia, V.; Hirzel, J.; Figueroa, C.R. Alkane biosynthesis is promoted in methyl jasmonate-treated sweet cherry (Prunus avium) fruit cuticles. J. Sci. Food Agric. 2024, 104, 530–535. [Google Scholar] [CrossRef]
- Katavic, V.; Mietkiewska, E.; Barton, D.L.; Giblin, E.M.; Reed, D.W.; Taylor, D.C. Restoring enzyme activity in nonfunctional low erucic acid Brassica napus fatty acid elongase 1 by a single amino acid substitution. Eur. J. Biochem. 2002, 269, 5625–5631. [Google Scholar] [CrossRef]
- Paul, S.; Gable, K.; Beaudoin, F.; Cahoon, E.; Jaworski, J.; Napier, J.A.; Dunn, T.M. Members of the Arabidopsis FAE1-like 3-Ketoacyl-CoA Synthase Gene Family Substitute for the Elop Proteins of Saccharomyces cerevisiae. J. Biol. Chem. 2006, 281, 9018–9029. [Google Scholar] [CrossRef] [PubMed]
- González Mellado, D.; Salas, J.J.; Venegas Calerón, M.; Moreno Pérez, A.J.; Garcés, R.; Martínez Force, E. Functional characterization and structural modelling of Helianthus annuus (sunflower) ketoacyl-CoA synthases and their role in seed oil composition. Planta 2019, 249, 1823–1836. [Google Scholar] [CrossRef] [PubMed]
- Stenback, K.E.; Flyckt, K.S.; Hoang, T.; Campbell, A.A.; Nikolau, B.J. Modifying the yeast very long chain fatty acid biosynthetic machinery by the expression of plant 3-ketoacyl CoA synthase isozymes. Sci. Rep. 2022, 12, 13235. [Google Scholar] [CrossRef] [PubMed]
- Haslam, T.M.; Kunst, L. Extending the story of very-long-chain fatty acid elongation. Plant Sci. 2013, 210, 93–107. [Google Scholar] [CrossRef]
- Haslam, T.M.; Mañas-Fernández, A.; Zhao, L.; Kunst, L. Arabidopsis ECERIFERUM2 Is a Component of the Fatty Acid Elongation Machinery Required for Fatty Acid Extension to Exceptional Lengths. Plant Physiol. 2012, 160, 1164–1174. [Google Scholar] [CrossRef]
- Wang, X.; Guan, Y.; Zhang, D.; Dong, X.; Tian, L.; Qu, L.Q. A β-Ketoacyl-CoA Synthase Is Involved in Rice Leaf Cuticular Wax Synthesis and Requires a CER2-LIKE Protein as a Cofactor. Plant Physiol. 2017, 173, 944–955. [Google Scholar] [CrossRef]
- Kim, J.; Kim, R.J.; Lee, S.B.; Suh, M.C. Protein–protein interactions in fatty acid elongase complexes are important for very-long-chain fatty acid synthesis. J. Exp. Bot. 2022, 73, 3004–3017. [Google Scholar] [CrossRef]
- Huang, H.; Yang, X.; Zheng, M.; Chen, Z.; Yang, Z.; Wu, P.; Jenks, M.A.; Wang, G.; Feng, T.; Liu, L.; et al. An ancestral role for 3-KETOACYL-COA SYNTHASE3 as a negative regulator of plant cuticular wax synthesis. Plant Cell 2023, 35, 2251–2270. [Google Scholar] [CrossRef]
- Lee, S.B.; Suh, M.C. Regulatory mechanisms underlying cuticular wax biosynthesis. J. Exp. Bot. 2022, 73, 2799–2816. [Google Scholar] [CrossRef]
- Lian, X.Y.; Gao, H.N.; Jiang, H.; Liu, C.; Li, Y.Y. MdKCS2 increased plant drought resistance by regulating wax biosynthesis. Plant Cell Rep. 2021, 40, 2357–2368. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).