Abstract
Background: The late embryogenesis abundant (LEA) gene family plays a critical role in abiotic stress tolerance during plant growth and development. Myricaria laxiflora, as a key pioneer species in the extreme hydrological fluctuation zone of the Yangtze River, has evolved unique adaptation mechanisms potentially linked to gene family evolution. However, the molecular mechanisms underlying how the LEA gene family responds to alternating flooding–drought cycles remain unclear. Methods and Results: In this study, we identified 31 LEA genes through whole-genome and transcriptome analyses using bioinformatics approaches, and classified them into nine subfamilies based on protein sequence similarity. These genes were distributed across 12 chromosomes. Our analysis revealed that LEA promoters contain cis-acting elements associated with anaerobic induction, abscisic acid (ABA) response, and combined low-temperature/light stress, suggesting their role in a multi-tiered environmental signal integration network. Spatio-temporal expression profiling further indicated that root-specific LEA genes maintain cellular integrity via membrane lipid binding, while leaf-predominant members cooperate with the antioxidant system to mitigate photoinhibition damage. Conclusions: This study elucidates the dynamic regulatory mechanisms of the LEA gene family during flooding-drought adaptation in M. laxiflora, providing molecular targets for ecological restoration in the Yangtze River Basin.
1. Introduction
As an endemic species in the Yangtze River Basin, Myricaria laxiflora (Tamaricaceae) is the flagship species of fragile habitats in this area. Its natural population is concentrated in the water-level-fluctuating zone of the main stream and tributaries of the Yangtze River (altitude of 145–175 m). It will suffer from 8 months of submergence and serious sediment erosion and deposition in a year. By relying on floods to achieve seed dispersal and germination, M. laxiflora can rapidly colonize the new tidal flat after the seasonal floods subside and become a constructive species of pioneer plant communities in the hydro-fluctuation belt [1,2].
M. laxiflora shows unique morphological and physiological adaptation characteristics: scaly leaves can reduce transpiration, and well-developed aerenchyma roots support oxygen diffusion during submergence. These characteristics make M. laxiflora play a key role in retaining sediments, enriching heavy metals, and providing microhabitats for endemic animals (such as the Chinese merganser). The ecological monitoring after the completion of the Three Gorges Dam shows that M. laxiflora has irreplaceable ecological functions in maintaining biodiversity and sediment retention in the reservoir area [3,4,5]. The Three Gorges Project (impoundment in 2003) caused about 70% of the native habitats of M. laxiflora to be permanently submerged, and the remaining populations were fragmented and distributed in the tributary hydro-fluctuation belt, facing the loss of genetic diversity and renewal obstacles due to the change in flood rhythm. Currently, M. laxiflora has been listed as an endangered (EN) species by the International Union for Conservation of Nature (IUCN) and included in China’s List of National Key Protected Wild Plants (Class II) [6].
Since 2010, China has launched ex situ conservation initiatives (e.g., the Wuhan Botanical Garden Germplasm Repository) and in situ restoration trials. Genomic research has identified candidate genes associated with flooding stress response in M. laxiflora [7,8], yet practical bottlenecks remain, including the low survival rate of transplanted seedlings in hydro-fluctuation zones and the unclear coupling mechanism between reservoir operations and population dynamics.
Late embryogenesis abundant (LEA) proteins constitute a class of small hydrophilic proteins ubiquitously present in plants. Their nomenclature derives from high abundance during late seed development and enrichment under dehydration conditions [9]. Based on conserved domain features, the LEA protein family is classified into seven subfamilies (LEA_2, LEA_3, and LEA_5). Notably, LEA_2 (dehydrins) and LEA_3 (seed maturation proteins) are extensively studied due to their broad involvement in stress responses [10]. Since initial isolation from cotton embryos in the 1980s, this protein family has been demonstrated to play critical roles in plant tolerance to abiotic stresses, including drought, high salinity, low temperature, and oxidative stress [11]. Advances in genomics and molecular biology have propelled research on LEA proteins’ functional mechanisms, evolutionary traits, and potential applications in enhancing crop stress resistance. For instance, SlLEA6 knockout in tomato compromised antioxidant defenses and reactive oxygen species (ROS)-scavenging systems, thereby reducing cellular damage resistance and drought tolerance [12]. Heterologous expression of the ginseng PgLEA2-50 gene in tobacco enhanced osmotic regulation capacity and antioxidant activity in transgenic lines [13].
However, current research still faces challenges; the functional conservation and differentiation mechanisms of LEA proteins in M. laxiflora’s response to flooding stress remain unclear. To address this knowledge gap, this study employs bioinformatics approaches to identify the LEA gene family through a genome-wide analysis of M. laxiflora. We further characterize the sequence features and potential biological functions of these genes, thereby establishing a theoretical foundation for investigating the LEA gene family’s role in abiotic stress responses.
2. Materials and Methods
2.1. Plant Material and Experiment Design
The experimental materials comprised uniformly grown 1-year-old seedlings of identical genetic background, cultivated at the experimental farm of the Yichang Yangtze River Rare Plant Research Institute, China (March 2020). The seedlings were subjected to flooding in test pools under ambient temperatures averaging 19–27 °C daily. Root samples were collected at 0, 6, 12, 18, 24, 30, 36, and 48 h post-flooding initiation. Following treatment, the seedlings were transferred to control conditions for 12 h recovery (RR12) with subsequent root sampling. All samples were flash-frozen in liquid nitrogen and stored at −80 °C pending analysis [7].
2.2. Search and Identification of LEA Gene Sequence in M. laxiflora
To identify MlLEA genes, whole-genome sequencing data on M. laxiflora were obtained from Figshare (Dataset ID: 25375366). Candidate sequences were screened using the Pfam database to detect LEA-conserved domains, followed by a removal of sequences lacking these domains. The resulting MlLEA genes were submitted to NCBI. AtLEA reference sequences were retrieved from The Arabidopsis Information Resource (TAIR) and NCBI. Finally, protein physicochemical properties were analyzed via ExPASy ProtParam, while subcellular localization predictions were performed using Cell-PLoc 2.0.
2.3. Nomenclature and Phylogenetic Tree Analysis of LEA Family Genes in M. laxiflora
Based on the conserved domain architecture, the LEA protein family was classified into subfamilies with a systematic nomenclature reflecting the subfamily characteristics. Reference protein sequences of Arabidopsis thaliana LEA family members were retrieved from TAIR. Phylogenetic analysis was then conducted using MEGA7.0 for tree construction and the iTOL platform for visualization, comparing the LEA proteins from A. thaliana and M. laxiflora.
2.4. Conserved Domain and Gene Structure Analysis and Cis-Element Analysis of LEA Gene in M. laxiflora
Conserved motifs in LEA proteins were predicted using the MEME suite with the motif count set to 10 and default parameters. The motif distributions were integrated with the M. laxiflora LEA gene phylogeny and visualized via TBtools (v2.030). Chromosomal locations were mapped using the M. laxiflora genome annotation files. Promoter regions (defined as 2 kb upstream of the initiation codon ATG) were extracted from the genomic data using TBtools (v2.030) and analyzed in PlantCARE for cis-acting element identification. Elements were filtered to retain abiotic stress-responsive cis-regulatory elements, excluding unannotated/blank entries.
2.5. Chromosomal Mapping, Gene Duplication, and Collinearity Analysis
Chromosomal locations of M. laxiflora LEA genes, including chromosome numbers, gene start/stop positions, and chromosome lengths, were extracted from the genome annotation files. Using TBtools (v2.030), the intragenomic collinearity within M. laxiflora and the interspecific collinearity between M. laxiflora and A. thaliana genomes were analyzed.
2.6. Differential Expression Analysis of LEA Gene Family in Myricaria laxiflora
Total RNA extraction, library construction, and RNA-seq were conducted by Shanghai OE Biotech Co., Ltd. (Shanghai, China). RNA integrity was assessed with an Agilent 2100 Bioanalyzer (Santa Clara, CA, USA). Libraries were prepared with the TruSeq Stranded mRNA LT Kit (Illumina, CA, USA) and sequenced on an Illumina HiSeq 2500 platform in 150 bp paired-end mode. Three biological replicates were included per sample.
Clean reads were generated by processing raw data with Trimmomatic to remove adapters and low-quality sequences. De novo transcriptome assembly was performed using Trinity with paired-end reads. The longest isoform per gene locus was designated as the unigene based on transcript length for downstream analysis. Gene expression levels were quantified with FPKM (fragments per kilobase of transcript per million mapped reads) [14,15].
2.7. Statistical Analysis
Differentially expressed genes (DEGs) were identified using DESeq2 with thresholds of false discovery rate (FDR) < 0.05 and |log2 (fold change)| > 1. Differentially expressed proteins (DEPs) were detected by Student’s t-test with |fold change| > 2 and p-value < 0.05. Three biological replicates were analyzed per sample. Protein–gene expression correlations were assessed via Pearson’s correlation coefficient.
3. Results
3.1. Identification of LEA Gene in M. laxiflora
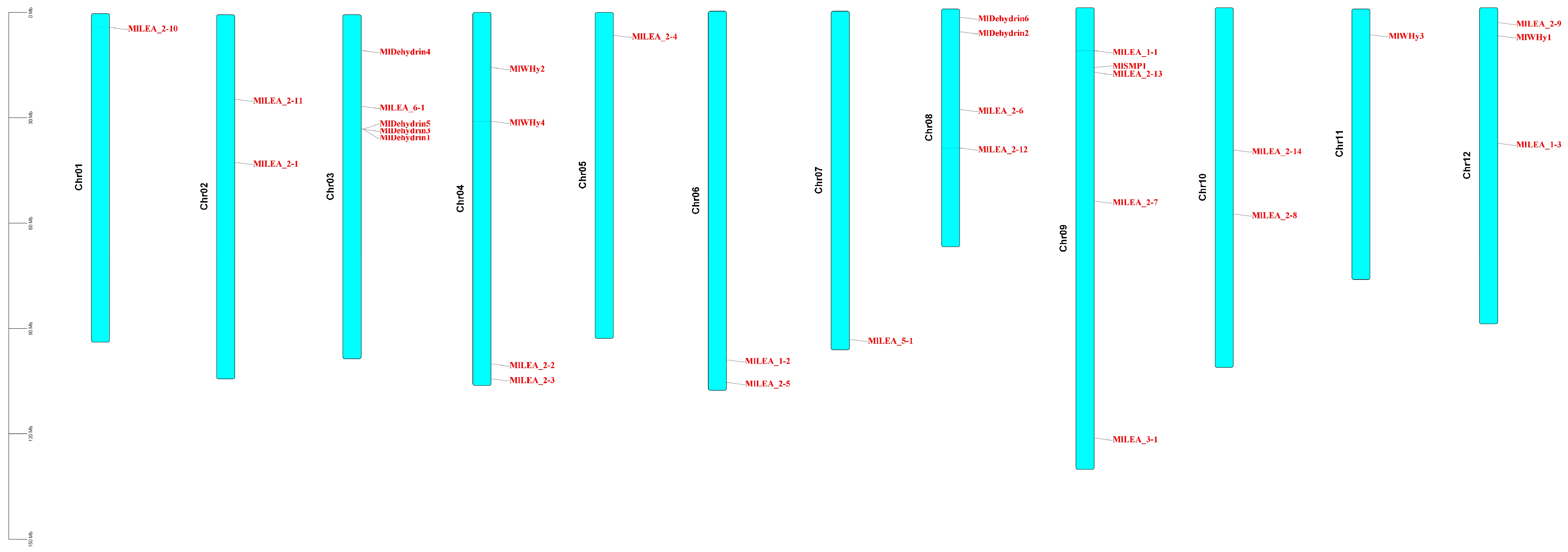
Chromosomal localization of 31 LEA proteins was mapped using M. laxiflora gene annotation files in TBtools. Analysis revealed scattered gene distribution across chromosomes (Figure 1), with 1–5 genes per chromosome. Tandem gene clusters were observed in specific subfamilies, notably dehydrins, showing localized distribution on chromosomes 3 and 8.
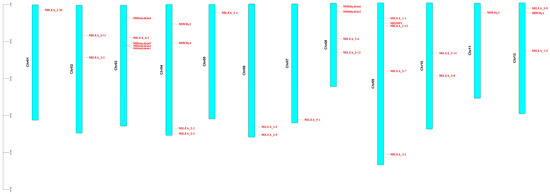
Figure 1.
Chromosomal localization of LEA gene family in Myricaria laxiflora.
3.2. Analysis of Structural Domains and Physicochemical Properties of LEA Gene Family Members in M. laxiflora
Using Clustal W, we performed a multiple sequence alignment, which revealed conserved domains and enabled classification of the LEA genes into nine subfamilies: Dehydrin, LEA_1, LEA_3, LEA_5, LEA_6, WHy, SMP, and LEA_2 superfamily (Table 1, Figure S1). The physicochemical properties and subcellular localization of the 31 identified LEA genes were analyzed (Table 2). Predicted localizations indicated that all LEA family members were predicted to localize to chloroplasts. The protein characteristics showed that the amino acid length ranged from 76 aa (MlLEA_6-1, MlLEA_1-3) to 420 aa (MlLEA_2-11); the molecular weight ranged from 8.08 kDa (MlLEA_6-1) to 45.91 kDa (MlLEA_2-11); and the isoelectric point (pI) ranged from 4.68 (MlWHy1) to 10.25 (MlLEA_2-6).

Table 1.
Structural domain identification and family classification of LEA gene family in M. laxiflora.

Table 2.
31 LEA family genes and related information of M. laxiflora.
3.3. Phylogenetic Tree Analysis of LEA Gene Family Members in M. laxiflora
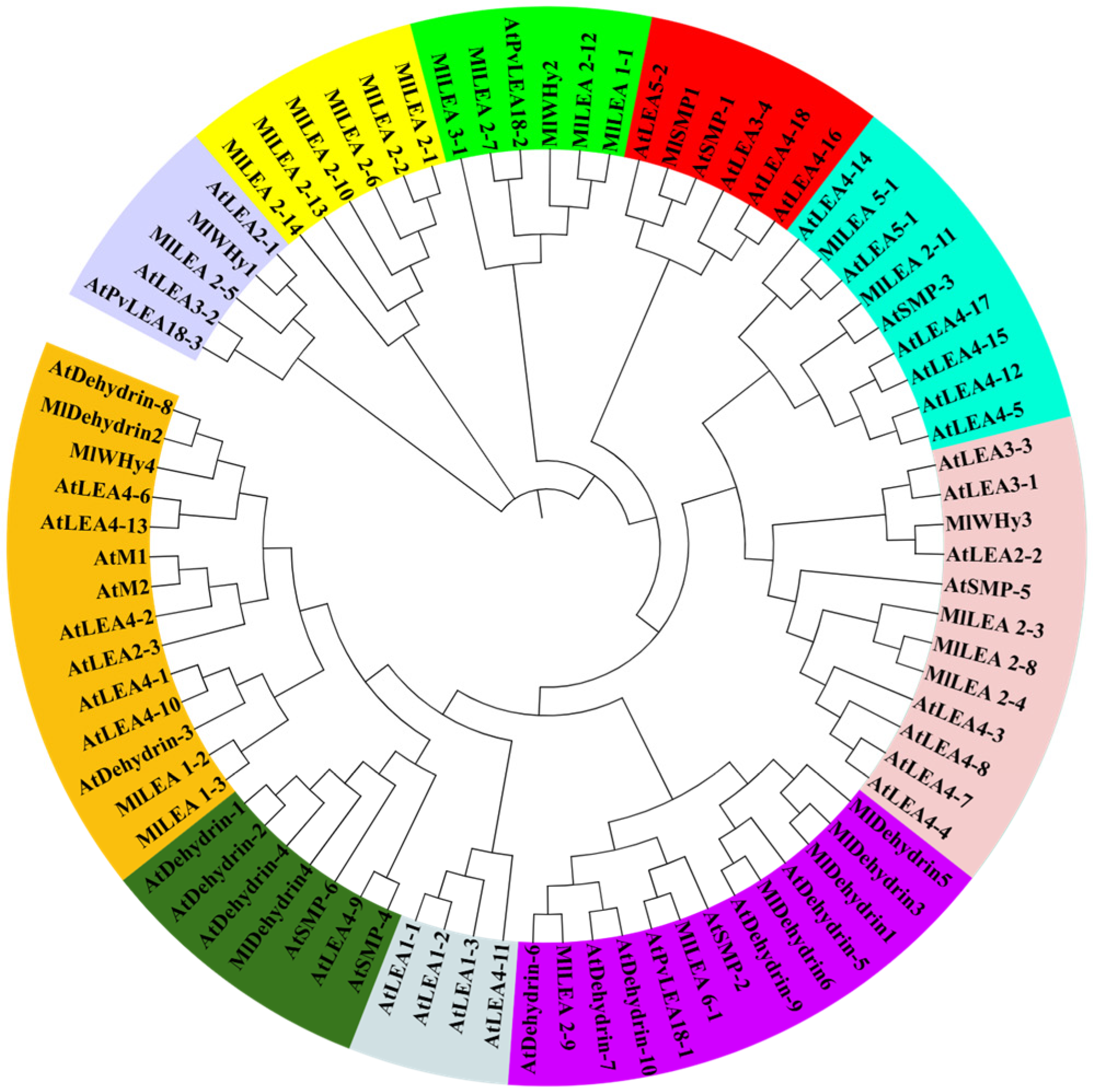
A phylogenetic tree was reconstructed using MEGA7 with the 31 identified MlLEA genes and AtLEA reference sequences. Based on evolutionary relationships, the LEA family members were classified into 10 clades, each demarcated by distinct colored backgrounds (Figure 2).
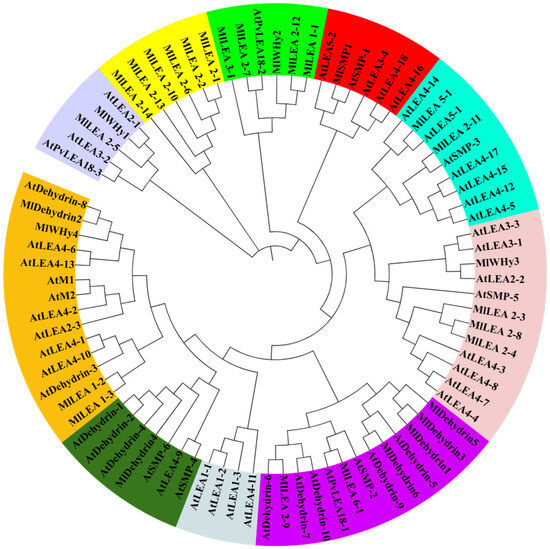
Figure 2.
Phylogenetic tree of LEA family genes in M. laxiflora and A. thaliana. Different colors represent different groups.
3.4. The Domain, Motif Distribution Pattern, and Gene Structure of LEA Gene Family Members in M. laxiflora
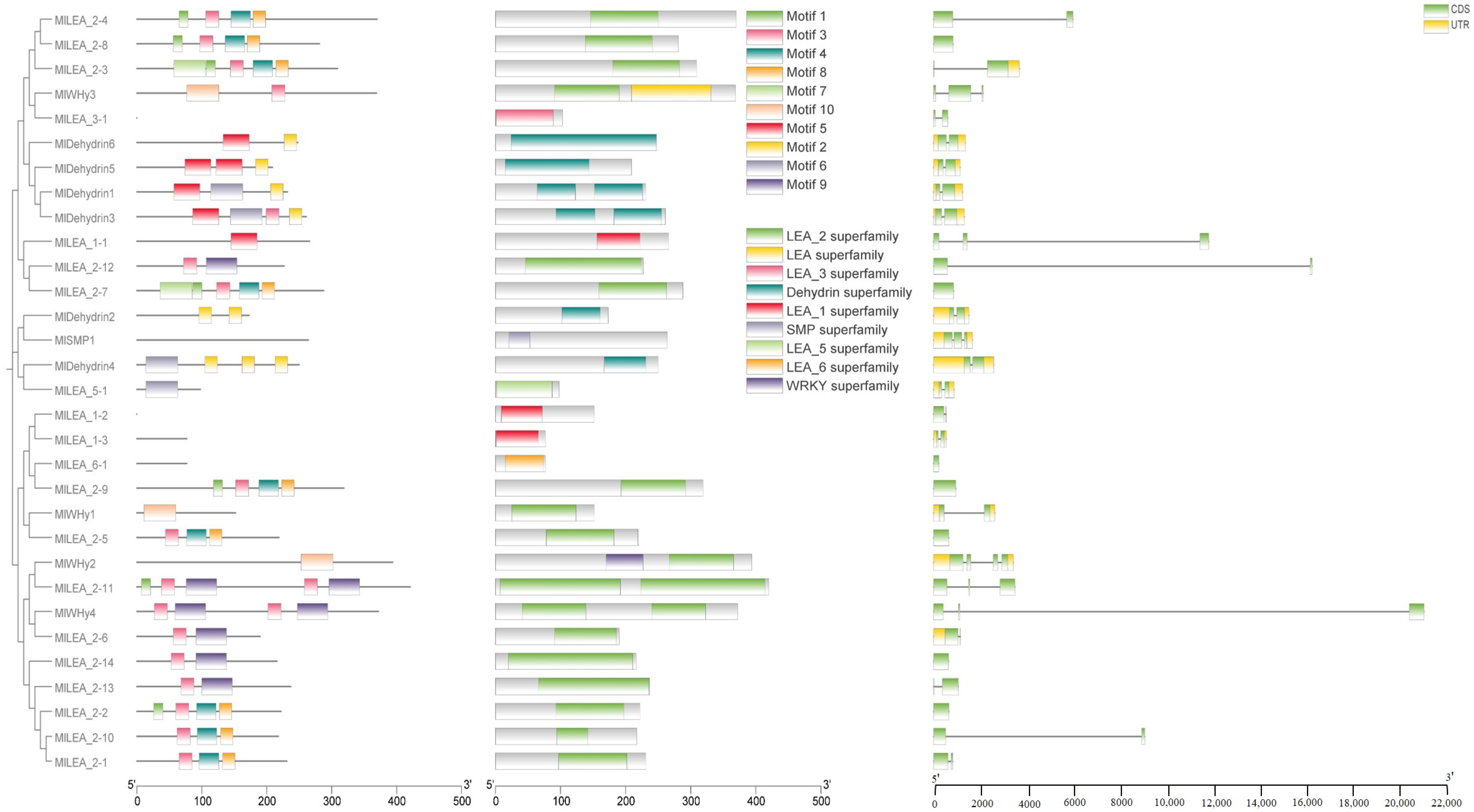
Conserved motifs in LEA proteins typically correlate with specific functions. In this study, ten conserved motifs (motif1–motif10) were identified in M. laxiflora LEA proteins (Figure 3). An analysis revealed that these motifs exhibit subfamily-specific distribution patterns. Further characterization of the gene structures and domains demonstrated that the Dehydrin, LEA_1, LEA_2, LEA_3, LEA_5, LEA_6, and SMP subfamilies all contain stress-responsive domains, consistent with their established biological roles.
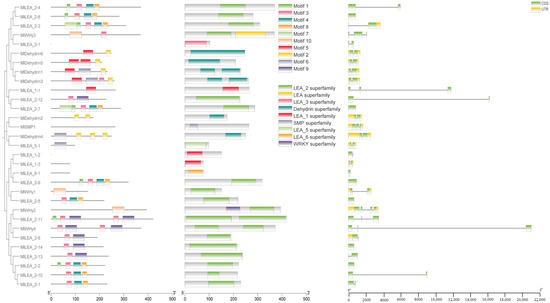
Figure 3.
LEA gene evolution, conserved domain, and gene structure analysis of M. laxiflora.
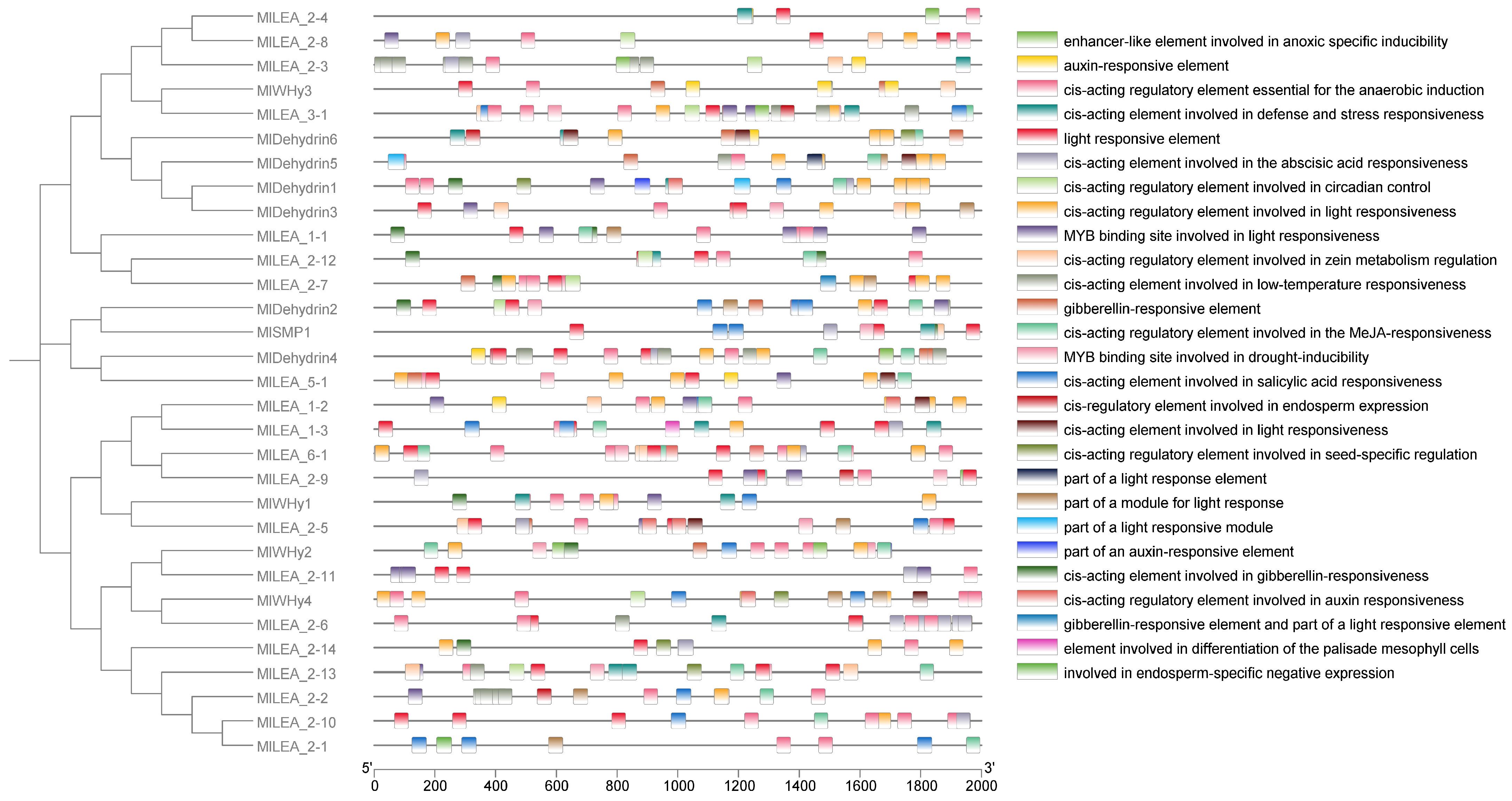
3.5. The Cis-Acting Elements of LEA Gene in M. laxiflora
To elucidate the transcriptional regulation mechanisms of LEA genes, we systematically characterized cis-acting regulatory elements within the promoter regions of the MlLEA gene family in M. laxiflora (Figure 4) and comprehensively characterized cis-acting regulatory elements in the promoter regions of 31 MlLEA genes. A bioinformatics analysis revealed 27 functionally distinct cis-regulatory elements categorized into three core functional modules: (1) abiotic stress response: low-temperature responsiveness (e.g., MBS elements), anaerobic induction (e.g., ARE motifs), and drought inducibility (e.g., MYB binding sites); (2) hormone signaling: abscisic acid (ABRE), gibberellin (GARE-motif), auxin (AuxRR-core/TGA-element), jasmonic acid (CGTCA-motif) responsiveness, and salicylic acid (TCA-element) regulation; and (3) growth and development: light responsiveness (multiple G-box variants), endosperm-specific expression (GCN4_motif), palisade mesophyll differentiation cis-elements, and seed-specific regulation (RY-element). Notably, stress-responsive elements dominate promoter architectures, particularly in dehydrin subfamilies (e.g., MIDehydrin1-7), where >80% of genes contain ABA/drought-responsive motifs. Conversely, MlLEA_2 and MlWHy members exhibit combinatorial enrichment of light-responsive and circadian control elements. These findings establish a molecular basis for MlLEA genes in mediating coordinated stress adaptation and developmental plasticity in M. laxiflora.
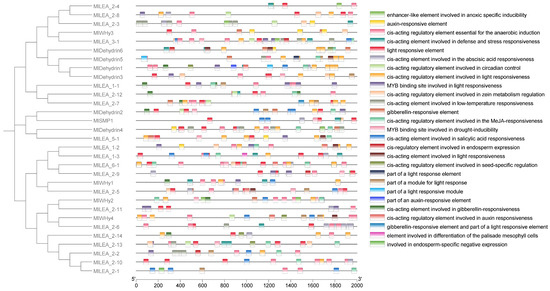
Figure 4.
Analysis of promoter elements of LEA family genes in Myricaria laxiflora.
3.6. Chromosomal Distribution and Synteny Analysis of LEA Genes
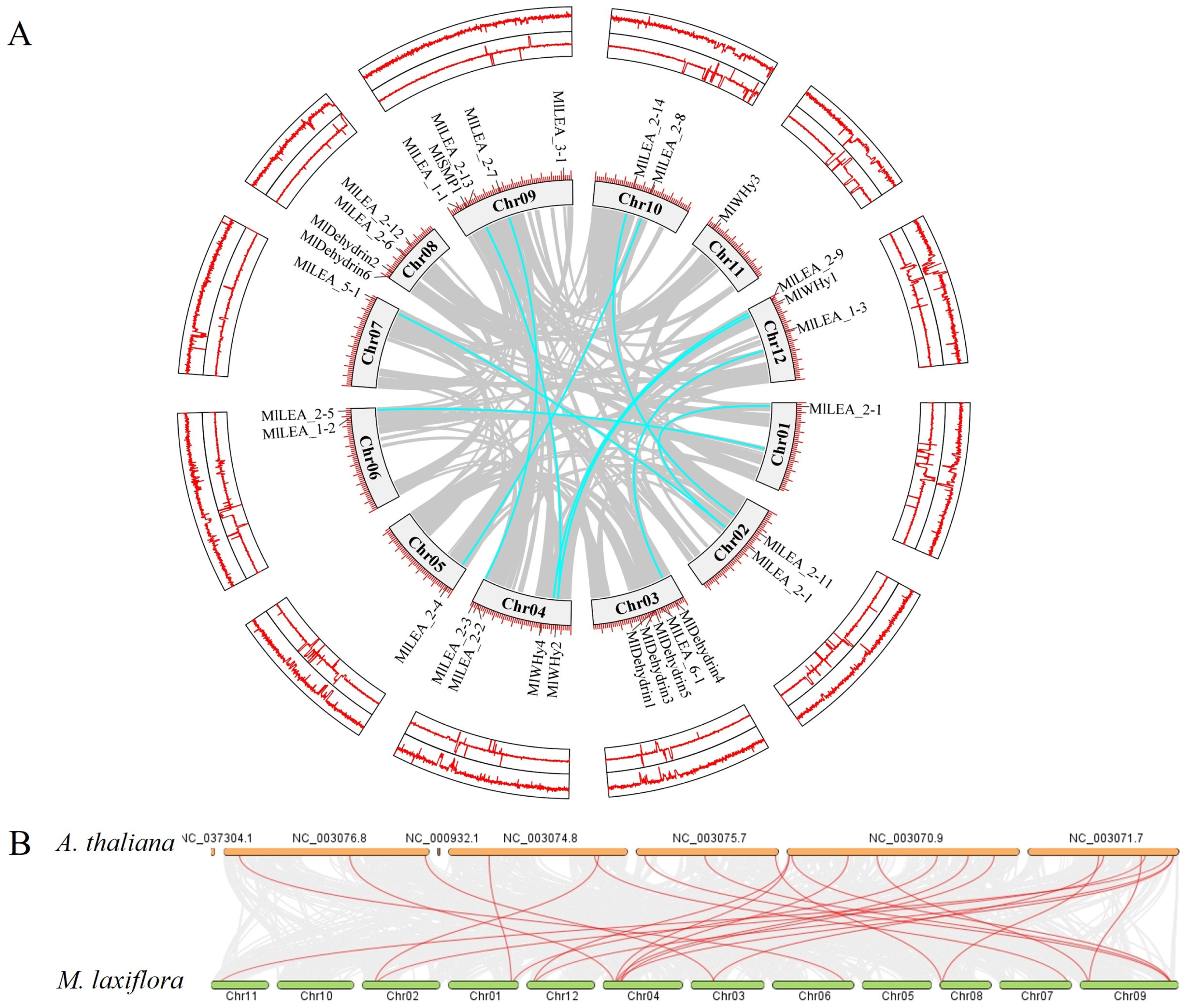
LEA genes were unevenly distributed across 12 chromosomes, with chromosome 3 (Chr03) and chromosome 9 (Chr09) harboring the highest number (five genes each). In contrast, Chr01, Chr05, Chr07, and Chr11 contained only one gene each (Figure 5A). No significant correlation was observed between chromosome length and LEA gene density. A microsynteny analysis between M. laxiflora and Arabidopsis thaliana genomes revealed collinear blocks with a conserved gene order (Figure 5B), suggesting purifying selection on functionally critical regions. Notably, chromosomal rearrangements, including the inversion of Chr01, Chr02 and disrupted synteny in segmental regions, indicate potential evolutionary events such as chromosome fission/fusion or local inversions, providing insights into species-specific genome evolution.
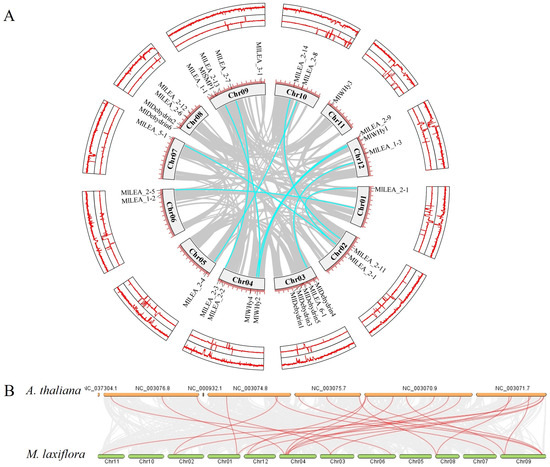
Figure 5.
Linearization analysis of LEA family genes within and between species (A. thaliana) in M. laxiflora. (A) Chromosome distribution and collinearity analysis of LEA gene family in the genome of M. laxiflora. (B) Interspecific collinearity analysis of LEA gene family between M. laxiflora and A. thaliana. Note: (B) is the complete Arabidopsis genome.
3.7. Expression Profiling of M. laxiflora LEA Genes with RNA-seq
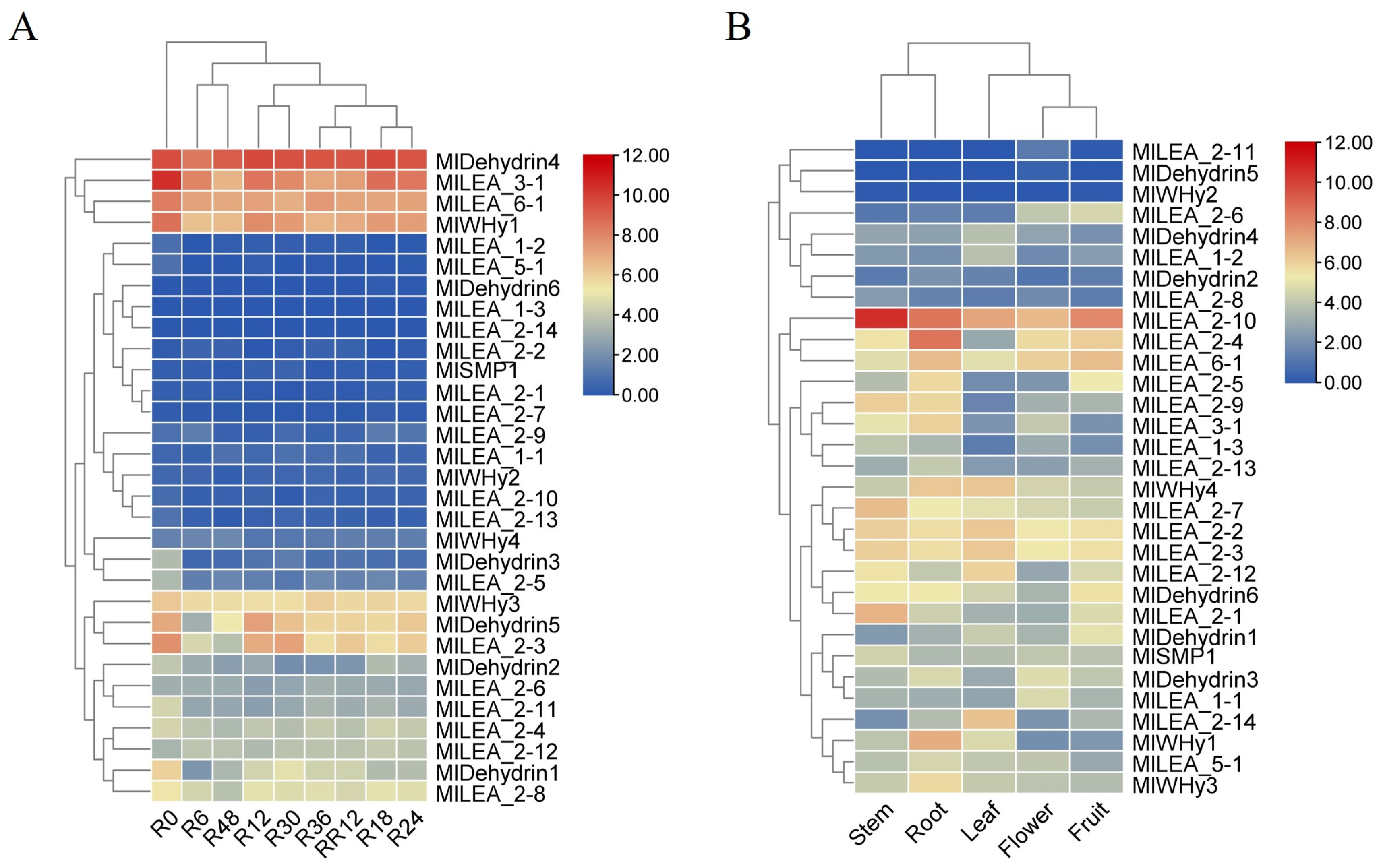
Transcriptomic analysis of published data revealed a high sensitivity of LEA genes in M. laxiflora to flooding stress. Significant differential expression of multiple LEA genes (e.g., MIDehydrin4, MILEA_3-1, MILEA_6-1, and MWHy1y1) was detected as early as 6 h (R6) post-flooding initiation compared to untreated controls (R0) (Figure 6A). The magnitude and pattern of this differential expression continued to evolve throughout the flooding period (R6 to R48), suggesting potential stage-specific roles for LEA genes in distinct phases of the stress response, including initial perception, adaptation, and maintenance. A key finding was the high reversibility of LEA gene expression. Following stress removal and entry into the recovery phase, the expression levels of nearly all LEA genes altered during flooding rapidly returned to baseline levels that were comparable to or equivalent with the R0 controls within just 12 h of recovery (RR12). This indicates that the observed expression changes are stress-specific and that the plant possesses a robust capacity for rapid transcriptional reprogramming to resume normal growth upon the cessation of the stress.
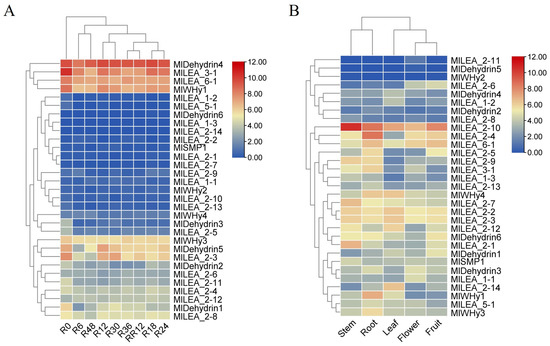
Figure 6.
The expression pattern of LEA gene in Myricaria laxiflora. (A) The expression of LEA family genes at different times under flooding stress. (B) The expression pattern of LEA gene in different tissues. The data were converted to log2 (FPKM+1), and the responsiveness of the LEA gene was represented by a heat map. Blue and red represented down-regulated and up-regulated genes under different tissues and treatments, respectively. Each point represents the average of three independent biological replicates.
Spatiotemporal expression profiling of all 31 identified LEA genes (MILEA) in M. laxiflora revealed their presence across various plant organs. Transcripts for the vast majority of genes, including but not limited to MILEA_1-1, MILEA_1-2, MILEA_1-3, most MILEA_2 subgroup members (MILEA_2-1 to MILEA_2-14), MILEA_3-1, MILEA_5-1, MlLEA6_1, MlSMP1, MlWHy1, MlWHy3, MlWHy4, MIDehydrin1-4, and MIDehydrin6, were detectable at certain levels in all examined tissues (Figure 6B). This broad, non-organ-restricted expression pattern strongly suggests that LEA genes perform universal functions throughout the M. laxiflora lifecycle and in diverse fundamental physiological processes, extending beyond specific stress responses. They likely play fundamental roles in maintaining cellular homeostasis, protecting macromolecular structures, and participating in developmental regulation. In contrast to these widely expressed genes, a small subset, notably MlWHy2 and MIDehydrin5, exhibited highly restricted expression patterns. This indicates that these genes may perform more specialized functions.
4. Discussion
M. laxiflora, a pioneer species in the hydrologically extreme water-level fluctuation zone of the Yangtze River, represents a critical model for studying flooding tolerance mechanisms. As a key scientific challenge in ecological restoration, we present the first systematic genomic analysis of the LEA gene family in this species. Through integrated bioinformatics approaches, we characterized the evolutionary dynamics and functional divergence of LEA proteins, revealing their putative roles in flooding adaptation strategies.
4.1. The Significant Expansion and Adaptive Evolution of LEA Gene Family
The LEA gene family is ubiquitous in plants and plays crucial roles in regulating growth and development under abiotic stress [16,17,18]. In M. laxiflora, we identified 31 MlLEA genes. Chromosomal localization revealed their distribution across all 12 chromosomes (Figure 1), with phylogenetic analysis classifying them primarily into the LEA_2, dehydrin, and WHy subfamilies (Table 1; Figure S1). These subfamilies exhibit evolutionarily conserved functions in osmotic stress responses [11,19,20,21]. Recent comparative genomic analyses have elucidated the evolutionary trajectories of the LEA gene family. In terrestrial plants, this family has undergone significant expansion via whole-genome duplication (WGD) and tandem duplication events. These genomic innovations correlate strongly with plant adaptation to xeric environments following terrestrial colonization [22]. Notably, dehydrin genes form clustered tandem arrays (Figure 5A), with their promoter regions enriched in cis-elements responsive to methyl jasmonate (MeJA), abscisic acid (ABA), low temperature, and light (Figure 4). This suggests that functional diversification post-duplication may enhance adaptation to periodic abiotic stresses [23,24,25]. These genomic features indicate that M. laxiflora’s duplication events likely prioritize hypoxia adaptation, reflecting a species-specific evolutionary strategy.
4.2. Diversified Regulation of Promoter Cis-Acting Elements
A promoter analysis revealed that MlLEA_2 genes harbor anaerobic-inducible elements (e.g., ARE motifs), with significant enrichment observed in MlLEA_2-1 and MlDehydrin4 (Figure 4). These cis-elements may be activated through plant hypoxia signaling pathways (e.g., HIF-1) to mitigate root hypoxia damage during flooding [26]. LEA gene expression is primarily regulated by the ABA signaling cascade, with core transcription factors (e.g., ABI3/ABI5) directly binding the ABRE motifs in promoter regions [27]. This regulatory mechanism is evolutionarily conserved, as evidenced by exogenous ABA-inducing ZmLEA_4 expression to enhance abiotic stress tolerance in maize [28]. In M. laxiflora, ABRE enrichment in MlLEA_2-10 and MlLEA_5-1 promoters confirms the central role of ABA signaling in stress perception and LEA transcriptional control [28,29]. Hormone crosstalk elements (e.g., MeJA- and GA-responsive motifs) may integrate multiple environmental signals through transcriptional regulatory networks, enabling spatiotemporal precision in stress responses [30]. Notably, MlLEA_2-3 and MlLEA_2-6 promoters co-localize low-temperature (LTR) and drought-responsive (MBS) cis-elements, suggesting potential dual stress-protection functions during compound stressors in riparian zones (e.g., winter flooding with concomitant hypothermia).
4.3. Relationship Between Expression Pattern and Flooding Stress Response
Studies have shown that LEA proteins enhance plant stress resistance through multiple molecular mechanisms by (1) functioning as molecular chaperones to prevent stress-induced protein misfolding and aggregation (e.g., Arabidopsis AtLEA4-5 stabilizes client proteins via hydrophobic domain interactions) [31]; (2) maintaining membrane integrity through phospholipid bilayer binding (e.g., rice OsLEA3-1 anchors to membranes, reducing drought-induced lipid peroxidation) [32]; and (3) Synergizing with antioxidant systems to scavenge reactive oxygen species (ROS) and mitigate oxidative damage [33].
Under continuous flooding stress, multiple MlLEA genes exhibited distinct temporal expression dynamics. For instance, MlLEA_2-1 and MlDehydrin4 showed rapid up-regulation (>5-fold) within 6–12 h (Figure 6A). As established molecular chaperones, these proteins likely mitigate hypoxia-driven protein misfolding by stabilizing native conformations during early stress phases [34,35]. This rapid response resembled the early dehydration-induced expression pattern of A. thaliana AtLEA4-5 [36]. However, the activation threshold of M. laxiflora LEA genes was lower, potentially reflecting pre-adaptation to periodic flooding. Notably, MILEA_3-1 and MILEA_5-1 maintained high expression levels at 48 h but rapidly downregulated during the recovery phase (RR12) (Figure 6A). This suggests that prolonged flooding induces sustained cellular protection needs, while rapid gene expression resetting post-stress avoids energy waste. This dynamic regulation aligns with the expression attenuation observed in sweet potato [10,37,38], yet M. laxiflora exhibits a more rapid recovery, which may correlate with frequent water-level fluctuations in its habitat. MlLEA genes exhibited significant tissue-specific expression divergence (Figure 6B). For instance, MlLEA_2-1 showed root-specific high expression, whereas MlLEA_5-1 was predominantly expressed in roots. Functional predictions suggest that root-localized LEA proteins (e.g., MlDehydrin) [39,40] may maintain plasma membrane integrity via phospholipid bilayer anchoring, while leaf-enriched LEA proteins (e.g., MlLEA_5-1) potentially cooperate with antioxidant enzymes (e.g., SOD/POD) to mitigate photoinhibition damage [12,41]. This organ-specific functional partitioning underscores M. laxiflora’s adaptation to riparian ecotones through compartmentalized stress-response strategies.
5. Conclusions
This study presents a systematic genomic analysis of the M. laxiflora LEA gene family. We identified 31 MlLEA genes classified into 9 subfamilies based on protein sequence phylogeny, distributed across all 12 chromosomes. Our findings reveal adaptive evolutionary mechanisms underlying LEA family diversification in the Yangtze River’s water-level fluctuation zone, characterized by (1) significant expansion of the MlLEA_2 subfamily with retention of conserved hydrophilic domains, enabling periodic flooding-drought adaptation through neofunctionalization; (2) promoter architecture enriched with anaerobic-inducible, ABRE, and hormone-crosstalk cis-elements, forming integrated environmental sensing networks; and (3) organ-specific functional divergence: root-localized genes (e.g., MlDehydrin) maintain membrane integrity via phospholipid anchoring, while leaf-predominant genes (e.g., MlLEA_5-1) mitigate photoinhibition through antioxidant coordination. These results provide molecular targets for ecological restoration in the Three Gorges Reservoir riparian zone and establish a framework for deciphering plant stress resilience mechanisms.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/genes16070763/s1; Figure S1: Sequence alignment of LEA gene family subfamily in Myricaria laxiflora. (A) Sequence alignment of Dehydrin subfamily. (B) Sequence alignment of LEA_1 subfamily. (C) Sequence alignment of LEA_2 subfamily. (D) Sequence alignment of LEA_2 superfamily subfamily. (E) Sequence alignment of WHy subfamily. (F) Sequence alignment of LEA_3, LEA_5, LEA_6 and SMP subfamily, the yellow marker is the domain sequence.
Author Contributions
G.H. conceived this project. L.L., H.Z., Y.S., J.W. (Jinhua Wu), and J.W. (Juncheng Wang) provided ideas and methods for article data analysis. C.L. puts forward valuable opinions on the structure and idea of the article. D.W. and T.Z. wrote this manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Ecological Environment Protection Fund of China Three Gorges Group (NBZZ202300130), the Hubei Provincial Government’s Special Project on Guiding Local Science and Technology Development (2024BS019). and the Discovery and identification of functional genes for the formation of important traits of ecological adaptability of three excellent plants, such as A. nelumboides, Plantago fengdouensis, and M. laxiflora (GCZX-2024-03-044).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The whole genome sequencing data of M. laxiflora Download (https://figshare.com/articles/dataset/Myricaria_laxiflora_genome_and_annotation_files/25375366); Transcriptome data download (PRJNA1164124 and PRJNA840865). All data in this manuscript are based on genome and transcriptome analysis, and no new data are generated.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Tian, W.; Bi, Y.H.; Zeng, W.; Jiang, W.; Xue, Y.H.; Wang, G.X.; Liu, S.P. Diversity of Endophytic Fungi of Myricaria laxiflora Grown Under Pre- and Post-Flooding Conditions. Genet. Mol. Res. 2015, 14, 10849–10862. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Wang, N.; Zhou, J.; Zhao, Z.; Lv, K.; Huang, Y.; Huang, G.; Qiu, L. Summer Dormancy of Myricaria laxiflora to Escape Flooding Stress: Changes in Phytohormones and Enzymes Induced by Environmental Factors. Plant Physiol. Biochem. 2022, 193, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, Y.; Huang, H. High Interpopulation Genetic Differentiation and Unidirectional Linear Migration Patterns in Myricaria laxiflora (Tamaricaceae), an Endemic Riparian Plant in the Three Gorges Valley of the Yangtze River. Am. J. Bot. 2006, 93, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wu, D.; Zhen, Q.; Zhang, J.; Qiu, L.; Huang, G.; Yang, C. Morphological Structures and Histochemistry of Roots and Shoots in Myricaria laxiflora (Tamaricaceae). Open Life Sci. 2021, 16, 455–463. [Google Scholar] [CrossRef]
- Peng, Y.; Yang, Y.-B.; Wang, J.-C.; Tian, M.-Y.; Yuan, X.-H.; Yang, Z.-J.; Zuo, Y.-W.; Deng, H.-P. Transcriptome Proffling, Physiological and Biochemical Analyses Reveal Comprehensive Insights into Cadmium Stress in Myricaria laxiflora. Plants 2024, 13, 3433. [Google Scholar] [CrossRef]
- Xie, Y. Progress and Application of IUCN Red List of Threatened Species. Biodivers. Sci. 2022, 30, 22445. [Google Scholar] [CrossRef]
- Li, L.; Huang, G.; Xiang, W.; Zhu, H.; Zhang, H.; Zhang, J.; Ding, Z.; Liu, J.; Wu, D. Integrated Transcriptomic and Proteomic Analyses Uncover the Regulatory Mechanisms of Myricaria laxiflora Under Flooding Stress. Front. Plant Sci. 2022, 13, 924490. [Google Scholar] [CrossRef]
- Li, L.; Su, Y.; Xiang, W.; Huang, G.; Liang, Q.; Dun, B.; Zhang, H.; Xiao, Z.; Qiu, L.; Zhang, J.; et al. Transcriptomic Network Underlying Physiological Alterations in the Stem of Myricaria laxiflora in Response to Waterlogging Stress. Ecotoxicol. Environ. Safety 2024, 284, 116991. [Google Scholar] [CrossRef]
- Battaglia, M.; Olvera-Carrillo, Y.; Garciarrubio, A.; Campos, F.; Covarrubias, A.A. The Enigmatic LEA Proteins and Other Hydrophilins. Plant Physiol. 2008, 148, 6–24. [Google Scholar] [CrossRef]
- Candat, A.; Paszkiewicz, G.; Neveu, M.; Gautier, R.; Logan, D.C.; Avelange-Macherel, M.-H.; Macherel, D. The Ubiquitous Distribution of Late Embryogenesis Abundant Proteins Across Cell Compartments in Arabidopsis Offers Tailored Protection Against Abiotic Stress. Plant Cell 2014, 26, 3148–3166. [Google Scholar] [CrossRef]
- Hundertmark, M.; Hincha, D.K. LEA (Late Embryogenesis Abundant) Proteins and Their Encoding Genes in Arabidopsis thaliana. BMC Genom. 2008, 9, 118. [Google Scholar] [CrossRef]
- Jia, C.; Guo, B.; Wang, B.; Li, X.; Yang, T.; Li, N.; Wang, J.; Yu, Q. The LEA Gene Family in Tomato and Its Wild Relatives: Genome-Wide Identification, Structural Characterization, Expression Profiling, and Role of SlLEA6 in Drought Stress. BMC Plant Biol. 2022, 22, 596. [Google Scholar] [CrossRef]
- Wang, Q.; Lei, X.; Wang, Y.; Di, P.; Meng, X.; Peng, W.; Rong, J.; Wang, Y. Genome-Wide Identification of the LEA Gene Family in Panax Ginseng: Evidence for the Role of PgLEA2-50 in Plant Abiotic Stress Response. Plant Physiol. Biochem. 2024, 212, 108742. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-Length Transcriptome Assembly from RNA-Seq Data Without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Altman, A. Plant Responses to Drought, Salinity and Extreme Temperatures: Towards Genetic Engineering for Stress Tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Cao, D.; Wang, Z.; Ma, L.; Tian, K.; Liu, Y.; Gong, Z.; Zhu, X.; Jiang, C.; Li, Y. Genome-Wide Identification and Expression Analyses of the LEA Protein Gene Family in Tea Plant Reveal Their Involvement in Seed Development and Abiotic Stress Responses. Sci. Rep. 2019, 9, 14123. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Li, S.; Chen, W.; Zhang, J.; Zhang, L.; Sun, W.; Wang, Z. Plant Dehydrins: Expression, Regulatory Networks, and Protective Roles in Plants Challenged by Abiotic Stress. Int. J. Mol. Sci. 2021, 22, 12619. [Google Scholar] [CrossRef] [PubMed]
- Mertens, J.; Aliyu, H.; Cowan, D.A. LEA Proteins and the Evolution of the WHy Domain. Appl. Environ. Microbiol. 2018, 84, e00539-18. [Google Scholar] [CrossRef] [PubMed]
- Abdul Aziz, M.; Sabeem, M.; Mullath, S.K.; Brini, F.; Masmoudi, K. Plant Group II LEA Proteins: Intrinsically Disordered Structure for Multiple Functions in Response to Environmental Stresses. Biomolecules 2021, 11, 1662. [Google Scholar] [CrossRef]
- Ghanmi, S.; Graether, S.P.; Hanin, M. The Halophyte Dehydrin Sequence Landscape. Biomolecules 2022, 12, 330. [Google Scholar] [CrossRef] [PubMed]
- Bies-Ethève, N.; Gaubier-Comella, P.; Debures, A.; Lasserre, E.; Jobet, E.; Raynal, M.; Cooke, R.; Delseny, M. Inventory, Evolution and Expression Profiling Diversity of the LEA (Late Embryogenesis Abundant) Protein Gene Family in Arabidopsis thaliana. Plant Mol. Biol. 2008, 67, 107–124. [Google Scholar] [CrossRef]
- Danquah, A.; de Zelicourt, A.; Colcombet, J.; Hirt, H. The Role of ABA and MAPK Signaling Pathways in Plant Abiotic Stress Responses. Biotechnol. Adv. 2014, 32, 40–52. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, W.; Zhang, Y.; Zhang, X.; Lang, D.; Zhang, X. The Roles of Methyl Jasmonate to Stress in Plants. Funct. Plant Biol. 2019, 46, 197–212. [Google Scholar] [CrossRef]
- Soliman, S.; Wang, Y.; Han, Z.; Pervaiz, T.; El-Kereamy, A. Strigolactones in Plants and Their Interaction with the Ecological Microbiome in Response to Abiotic Stress. Plants 2022, 11, 3499. [Google Scholar] [CrossRef] [PubMed]
- Licausi, F.; Kosmacz, M.; Weits, D.A.; Giuntoli, B.; Giorgi, F.M.; Voesenek, L.A.C.J.; Perata, P.; van Dongen, J.T. Oxygen Sensing in Plants Is Mediated by an N-End Rule Pathway for Protein Destabilization. Nature 2011, 479, 419–422. [Google Scholar] [CrossRef] [PubMed]
- Skubacz, A.; Daszkowska-Golec, A.; Szarejko, I. The Role and Regulation of ABI5 (ABA-Insensitive 5) in Plant Development, Abiotic Stress Responses and Phytohormone Crosstalk. Front. Plant Sci. 2016, 7, 1884. [Google Scholar] [CrossRef]
- Zamora-Briseño, J.A.; de Jiménez, E.S. A LEA 4 Protein Up-Regulated by ABA Is Involved in Drought Response in Maize Roots. Mol. Biol. Rep. 2016, 43, 221–228. [Google Scholar] [CrossRef]
- Král, D.; Šenkyřík, J.B.; Ondřej, V. Expression of Genes Involved in ABA and Auxin Metabolism and LEA Gene During Embryogenesis in Hemp. Plants 2022, 11, 2995. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant Hormone-Mediated Regulation of Stress Responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Nguyen, C.T.T.; Jung, C.; Cheong, J.-J. AtMYB44 Suppresses Transcription of the Late Embryogenesis Abundant Protein Gene AtLEA4-5. Biochem. Biophys. Res. Commun. 2019, 511, 931–934. [Google Scholar] [CrossRef]
- Xiao, B.; Huang, Y.; Tang, N.; Xiong, L. Over-Expression of a LEA Gene in Rice Improves Drought Resistance Under the Field Conditions. Theor. Appl. Genet. 2007, 115, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, Q.; Qin, J.; Xiao, G.; Zhu, S.; Hu, T. OsLEA1a Overexpression Enhances Tolerance to Diverse Abiotic Stresses by Inhibiting Cell Membrane Damage and Enhancing ROS Scavenging Capacity in Transgenic Rice. Funct. Plant Biol. 2021, 48, 860–870. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, D.; Agoston, B.; Tompa, P. Disordered Plant LEA Proteins as Molecular Chaperones. Plant Signal. Behav. 2008, 3, 710–713. [Google Scholar] [CrossRef]
- Chakrabortee, S.; Tripathi, R.; Watson, M.; Schierle, G.S.K.; Kurniawan, D.P.; Kaminski, C.F.; Wise, M.J.; Tunnacliffe, A. Intrinsically Disordered Proteins as Molecular Shields. Mol. Biosyst. 2012, 8, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Romero-Pérez, P.S.; Martínez-Castro, L.V.; Linares, A.; Arroyo-Mosso, I.; Sánchez-Puig, N.; Cuevas-Velazquez, C.L.; Sukenik, S.; Guerrero, A.; Covarrubias, A.A. Self-Association and Multimer Formation in AtLEA4-5, a Desiccation-Induced Intrinsically Disordered Protein from Plants. Protein Sci. 2024, 33, e5192. [Google Scholar] [CrossRef]
- Artur, M.A.S.; Rienstra, J.; Dennis, T.J.; Farrant, J.M.; Ligterink, W.; Hilhorst, H. Structural Plasticity of Intrinsically Disordered LEA Proteins from Xerophyta Schlechteri Provides Protection In Vitro and In Vivo. Front. Plant Sci. 2019, 10, 1272. [Google Scholar] [CrossRef]
- Hu, M.; Li, Z.; Lin, X.; Tang, B.; Xing, M.; Zhu, H. Comparative Analysis of the LEA Gene Family in Seven Ipomoea Species, Focuses on Sweet Potato (Ipomoea batatas L.). BMC Plant Biol. 2024, 24, 1256. [Google Scholar] [CrossRef]
- Andersson, J.M.; Pham, Q.D.; Mateos, H.; Eriksson, S.; Harryson, P.; Sparr, E. The Plant Dehydrin Lti30 Stabilizes Lipid Lamellar Structures in Varying Hydration Conditions. J. Lipid Res. 2020, 61, 1014–1024. [Google Scholar] [CrossRef]
- Mishra, M.K.; Tiwari, S.; Misra, P. Overexpression of WssgtL3.1 Gene from Withania somnifera Confers Salt Stress Tolerance in Arabidopsis. Plant Cell Rep. 2021, 40, 2191–2204. [Google Scholar] [CrossRef]
- Gao, C.; Wang, C.; Zheng, L.; Wang, L.; Wang, Y. A LEA Gene Regulates Cadmium Tolerance by Mediating Physiological Responses. Int. J. Mol. Sci. 2012, 13, 5468–5481. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).