Eight New Sedum Plastomes: Comprehensive Analyses and Phylogenetic Implications
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Material, DNA Extraction, and Sequencing
2.2. Genome Assembly and Annotation
2.3. Codon Usage, Putative RNA Editing Sites, and IR Junction Analysis
2.4. Microsatellite Repeat Analysis
2.5. Phylogenetic Analysis
3. Results
3.1. Plastid Genome Organization and Features
3.2. Comparative Analyses of Nucleotide Composition, Codon Usage, and Amino Acid Frequencies
3.3. Microsatellite Repeat Polymorphisms
3.4. Prediction of RNA Editing Sites
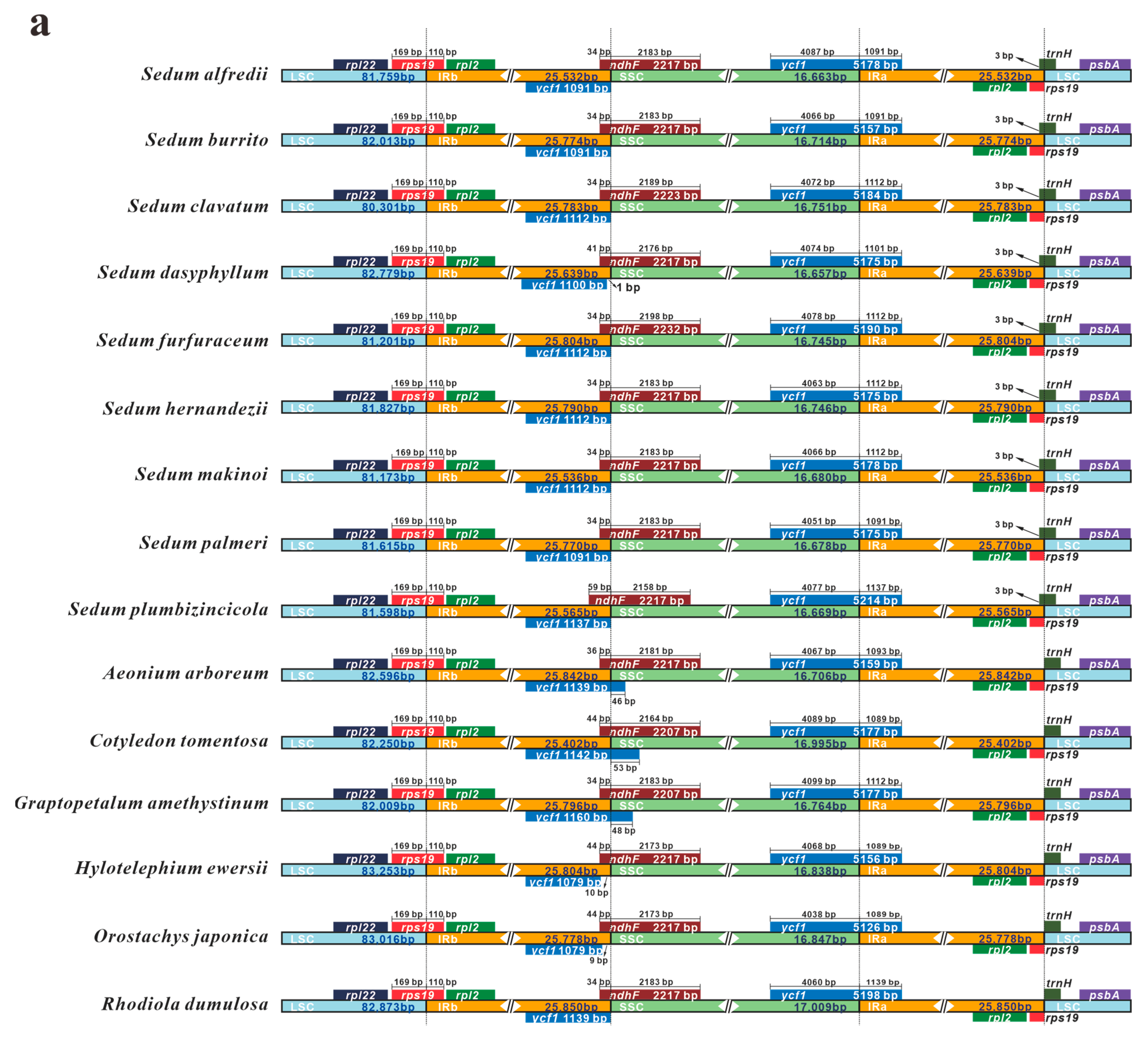
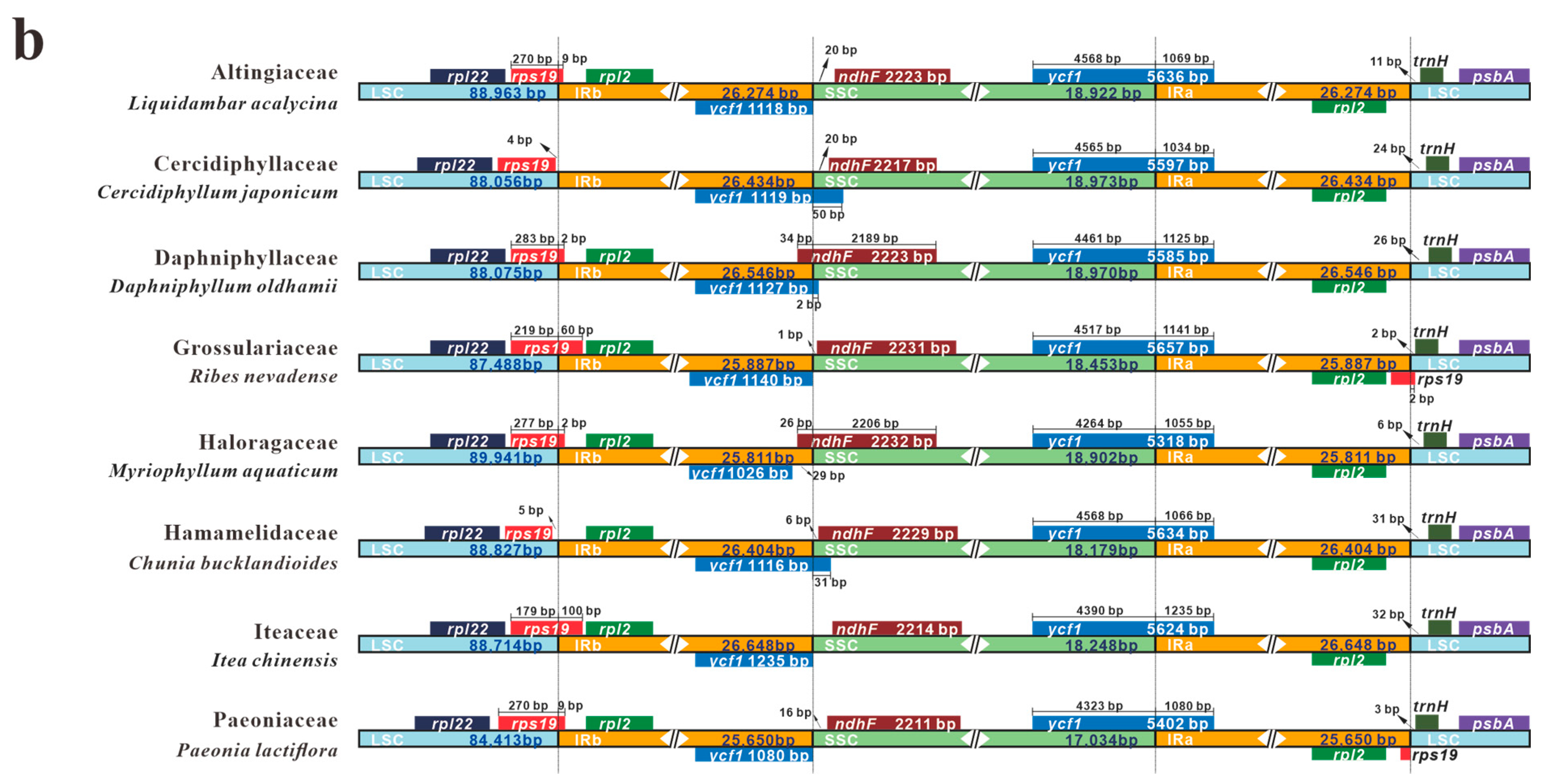
3.5. IR Contraction and Expansion
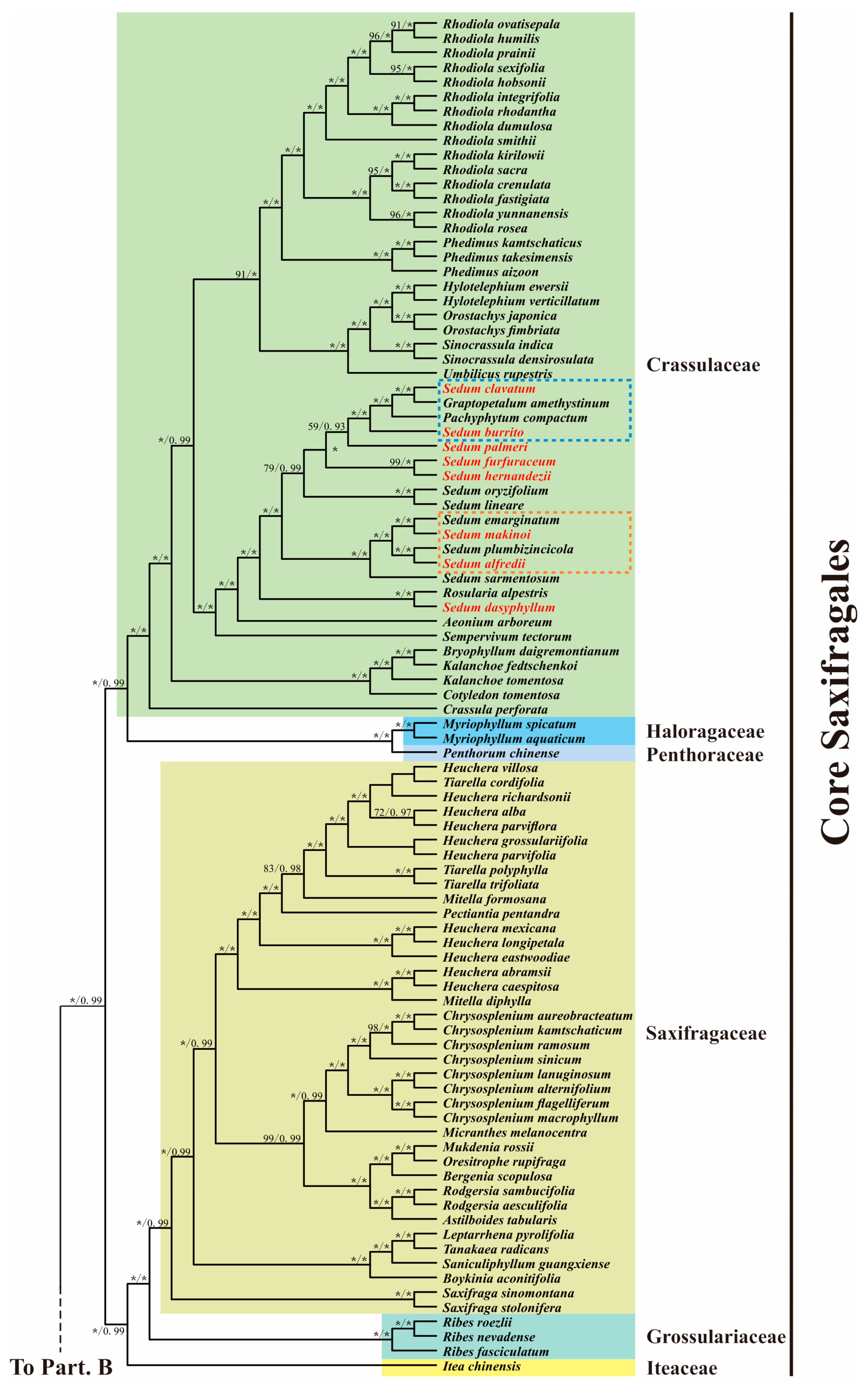
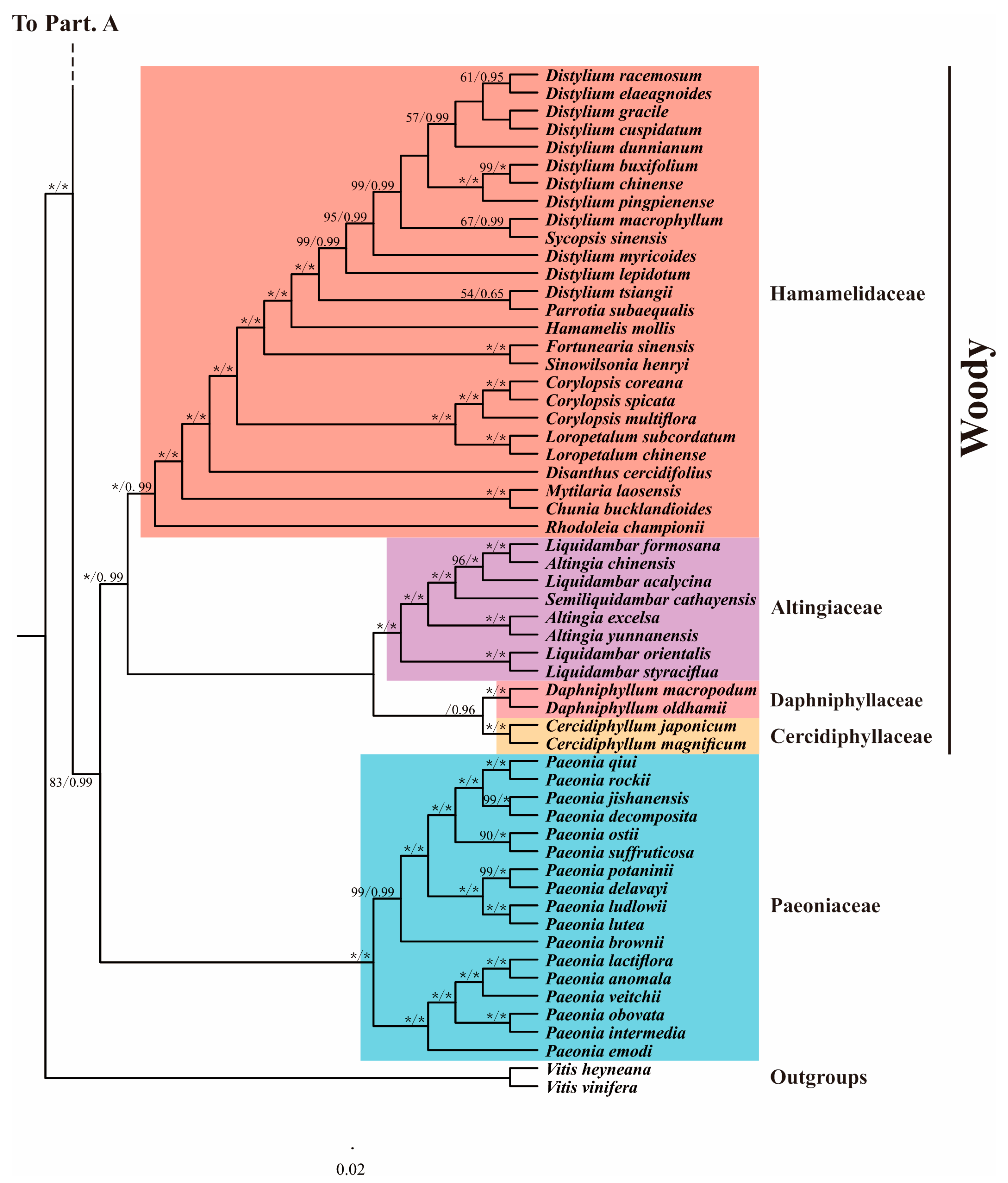
3.6. Phylogenetic Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Messerschmid, T.F.E.; Klein, J.T.; Kadereit, G.; Kadereit, J.W. Linnaeus’s folly—phylogeny, evolution and classification of Sedum (Crassulaceae) and Crassulaceae subfamily Sempervivoideae. Taxon 2020, 69, 892–926. [Google Scholar] [CrossRef]
- Nikulin, V.Y.; Gontcharova, S.B.; Stephenson, R.; Gontcharov, A.A. Phylogenetic relationships between Sedum L. and related genera (Crassulaceae) based on ITS rDNA sequence comparisons. Flora 2016, 224, 218–229. [Google Scholar] [CrossRef]
- Berger, A. Crassulaceae . Die Nat. Pflanzenfamilien 1930, 2, 352–483. [Google Scholar]
- Ohba, H. Generic and infrageneric classification of the Old World Sedoideae (Crassulaceae). J. Fac. Sci. Univ. Tokyo 3 1978, 12, 139–198. [Google Scholar]
- Thorne, R.F.; Reveal, J.L. An updated classification of the class Magnoliopsida (“Angiospermae”). Bot. Rev. 2007, 73, 67–181. [Google Scholar] [CrossRef]
- Thiede, J.; Eggli, U. Crassulaceae . In Flowering Plants Eudicots; Springer: Berlin, Heidelberg, 2007; pp. 83–118. [Google Scholar]
- Van Ham, R.C.; Hart, H.t. Phylogenetic relationships in the Crassulaceae inferred from chloroplast DNA restriction-site variation. Am. J. Bot. 1998, 85, 123–134. [Google Scholar] [CrossRef]
- Mort, M.E.; Soltis, D.E. Phylogenetics and evolution of the Macaronesian clade of Crassulaceae inferred from nuclear and chloroplast sequence data. Syst. Bot. 2002, 27, 271–288. [Google Scholar]
- Mort, M.E.; Soltis, D.E. Phylogenetic relationships and evolution of Crassulaceae inferred from matK sequence data. Am. J. Bot. 2001, 88, 76–91. [Google Scholar] [CrossRef]
- Mayuzumi, S.; Ohba, H. The phylogenetic position of eastern Asian Sedoideae (Crassulaceae) inferred from chloroplast and nuclear DNA sequences. Syst. Bot. 2004, 29, 587–598. [Google Scholar] [CrossRef]
- Gontcharova, S.B.; Artyukova, E.V.; Gontcharov, A.A. Phylogenetic relationships among members of the subfamily Sedoideae (Crassulaceae) inferred from the ITS region sequences of nuclear rDNA. Russ. J. Genet. 2006, 42, 654–661. [Google Scholar] [CrossRef]
- Carrillo-Reyes, P.; Sosa, V.; Mort, M.E. Molecular phylogeny of the Acre clade (Crassulaceae): Dealing with the lack of definitions for Echeveria and Sedum. Mol. Phylogenet Evol. 2009, 53, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Mort, M.; O’Leary, T.; Carrillo-Reyes, P.; Nowell, T.; Archibald, J.; Randle, C. Phylogeny and evolution of Crassulaceae: Past, present, and future. Schumannia 2010, 6, 69–86. [Google Scholar]
- Daniell, H.; Lin, C.-S.; Yu, M.; Chang, W.-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Oldenburg, D.J.; Bendich, A.J. The linear plastid chromosomes of maize: Terminal sequences, structures, and implications for DNA replication. Curr. Genet. 2015, 62, 431–442. [Google Scholar] [CrossRef]
- Oldenburg, D.J.; Bendich, A.J. Most chloroplast DNA of maize seedlings in linear molecules with defined ends and branched forms. J. Mol. Biol. 2004, 335, 953–970. [Google Scholar] [CrossRef]
- Bendich, A.J. Circular chloroplast chromosomes: The grand illusion. Plant Cell 2004, 16, 1661–1666. [Google Scholar] [CrossRef]
- Palmer, J.D. Plastid chromosomes: Structure and evolution. Cell Cult. Som. Cell Genet. Plants 1991, 7A, 5–53. [Google Scholar]
- Palmer, J.D. Comparative organization of chloroplast genomes. Annu. Rev. Genet. 1985, 19, 325–354. [Google Scholar] [CrossRef]
- Kan, J.; Zhang, S.; Wu, Z.; Bi, D. Exploring plastomic resources in Sempervivum (Crassulaceae): Implications for phylogenetics. Genes 2024, 15, 441. [Google Scholar] [CrossRef]
- Parks, M.; Cronn, R.; Liston, A. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 2009, 7, 84. [Google Scholar] [CrossRef]
- Wu, Y.; Liu, F.; Yang, D.; Li, W.; Zhou, X.; Pei, X.; Liu, Y.; He, K.; Zhang, W.; Ren, Z.; et al. Comparative chloroplast genomics of Gossypium species: Insights into repeat sequence variations and phylogeny. Front. Plant Sci. 2018, 9, 376. [Google Scholar] [CrossRef] [PubMed]
- Henriquez, C.L.; Abdullah; Ahmed, I.; Carlsen, M.M.; Zuluaga, A.; Croat, T.B.; McKain, M.R. Evolutionary dynamics of chloroplast genomes in subfamily Aroideae (Araceae). Genomics 2020, 112, 2349–2360. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Bi, D.; Yi, R.; Ding, H.; Wu, L.; Kan, X. Plastome evolution of Aeonium and Monanthes (Crassulaceae): Insights into the variation of plastomic tRNAs, and the patterns of codon usage and aversion. Planta 2022, 256, 35. [Google Scholar] [CrossRef]
- Han, S.; Wang, R.; Hong, X.; Wu, C.; Zhang, S.; Kan, X. Plastomes of Bletilla (Orchidaceae) and Phylogenetic Implications. Int. J. Mol. Sci. 2022, 23, 10151. [Google Scholar] [CrossRef]
- Bi, D.; Han, S.; Zhou, J.; Zhao, M.; Zhang, S.; Kan, X. Codon usage analyses reveal the evolutionary patterns among plastid genes of Saxifragales at a larger-sampling scale. Genes 2023, 14, 694. [Google Scholar] [CrossRef]
- Ding, H.; Bi, D.; Han, S.; Yi, R.; Zhang, S.; Ye, Y.; Gao, J.; Yang, J.; Kan, X. Mitogenomic codon usage patterns of superfamily Certhioidea (Aves, Passeriformes): Insights into asymmetrical bias and phylogenetic implications. Animals 2022, 13, 96. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 26 June 2025).
- Jin, J.-J.; Yu, W.-B.; Yang, J.-B.; Song, Y.; DePamphilis, C.W.; Yi, T.-S.; Li, D.-Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Ding, H.; Zhu, R.; Dong, J.; Bi, D.; Jiang, L.; Zeng, J.; Huang, Q.; Liu, H.; Xu, W.; Wu, L.; et al. Next-Generation genome sequencing of Sedum plumbizincicola sheds light on the structural evolution of plastid rRNA operon and phylogenetic implications within Saxifragales. Plants 2019, 8, 386. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Zheng, S.; Poczai, P.; Hyvönen, J.; Tang, J.; Amiryousefi, A. Chloroplot: An online program for the versatile plotting of organelle genomes. Front. Genet. 2020, 11, 576124. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Mower, J.P. The PREP suite: Predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res. 2009, 37, W253–W259. [Google Scholar] [CrossRef] [PubMed]
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef]
- Chateigner-Boutin, A.-L.; Small, I. Plant RNA editing. RNA Biol. 2010, 7, 213–219. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Morton, B.R.; Maier, U.G. The evolution of chloroplast RNA editing. Mol. Biol. Evol. 2006, 23, 1912–1921. [Google Scholar] [CrossRef]
- Chase, M.W.; Christenhusz, M.; Fay, M.; Byng, J.; Judd, W.S.; Soltis, D.; Mabberley, D.; Sennikov, A.; Soltis, P.S.; Stevens, P.F. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar]
- Fishbein, M.; Hibsch-Jetter, C.; Soltis, D.E.; Hufford, L. Phylogeny of Saxifragales (angiosperms, eudicots): Analysis of a rapid, ancient radiation. Syst. Biol. 2001, 50, 817–847. [Google Scholar] [CrossRef] [PubMed]
- Soltis, D.E.; Mort, M.E.; Latvis, M.; Mavrodiev, E.V.; O’Meara, B.C.; Soltis, P.S.; Burleigh, J.G.; Rubio de Casas, R. Phylogenetic relationships and character evolution analysis of Saxifragales using a supermatrix approach. Am. J. Bot. 2013, 100, 916–929. [Google Scholar] [CrossRef]
- Mehmood, F.; Abdullah; Shahzadi, I.; Ahmed, I.; Waheed, M.T.; Mirza, B. Characterization of Withania somnifera chloroplast genome and its comparison with other selected species of Solanaceae. Genomics 2020, 112, 1522–1530. [Google Scholar] [CrossRef]
- Shen, X.; Wu, M.; Liao, B.; Liu, Z.; Bai, R.; Xiao, S.; Li, X.; Zhang, B.; Xu, J.; Chen, S. Complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Artemisia annua. Molecules 2017, 22, 1330. [Google Scholar] [CrossRef]
- Cui, Y.; Nie, L.; Sun, W.; Xu, Z.; Wang, Y.; Yu, J.; Song, J.; Yao, H. Comparative and phylogenetic analyses of ginger (Zingiber officinale) in the family Zingiberaceae based on the complete chloroplast genome. Plants 2019, 8, 283. [Google Scholar] [CrossRef]
- Abdullah; Mehmood, F.; Shahzadi, I.; Waseem, S.; Mirza, B.; Ahmed, I.; Waheed, M.T. Chloroplast genome of Hibiscus rosa-sinensis (Malvaceae): Comparative analyses and identification of mutational hotspots. Genomics 2020, 112, 581–591. [Google Scholar]
- Cao, J.; Wang, H.; Cao, Y.; Kan, S.; Li, J.; Liu, Y. Extreme reconfiguration of plastid genomes in Papaveraceae: Rearrangements, gene loss, pseudogenization, IR expansion, and repeats. Int. J. Mol. Sci. 2024, 25, 2278. [Google Scholar] [CrossRef]
- Guo, X.; Liu, J.; Hao, G.; Zhang, L.; Mao, K.; Wang, X.; Zhang, D.; Ma, T.; Hu, Q.; Al-Shehbaz, I.A. Plastome phylogeny and early diversification of Brassicaceae. BMC Genomics 2017, 18, 176. [Google Scholar] [CrossRef]
- Ueda, M.; Fujimoto, M.; Arimura, S.-I.; Murata, J.; Tsutsumi, N.; Kadowaki, K.-I. Loss of the rpl32 gene from the chloroplast genome and subsequent acquisition of a preexisting transit peptide within the nuclear gene in Populus. Gene 2007, 402, 51–56. [Google Scholar] [CrossRef]
- Dong, W.; Xu, C.; Wu, P.; Cheng, T.; Yu, J.; Zhou, S.; Hong, D. Resolving the systematic positions of enigmatic taxa: Manipulating the chloroplast genome data of Saxifragales. Mol. Phylogenet Evol. 2018, 126, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Yengkhom, S.; Uddin, A.; Chakraborty, S. Deciphering codon usage patterns and evolutionary forces in chloroplast genes of Camellia sinensis var. assamica and Camellia sinensis var. sinensis in comparison to Camellia pubicosta. J. Integr. Agric. 2019, 18, 2771–2785. [Google Scholar]
- Leffler, E.M.; Bullaughey, K.; Matute, D.R.; Meyer, W.K.; Segurel, L.; Venkat, A.; Andolfatto, P.; Przeworski, M. Revisiting an old riddle: What determines genetic diversity levels within species? PLoS Biol. 2012, 10, e1001388. [Google Scholar] [CrossRef]
- Zhou, J.; Ding, Y.; He, Y.; Chu, Y.; Zhao, P.; Ma, L.; Wang, X.; Li, X.; Liu, Y. The effect of multiple evolutionary selections on synonymous codon usage of genes in the Mycoplasma bovis genome. PLoS ONE 2014, 9, e108949. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Yengkhom, S.; Uddin, A. Analysis of codon usage bias of chloroplast genes in Oryza species: Codon usage of chloroplast genes in Oryza species. Planta 2020, 252, 67. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Yang, C.; Zhao, X.; Chen, S.; Qu, G. Complete chloroplast genome sequence of Betula platyphylla: Gene organization, RNA editing, and comparative and phylogenetic analyses. BMC Genomics 2018, 19, 950. [Google Scholar] [CrossRef]
- Park, I.; Yang, S.; Kim, W.J.; Song, J.H.; Lee, H.S.; Lee, H.O.; Lee, J.H.; Ahn, S.N.; Moon, B.C. Sequencing and comparative analysis of the chloroplast genome of Angelica polymorpha and the development of a novel indel marker for species identification. Molecules 2019, 24, 1038. [Google Scholar] [CrossRef]
- Shapiro, J.A.; von Sternberg, R. Why repetitive DNA is essential to genome function. Biol. Rev. Camb. Philos. Soc. 2005, 80, 227–250. [Google Scholar] [CrossRef] [PubMed]
- Schlötterer, C.; Vogl, C.; Tautz, D. Polymorphism and locus-specific effects on polymorphism at microsatellite loci in natural Drosophila melanogaster populations. Genetics 1997, 146, 309–320. [Google Scholar] [CrossRef]
- Layton, K.K.; Dempson, B.; Snelgrove, P.V.; Duffy, S.J.; Messmer, A.M.; Paterson, I.G.; Jeffery, N.W.; Kess, T.; Horne, J.B.; Salisbury, S.J. Resolving fine-scale population structure and fishery exploitation using sequenced microsatellites in a northern fish. Evol. Appl. 2020, 13, 1055–1068. [Google Scholar] [CrossRef] [PubMed]
- González-Castellano, I.; González-López, J.; González-Tizón, A.M.; Martínez-Lage, A. Genetic diversity and population structure of the rockpool shrimp Palaemon elegans based on microsatellites: Evidence for a cryptic species and differentiation across the Atlantic–Mediterranean transition. Sci. Rep. 2020, 10, 10784. [Google Scholar] [CrossRef]
- Liu, Z.; Biyashev, R.; Maroof, M.S. Development of simple sequence repeat DNA markers and their integration into a barley linkage map. Theor. Appl. Genet. 1996, 93, 869–876. [Google Scholar] [CrossRef]
- Schug, M.D.; Hutter, C.M.; Wetterstrand, K.A.; Gaudette, M.S.; Mackay, T.; Aquadro, C.F. The mutation rates of di-, tri-and tetranucleotide repeats in Drosophila melanogaster. Mol. Biol. Evol. 1998, 15, 1751–1760. [Google Scholar] [CrossRef]
- Dong, W.; Xu, C.; Cheng, T.; Zhou, S. Complete chloroplast genome of Sedum sarmentosum and chloroplast genome evolution in Saxifragales. PLoS ONE 2013, 8, e77965. [Google Scholar] [CrossRef]
- Zhang, H.; Li, C.; Miao, H.; Xiong, S. Insights from the complete chloroplast genome into the evolution of Sesamum indicum L. PLoS ONE 2013, 8, e80508. [Google Scholar]
- Han, S.; Ding, H.; Bi, D.; Zhang, S.; Yi, R.; Gao, J.; Yang, J.; Ye, Y.; Wu, L.; Kan, X. Structural diversities and phylogenetic signals in plastomes of the early-divergent Angiosperms: A case study in Saxifragales. Plants 2022, 11, 3544. [Google Scholar] [CrossRef]
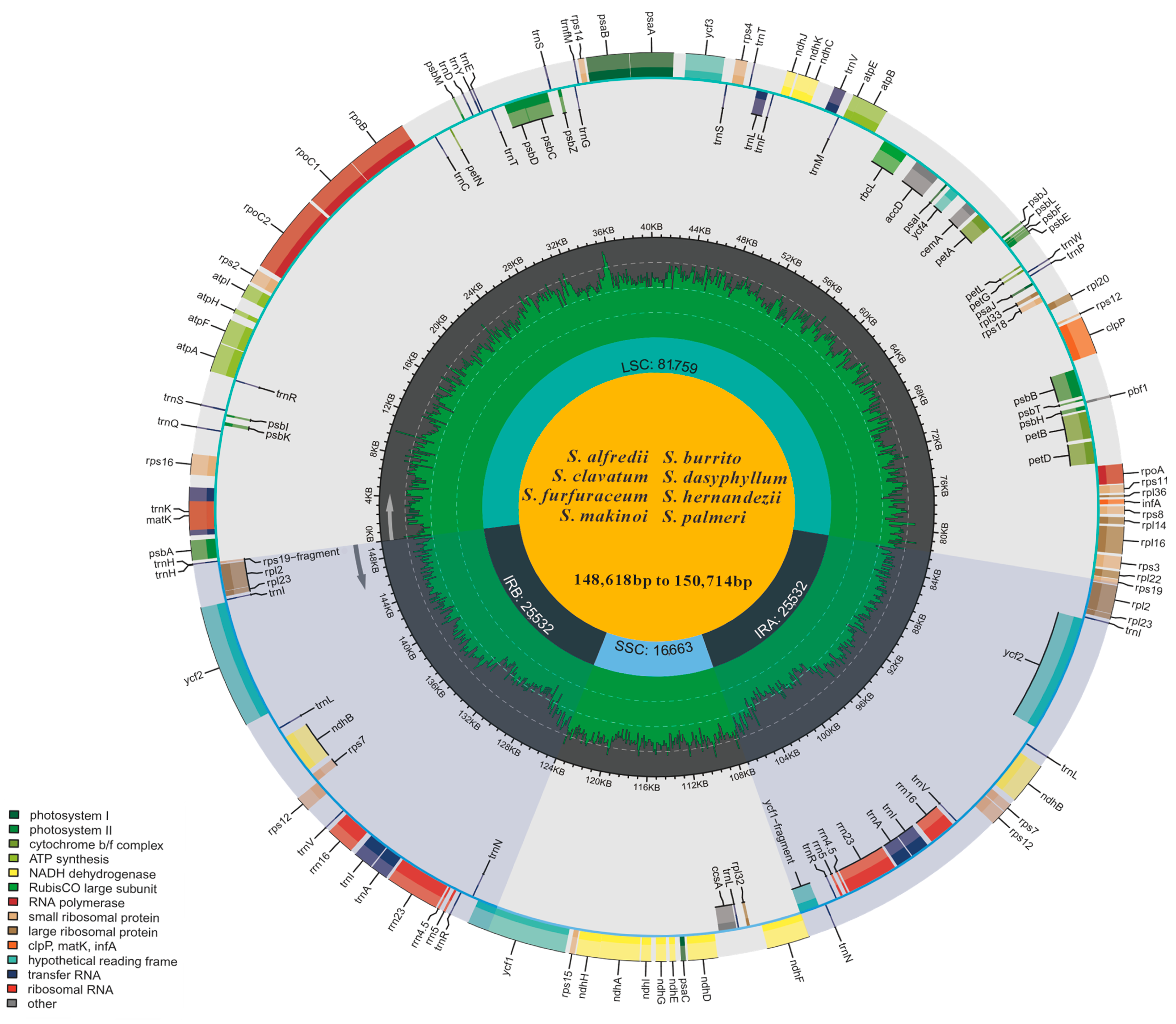
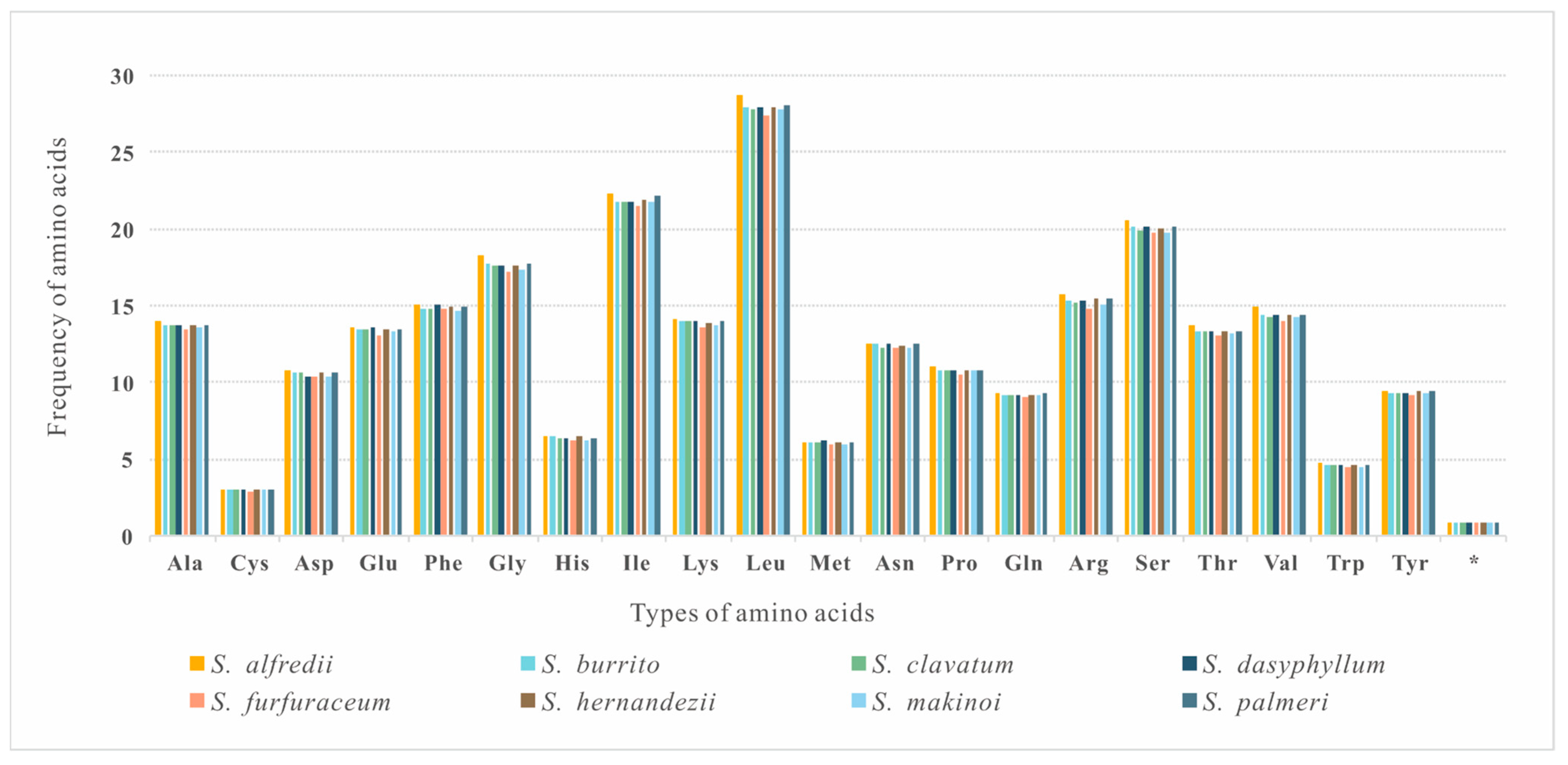
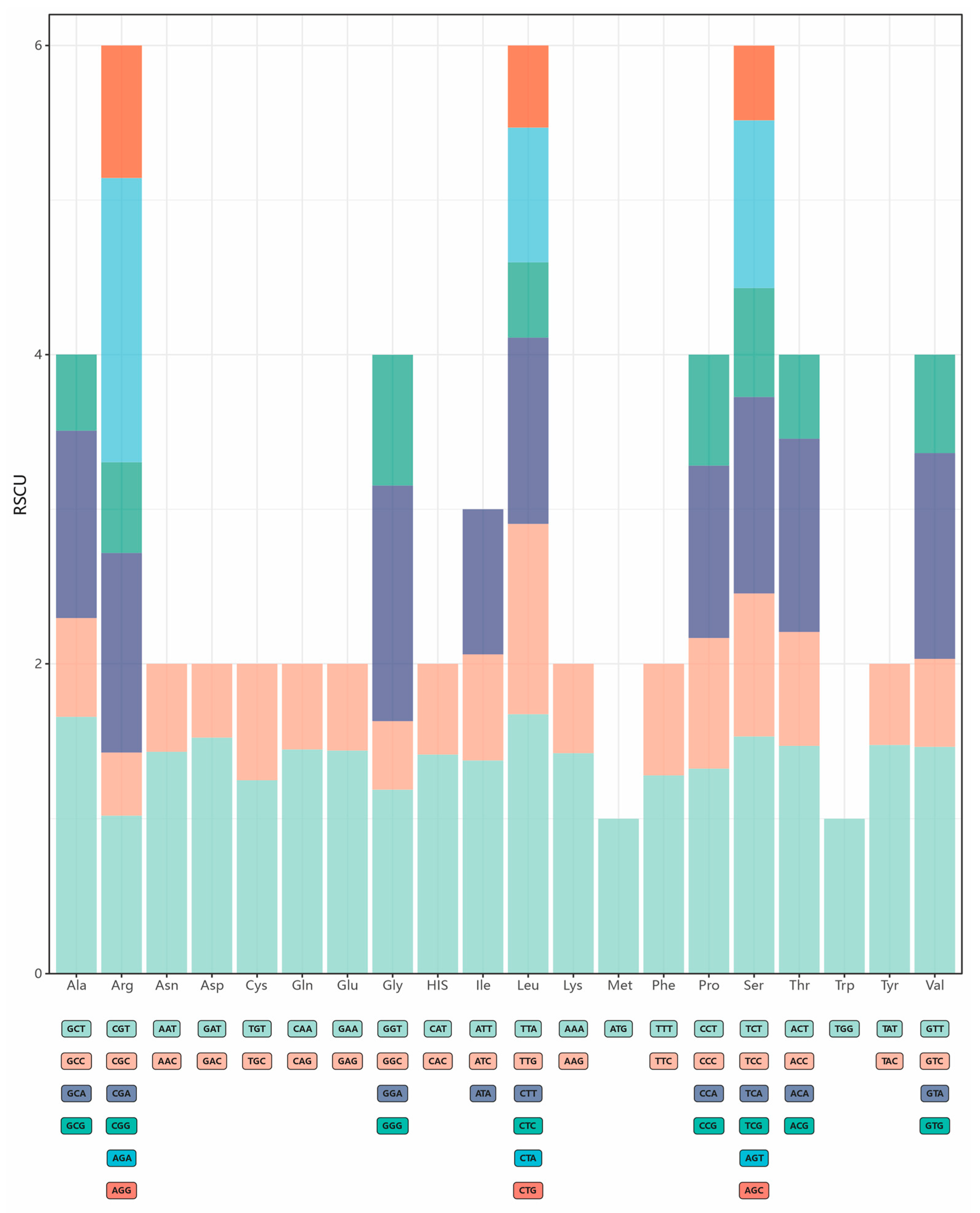

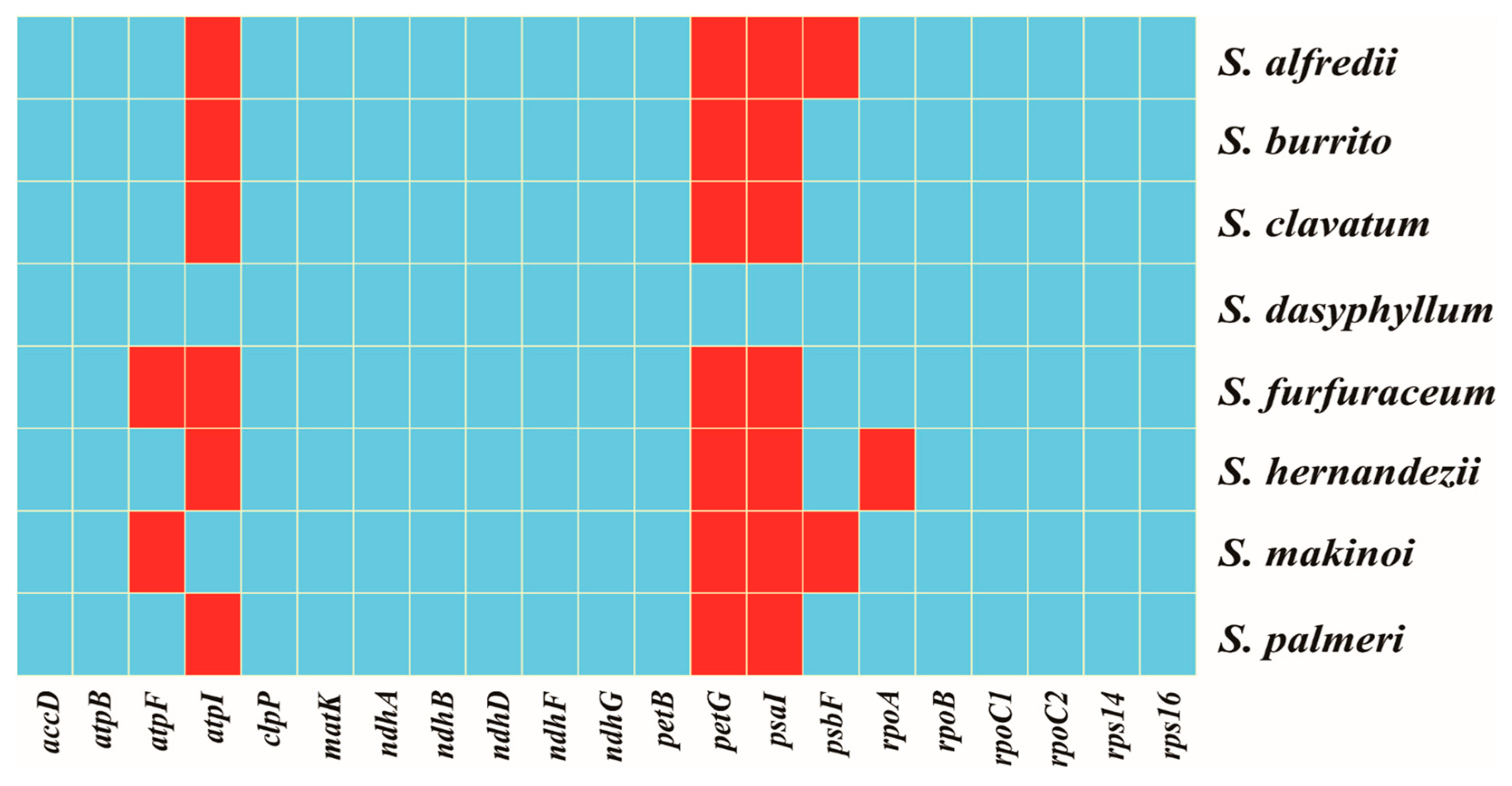
| Features | S. alfredii | S. burrito | S. clavatum | S. dasyphyllum | S. furfuraceum | S. hernandezii | S. makinoi | S. palmeri | |
|---|---|---|---|---|---|---|---|---|---|
| Voucher | AHNU-KL00119 | AHNU-KPXY016 | AHNU-KPLE014 | AHNU-KPDJ018 | AHNU-KPQM017 | AHNU-KPLG019 | AHNU-KL00214 | AHNU-KPBH015 | |
| Plastome size (bp) | 149,486 | 150,275 | 148,618 | 150,714 | 149,554 | 150,153 | 148,925 | 149,833 | |
| LSC length (bp) | 81,759 | 82,013 | 80,301 | 82,779 | 81,201 | 81,827 | 81,173 | 81,615 | |
| SSC length (bp) | 16,663 | 16,714 | 16,751 | 16,657 | 16,745 | 16,746 | 16,680 | 16,678 | |
| IR length (bp) | 25,532 | 25,774 | 25,783 | 25,639 | 25,804 | 25,790 | 25,536 | 25,770 | |
| Gene number | Total | 133 | 133 | 133 | 133 | 133 | 133 | 133 | 133 |
| IR regions | 21 | 21 | 21 | 21 | 21 | 21 | 21 | 21 | |
| CDS | 85 | 85 | 85 | 85 | 85 | 85 | 85 | 85 | |
| rRNAs | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | |
| tRNAs | 36 | 36 | 36 | 36 | 36 | 36 | 36 | 36 | |
| Pseudogenes | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | |
| GC content (%) | Overall | 37.7 | 37.9 | 38 | 37.8 | 37.9 | 37.8 | 37.8 | 37.8 |
| LSC | 35.7 | 35.8 | 36 | 35.8 | 35.9 | 35.8 | 35.7 | 35.8 | |
| SSC | 31.6 | 32 | 31.9 | 31.9 | 32 | 31.8 | 31.8 | 31.9 | |
| IR | 43 | 43 | 43 | 43 | 43 | 43 | 43 | 43 | |
| CDS | 37.7 | 37.8 | 37.7 | 38 | 37.7 | 37.8 | 37.6 | 37.7 | |
| rRNA | 55.4 | 55.4 | 55.4 | 55.4 | 55.4 | 55.4 | 55.4 | 55.4 | |
| tRNA | 52.9 | 52.8 | 52.9 | 53 | 52.8 | 52.8 | 53 | 52.8 | |
| Species | Length (bp) | T% | C% | A% | G% | AT% | AT-Skew | GC-Skew |
|---|---|---|---|---|---|---|---|---|
| S. alfredii | 149,486 | 31.37 | 19.23 | 30.88 | 18.51 | 62.25 | −0.00787 | −0.01908 |
| S. burrito | 150,275 | 31.29 | 19.30 | 30.83 | 18.58 | 62.12 | −0.00741 | −0.01901 |
| S. clavatum | 148,618 | 31.25 | 19.32 | 30.79 | 18.63 | 62.04 | −0.00741 | −0.01818 |
| S. dasyphyllum | 150,714 | 31.31 | 19.24 | 30.87 | 18.58 | 62.18 | −0.00708 | −0.01745 |
| S. furfuraceum | 149,554 | 31.29 | 19.32 | 30.79 | 18.60 | 62.08 | −0.00805 | −0.01899 |
| S. hernandezii | 150,153 | 31.33 | 19.27 | 30.84 | 18.57 | 62.17 | −0.00788 | −0.01850 |
| S. makinoi | 148,925 | 31.34 | 19.22 | 30.89 | 18.55 | 62.23 | −0.00723 | −0.01774 |
| S. palmeri | 149,833 | 31.34 | 19.28 | 30.81 | 18.57 | 62.15 | −0.00853 | −0.01876 |
| Average | 149,662 | 31.32 | 19.26 | 30.84 | 18.57 | 62.17 | −0.00774 | −0.01842 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, L.; Han, S.; Xiao, Y.; Zhang, M.; Kan, X. Eight New Sedum Plastomes: Comprehensive Analyses and Phylogenetic Implications. Genes 2025, 16, 761. https://doi.org/10.3390/genes16070761
Xu L, Han S, Xiao Y, Zhang M, Kan X. Eight New Sedum Plastomes: Comprehensive Analyses and Phylogenetic Implications. Genes. 2025; 16(7):761. https://doi.org/10.3390/genes16070761
Chicago/Turabian StyleXu, Liying, Shiyun Han, Yingying Xiao, Mengsa Zhang, and Xianzhao Kan. 2025. "Eight New Sedum Plastomes: Comprehensive Analyses and Phylogenetic Implications" Genes 16, no. 7: 761. https://doi.org/10.3390/genes16070761
APA StyleXu, L., Han, S., Xiao, Y., Zhang, M., & Kan, X. (2025). Eight New Sedum Plastomes: Comprehensive Analyses and Phylogenetic Implications. Genes, 16(7), 761. https://doi.org/10.3390/genes16070761






