Identification of Solanum lycopersicum L. Casein Kinase I-like Gene Family and Analysis of Abiotic Stress Response
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Stress Treatment
2.2. RNA Extraction and Expression Analysis
2.3. Identification of SlCKL Gene Family Members
2.4. Analysis of Physicochemical Properties of SlCKL Family Genes
2.5. Chromosomal Localization of SlCKL Genes
2.6. Structural Analysis of SlCKL Genes and Proteins
2.7. Phylogenetic Analysis of SlCKL Genes
2.8. Promoter Cis-Acting Element Analysis of SlCKL Genes
2.9. Collinearity Analysis of SlCKL Genes
2.10. Statistical Analysis
3. Results
3.1. Basic Information of SlCKL Gene Family in Tomato
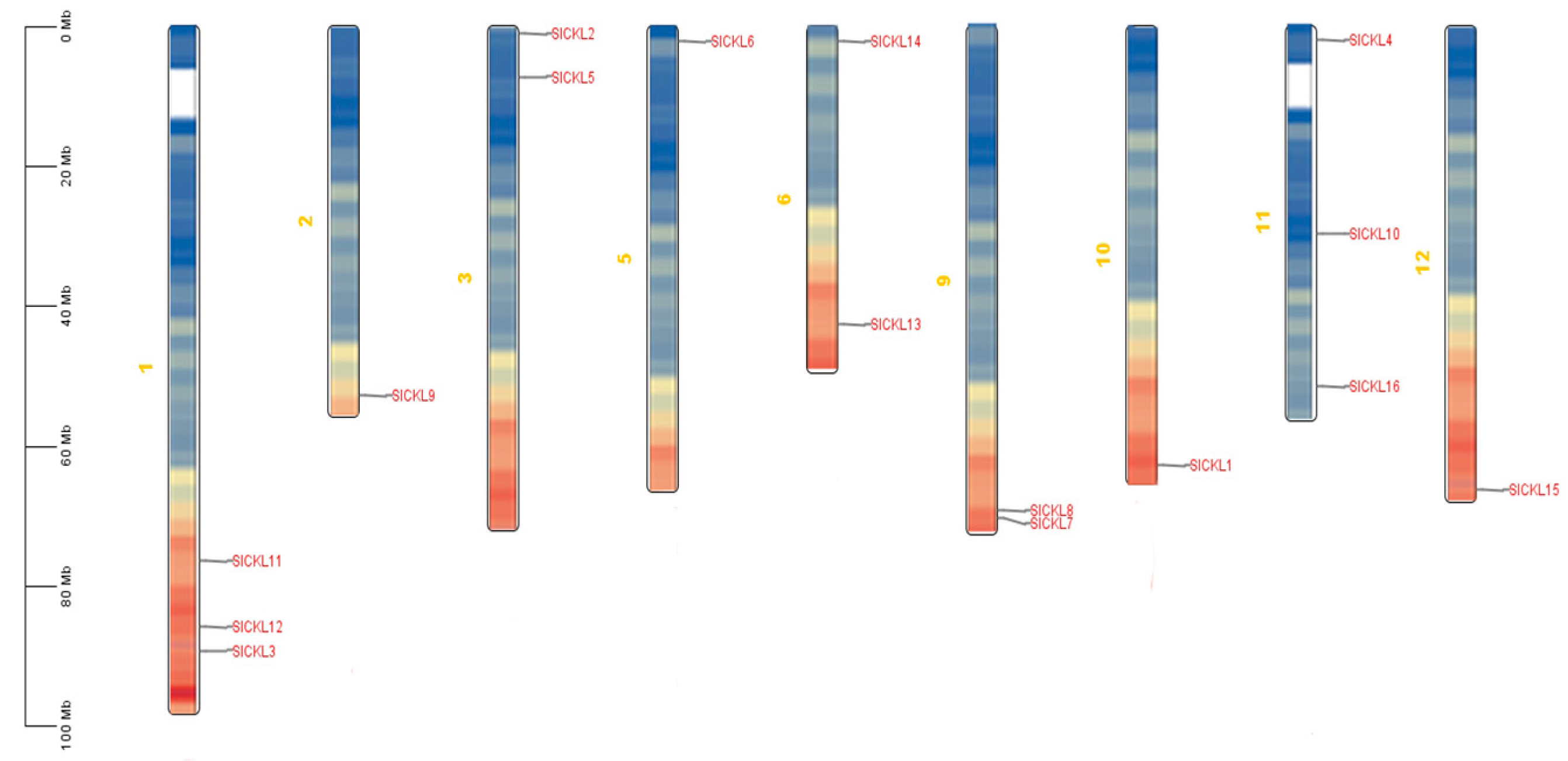
3.2. SlCKL Chromosome Distribution Analysis
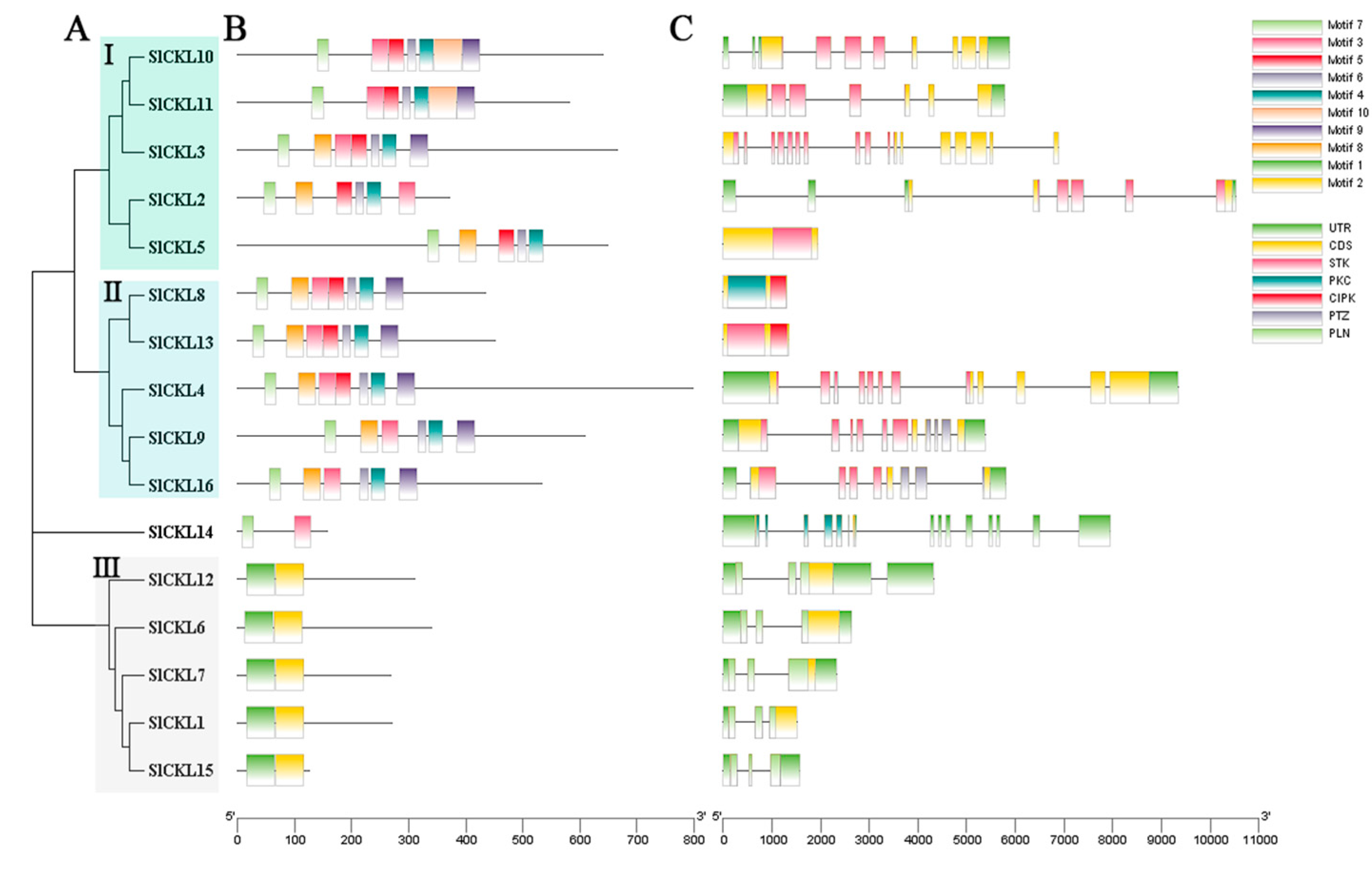
3.3. Structural Analysis of SlCKL Genes
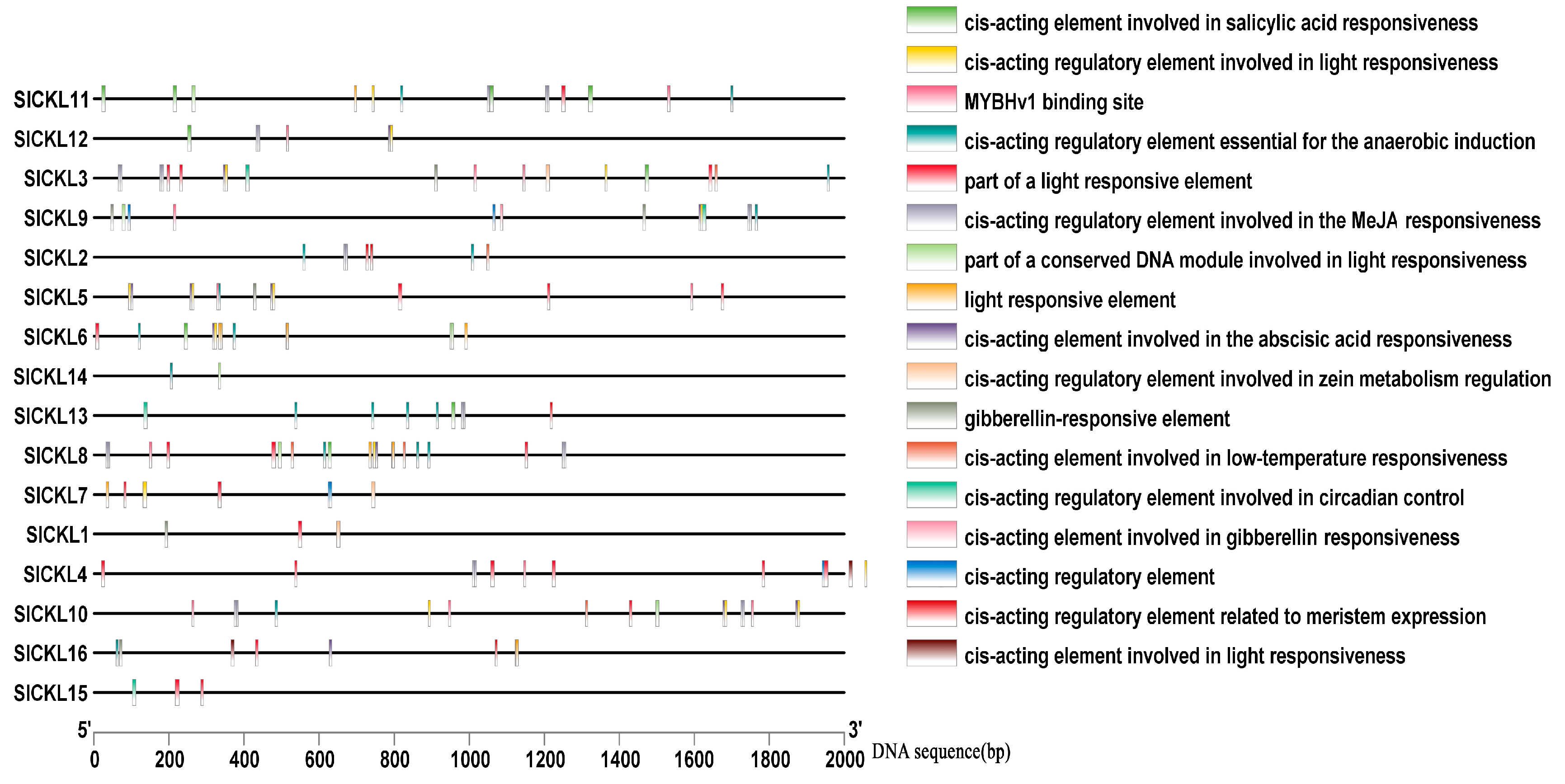
3.4. Cis-Acting Element Analysis of Promoter of SlCKL Genes
3.5. Multi-Species Collinearity Analysis of SlCKL Genes
3.6. Tissue Expression Analysis of SlCKL Gene
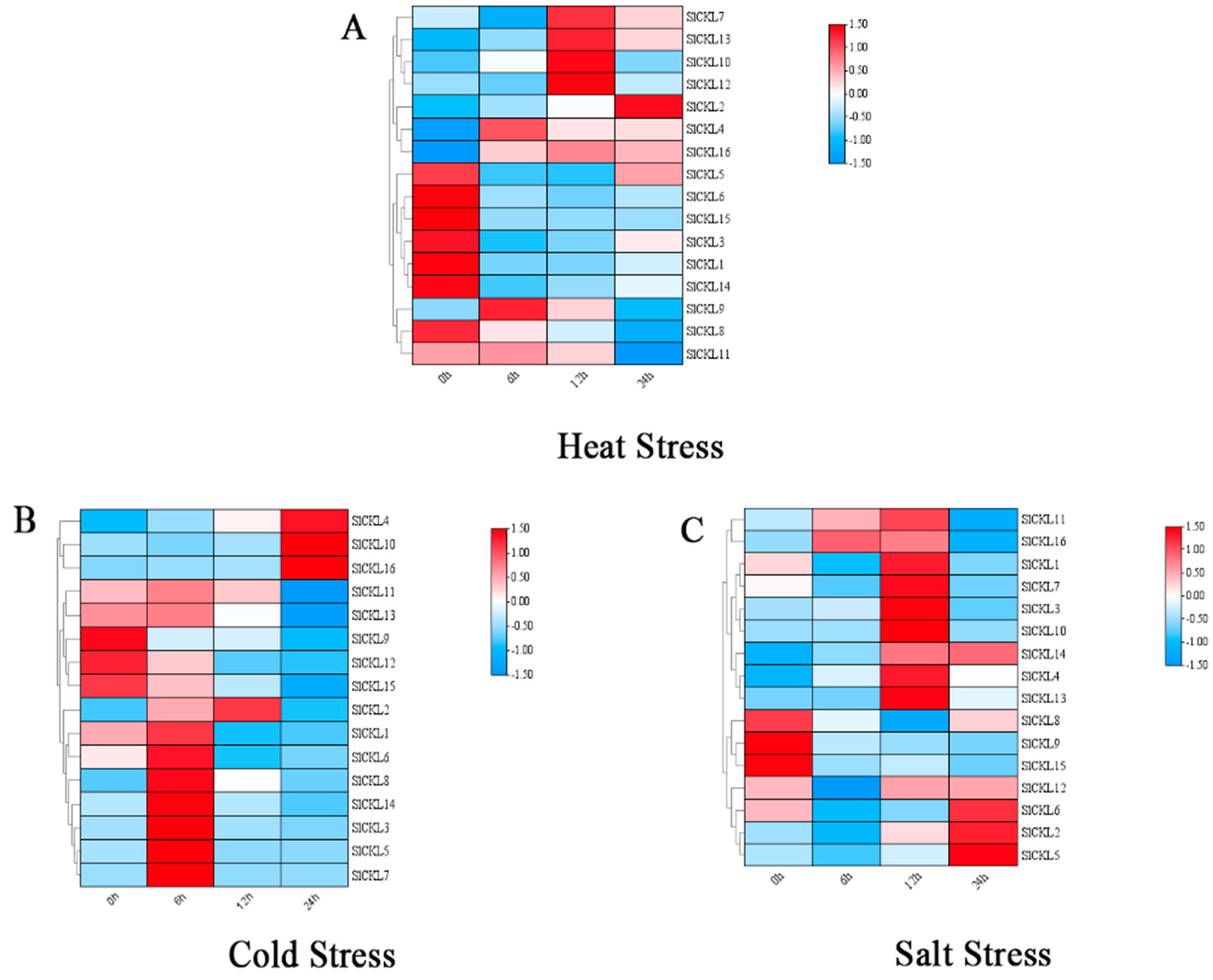
3.7. Expression Analysis of Tomato SlCKL Gene Under Abiotic Stress Conditions
4. Discussion
4.1. Identification of Tomato SlCKL Gene Family
4.2. Tomato SlCKL Gene Family Responds to Different Stresses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tuazon, P.T.; Traugh, J.A. Casein kinase I and II—Multipotential serine protein kinases: Structure, function, and regulation. Adv. Second. Messenger Phosphoprot. Res. 1991, 23, 123–164. [Google Scholar]
- Gross, S.D.; Anderson, R.A. Casein kinase I: Spatial organization and positioning of a multifunctional protein kinase family. Cell. Signal. 1998, 10, 699–711. [Google Scholar] [CrossRef]
- Knippschild, U.; Gocht, A.; Wolff, S.; Huber, N.; Löhler, J.; Stöter, M. The casein kinase 1 family: Participation in multiple cellular processes in eukaryotes. Cell. Signal. 2005, 17, 675–689. [Google Scholar] [CrossRef]
- Bidère, N.; Ngo, V.N.; Lee, J.; Collins, C.; Zheng, L.; Wan, F.; Davis, R.E.; Lenz, G.; Anderson, D.E.; Arnoult, D.; et al. Casein kinase 1α governs antigen-receptor-induced NF-κB activation and human lymphoma cell survival. Nature 2009, 458, 92–96. [Google Scholar] [CrossRef]
- Mehra, A.; Shi, M.; Baker, C.L.; Colot, H.V.; Loros, J.J.; Dunlap, J.C. A role for casein kinase 2 in the mechanism underlying circadian temperature compensation. Cell 2009, 137, 749–760. [Google Scholar] [CrossRef]
- Honaker, Y.; Piwnica-Worms, H. Casein kinase 1 functions as both penultimate and ultimate kinase in regulating Cdc25A destruction. Oncogene 2010, 29, 3324–3334. [Google Scholar] [CrossRef]
- Rumpf, C.; Cipak, L.; Dudas, A.; Benko, Z.; Pozgajova, M.; Riedel, C.G.; Ammerer, G.; Mechtler, K.; Gregan, J. Casein kinase 1 is required for efficient removal of Rec8 during meiosis I. Cell Cycle 2010, 9, 2657–2662. [Google Scholar] [CrossRef]
- Sugiyama, Y.; Hatano, N.; Sueyoshi, N.; Suetake, I.; Tajima, S.; Kinoshita, E.; Kinoshita-Kikuta, E.; Koike, T.; Kameshita, I. The DNA-binding activity of mouse DNA methyltransferase 1 is regulated by phosphorylation with casein kinase 1δ/ε. Biochem. J. 2010, 427, 489–497. [Google Scholar] [CrossRef]
- Vancura, A.; Sessler, A.; Leichus, B.; Kuret, J. A prenylation motif is required for plasma membrane localization and biochemical function of casein kinase I in budding yeast. J. Biol. Chem. 1994, 269, 19271–19278. [Google Scholar] [CrossRef]
- Liu, W.; Xu, Z.H.; Luo, D.; Xue, H.W. Roles of OsCKI1, a rice casein kinase I, in root development and plant hormone sensitivity. Plant J. 2003, 36, 189–202. [Google Scholar] [CrossRef]
- Graves, P.R.; Roach, P.J. Role of COOH-terminal Phosphorylation in the Regulation of Casein Kinase Iδ (∗). J. Biol. Chem. 1995, 270, 21689–21694. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Ye, J.; Guo, X.; Chang, H.; Yuan, C.; Wang, Y.; Hu, S.; Liu, X.; Li, X. Arabidopsis casein kinase 1-like 2 involved in abscisic acid signal transduction pathways. J. Plant Interact. 2014, 9, 19–25. [Google Scholar] [CrossRef]
- Lee, J.Y.; Taoka, K.; Yoo, B.C.; Ben-Nissan, G.; Kim, D.J.; Lucas, W.J. Plasmodesmal-associated protein kinase in tobacco and Arabidopsis recognizes a subset of non-cell-autonomous proteins. Plant Cell 2005, 17, 2817–2831. [Google Scholar] [CrossRef]
- Wang, M.; Yu, D.; Guo, X.; Li, X.; Zhang, J.; Zhao, L.; Chang, H.; Hu, S.; Zhang, C.; Shi, J.; et al. Casein kinase 1-Like 3 is required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Afr. J. Biotechnol. 2011, 10, 13219–13229. [Google Scholar]
- Lee, J.Y. Versatile casein kinase 1: Multiple locations and functions. Plant Signal. Behav. 2009, 4, 652–654. [Google Scholar] [CrossRef]
- Zhao, S.; Jiang, Y.; Zhao, Y.; Huang, S.; Yuan, M.; Zhao, Y.; Guo, Y. CASEIN KINASE1-LIKE PROTEIN2 regulates actin filament stability and stomatal closure via phosphorylation of actin depolymerizing factor. Plant Cell 2016, 28, 1422–1439. [Google Scholar] [CrossRef]
- Tan, S.T.; Xue, H.W. Casein kinase 1 regulates ethylene synthesis by phosphorylating and promoting the turnover of ACS5. Cell Rep. 2014, 9, 1692–1702. [Google Scholar] [CrossRef]
- Mizrahi, Y. Effect of salinity on tomato fruit ripening. Plant Physiol. 1982, 69, 966–970. [Google Scholar] [CrossRef]
- Mizrahi, Y.; Pasternak, D.O.V. Effect of salinity on quality of various agricultural crops. Plant Soil 1985, 89, 301–307. [Google Scholar] [CrossRef]
- Liu, H. Transcriptome Analysis and Related Gene Function Identification of Solanum lycopersicum L. Cold-Tolerant Germplasm Under Low Temperature Stress. Ph.D. Thesis, Zhonghua Agricultural University, Wuhan, China, 2012. [Google Scholar]
- Li, C.; Yan, J.M.; Li, Y.Z.; Zhang, Z.C.; Wang, Q.L.; Liang, Y. Silencing the SpMPK1, SpMPK2, and SpMPK3 genes in tomato reduces abscisic acid—Mediated drought tolerance. Int. J. Mol. Sci. 2013, 14, 21983–21996. [Google Scholar] [CrossRef]
- Huanwen, M.; Zhihui, C.; Yang, W. Impact of temperature stress on invertase expression and photosynthetic characteristic in tomato plant. J. Northwest Sci-Tech Univ. Agric. For. 2006, 34, 41–47. [Google Scholar]
- Bai, P.; Zou, Z.; Yang, Z.; Hu, X.; Zhao, Y. Effects of different temperature and light treatments on sugar content in different parts of Solanum lycopersicum L fruit. Northwest Agric. J. 2010, 19, 184–187. [Google Scholar]
- Li, Y.; Min, L.; Zhang, L.; Hu, Q.; Wu, Y.; Li, J.; Xie, S.; Ma, Y.; Zhang, X.; Zhu, L. Promoters of Arabidopsis Casein kinase I-like 2 and 7 confer specific high-temperature response in anther. Plant Mol. Biol. 2018, 98, 33–49. [Google Scholar] [CrossRef]
- Wang, Q. Study on Mechanism of FIM1 in Arabidopsis thaliana Regulating Stomatal Movement Through Dynamic Changes of Microfilaments. Ph.D. Thesis, Shandong Normal University, Jinan, China, 2022. [Google Scholar]
- Wang, Y.; Zhang, J.; Hu, Z.; Guo, X.; Tian, S.; Chen, G. Genome-wide analysis of the MADS-box transcription factor family in Solanum lycopersicum. Int. J. Mol. Sci. 2019, 20, 2961. [Google Scholar] [CrossRef]
- Chen, F.; Chen, Q.; Lin, J.; Wang, Y.; Liu, H.; Liang, B.; Deng, Y.; Ren, C.; Zhang, Y.; Yang, F.; et al. Identification of DIR gene family in Solanum lycopersicum L and analysis of its response to abiotic stress. China Agric. Sci. 2022, 55, 3807–3824. [Google Scholar]
- Kim, M.J.; Go, Y.S.; Lee, S.B.; Kim, Y.S.; Shin, J.S.; Min, M.K.; Hwang, I.; Suh, M.C. Seed-expressed casein kinase I acts as a positive regulator of the SeFAD2 promoter via phosphorylation of the SebHLH transcription factor. Plant Mol. Biol. 2010, 73, 425–437. [Google Scholar] [CrossRef]
- Dai, C.; Xue, H.W. Rice early flowering 1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J. 2010, 29, 1916–1927. [Google Scholar] [CrossRef]
- Menkens, A.E.; Schindler, U.; Cashmore, A.R. The G-box: A ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem. Sci. 1995, 20, 506–510. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Fujita, M.; Satoh, R.; Maruyama, K.; Parvez, M.M.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 2005, 17, 3470–3488. [Google Scholar] [CrossRef]
- Liu, S.; Chen, H.; Li, X.; Zhang, W. A low-temperature-responsive element involved in the regulation of the Arabidopsis thaliana At1g71850/At1g71860 divergent gene pair. Plant Cell Rep. 2016, 35, 1757–1767. [Google Scholar] [CrossRef]
- Shariatipour, N.; Heidari, B. Investigation of drought and salinity tolerance related genes and their regulatory mechanisms in Arabidopsis (Arabidopsis thaliana). Open Bioinform. J. 2018, 11, 12. [Google Scholar] [CrossRef]
- Zhao, M.; Liu, Z.; Gan, J.; Yang, C.; Lu, A.; Han, Q.; Yang, H.; Xu, Y.; Sun, G.; Wu, D. Identification and expression analysis of XIP gene family members in rice. Genetica 2024, 152, 83–100. [Google Scholar] [CrossRef]
- Li, H.L.; Sun, Z.Y.; Zhao, L.J.; Han, L.; Ju, G.S. Effect of jasmonic acid and methyl jasmonate on the plant development and resistance. Chin. Agric. Sci. Bull. 2009, 25, 167–172. [Google Scholar]
- Zhu, F.F.; Liu, Y.Q.; Chen, Z.X.; Lan, J.B. Effect of abscisic acid on physiological characteristics in Lonicera macranthoides seedlings under salt stress. J. Chin. Med. Mater. 2013, 36, 1043–1046. [Google Scholar]
- Tuteja, N. Abscisic acid and abiotic stress signaling. Plant Signal. Behav. 2007, 2, 135–138. [Google Scholar] [CrossRef]
- Mahajan, S.; Tuteja, N. Cold, salinity and drought stresses: An overview. Arch. Biochem. Biophys. 2005, 444, 139–158. [Google Scholar] [CrossRef]
- Swamy, P.M.; Smith, B.N. Role of abscisic acid in plant stress tolerance. Curr. Sci. 1999, 76, 1220–1227. [Google Scholar]
- Thomashow, M.F. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 2000, 3, 217–223. [Google Scholar] [CrossRef]
- Uno, Y.; Furihata, T.; Abe, H.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 2000, 97, 11632–11637. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef]
- Xu, L.; Lin, Z.; Tao, Q.; Liang, M.; Zhao, G.; Yin, X.; Fu, R. Multiple NUCLEAR FACTOR Y transcription factors respond to abiotic stress in Brassica napus L. PLoS ONE 2014, 9, e111354. [Google Scholar] [CrossRef]
- Dinneny, J.R.; Long, T.A.; Wang, J.Y.; Jung, J.W.; Mace, D.; Pointer, S.; Barron, C.; Brady, S.M.; Schiefelbein, J.; Benfey, P.N. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 2008, 320, 942–945. [Google Scholar] [CrossRef]



| Gene Name | Forword (5′–3′) | Reverse (5′–3′) |
|---|---|---|
| SlCKL1 | CTGGGTTGATGAGATGCAGC | GCTGCCCATTTGTTACCCAA |
| SlCKL2 | CCTAGAATCGGCATGGGAGT | TGTGTGCTGCTAAAACCACC |
| SlCKL3 | TTGGCGAAAGAACAGGAAGC | ACTCCTCCGTGTACTTCTGC |
| SlCKL4 | GGAAAGGCCGTTTGTCTGAA | TACATAAGCGGCTGTTCCCA |
| SlCKL5 | AACAGGACGAGCAAGCAAAG | GCATCACAATTCCCTGTCCC |
| SlCKL6 | GCCAAGCAGAACAGATAACGA | TCCCATTCAGCCATGTGACT |
| SlCKL7 | TGGGGTTAAAGAAAGGGCCA | CACATCGCAATAATCCGGCT |
| SlCKL8 | AGGGTACGATGGTGCTAAGG | ATCCTCCGGGCTGATTTTGA |
| SlCKL9 | GTCTCCAGCGAAACACATCC | GCCCTCTTCCAACTTCCTCT |
| SlCKL10 | GAAAGGCGTGGTACTGCTTC | CGCCTTCTTGCTTCCTCTTC |
| SlCKL11 | GATGACAATGCACGGCCTAG | GCCCAATCTTAGCCAGCTTC |
| SlCKL12 | GACCTTGGACCCCTGAAGAA | CCTCGTATGATGTCTGGCCT |
| SlCKL13 | CGAATGGGCGAAAAGGGAAA | CTTTAAGGGGAGGCCGTACA |
| SlCKL14 | AGGTCGCAAAGCAAATCAGG | CTCCTGCTTTGCTCAACTCC |
| SlCKL15 | TGCCTATGAATGCTGGACTTC | TCCGGCTATCAAGGACCATC |
| SlCKL16 | GATGTCTGGAGTGCTGGAGT | TCCGGGTCAAGCATCTTCTT |
| SleiF | ATCCTTCAGAGCGGTGTTCA | ATCTCAAGAGCCTCTGGTGG |
| Gene | Gene Number | Amino Acids Length | Chromosomes Positioning | Pl | Mw | Subcellular Positioning | Hydrophilicity | Unstable Index |
| SlCKL1 | Solyc10g081490.2.1 | 271 | 10 | 6.05 | 30.86 | nucl | −0.774 | 55.75 |
| SlCKL2 | Solyc03g006500.3.1 | 371 | 3 | 8.11 | 41.24 | chlo | −0.35 | 39.82 |
| SlCKL3 | Solyc01g098980.3.1 | 665 | 1 | 5.2 | 72.68 | chlo | −0.45 | 50.66 |
| SlCKL4 | Solyc11g007760.2.1 | 798 | 11 | 7.31 | 89.32 | extr | −0.352 | 38.87 |
| SlCKL5 | Solyc03g043710.1.1 | 648 | 3 | 5.68 | 71.93 | vacu | −0.097 | 27.68 |
| SlCKL6 | Solyc05g007710.3.1 | 339 | 5 | 8.60 | 38.65 | nucl | −0.712 | 47.85 |
| SlCKL7 | Solyc09g090130.3.1 | 269 | 9 | 5.76 | 30.21 | nucl | −0.751 | 53.54 |
| SlCKL8 | Solyc09g083100.1.1 | 435 | 9 | 8.73 | 48.97 | chlo | −0.306 | 39.75 |
| SlCKL9 | Solyc02g090510.3.1 | 608 | 2 | 9.14 | 67.92 | chlo | −0.401 | 52.33 |
| SlCKL10 | Solyc11g045610.2.1 | 641 | 11 | 9.03 | 71.16 | nucl | −0.545 | 50.64 |
| SlCKL11 | Solyc01g067640.3.1 | 581 | 1 | 9.51 | 64.26 | nucl | −0.657 | 56.96 |
| SlCKL12 | Solyc01g094360.3.1 | 310 | 1 | 7.05 | 34.62 | nucl | −0.707 | 57.9 |
| SlCKL13 | Solyc06g068450.2.1 | 451 | 6 | 9.03 | 51.05 | chlo | −0.363 | 35.61 |
| SlCKL14 | Solyc06g008320.3.1 | 158 | 6 | 9.21 | 17.94 | mito | −0.207 | 30.8 |
| SlCKL15 | Solyc12g096200.2.1 | 126 | 12 | 9.73 | 14.20 | nucl | −0.504 | 45.42 |
| SlCKL16 | Solyc11g065660.2.1 | 534 | 11 | 6.03 | 59.66 | cyto | −0.46 | 37.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, M.; Xie, X.; Wang, Q.; Wang, X.; Zhang, Y. Identification of Solanum lycopersicum L. Casein Kinase I-like Gene Family and Analysis of Abiotic Stress Response. Genes 2025, 16, 757. https://doi.org/10.3390/genes16070757
Jia M, Xie X, Wang Q, Wang X, Zhang Y. Identification of Solanum lycopersicum L. Casein Kinase I-like Gene Family and Analysis of Abiotic Stress Response. Genes. 2025; 16(7):757. https://doi.org/10.3390/genes16070757
Chicago/Turabian StyleJia, Miao, Xiaoxiao Xie, Quanhua Wang, Xiaoli Wang, and Yingying Zhang. 2025. "Identification of Solanum lycopersicum L. Casein Kinase I-like Gene Family and Analysis of Abiotic Stress Response" Genes 16, no. 7: 757. https://doi.org/10.3390/genes16070757
APA StyleJia, M., Xie, X., Wang, Q., Wang, X., & Zhang, Y. (2025). Identification of Solanum lycopersicum L. Casein Kinase I-like Gene Family and Analysis of Abiotic Stress Response. Genes, 16(7), 757. https://doi.org/10.3390/genes16070757







