Exploring Runs of Homozygosity and Heterozygosity in Sheep Breeds Maintained in Poland
Abstract
1. Introduction
2. Material and Methods
2.1. Study Material, DNA Isolation and Genotyping, and Filtering of Genotypic Data
2.2. Identification of Runs of Homozygosity and Heterozygosity, and Estimation of Genomic Inbreeding
Identification of ROH Hotspot Regions and Association with QTL Database
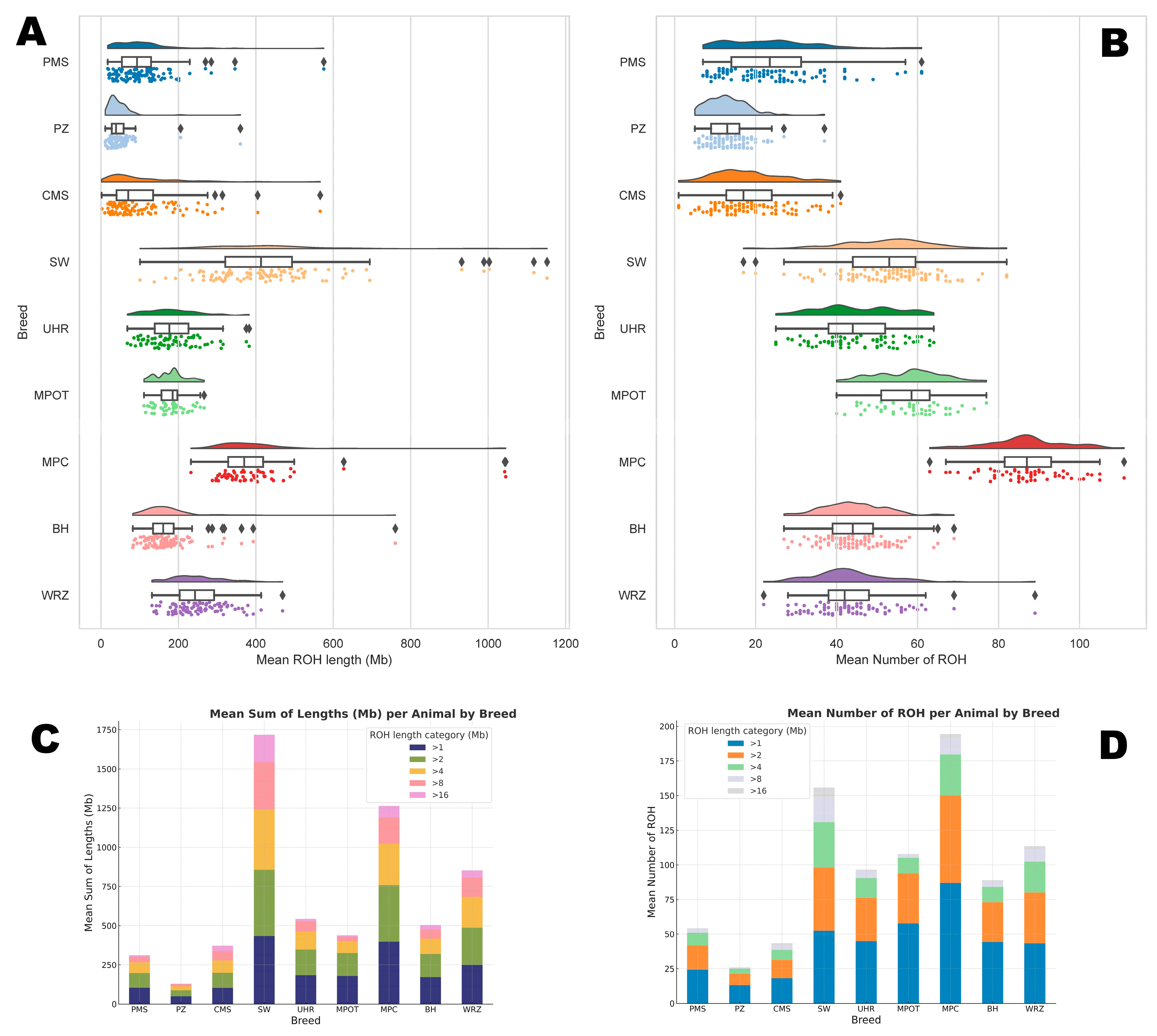
3. Results
3.1. Number of ROH per Animal
3.2. Sums of ROH Lengths per Animal
3.3. Assessment of Genomic Inbreeding Levels
4. Distribution of ROHet
4.1. ROHet Segment Counts per Animal
4.2. ROHet Segment Lengths per Animal
5. ROH Islands
ROH Island Distribution
6. Discussion
6.1. Genetic Diversity of Polish Sheep Breeds
6.2. Patterns of Runs of Homozygosity Across Polish Sheep Breeds in a Global Context
6.3. Runs of Heterozygosity Profiles
6.4. ROH Islands
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Purfield, D.C.; Berry, D.P.; McParland, S.; Bradley, D.G. Runs of homozygosity and population history in cattle. BMC Genet. 2012, 13, 70. [Google Scholar] [CrossRef] [PubMed]
- Curik, I.; Ferencakovic, M.; Solkner, J. Inbreeding and runs of homozygosity: A possible solution to an old problem. Livest. Sci. 2014, 166, 26–34. [Google Scholar] [CrossRef]
- Larsson, M.N.A.; Morell Miranda, P.; Pan, L.; Başak Vural, K.; Kaptan, D.; Rodrigues Soares, A.E.; Galnther, T. Ancient sheep genomes reveal four millennia of north european short-tailed sheep in the baltic sea region. Genome Biol. Evol. 2024, 16, evae114. [Google Scholar] [CrossRef] [PubMed]
- Kizilaslan, M.; Arzik, Y.; Behrem, S.; White, S.N.; Cinar, M.U. Comparative genomic characterization of indigenous fat-tailed akkaraman sheep with local and transboundary sheep breeds. Food Energy Secur. 2023, 13, e508. [Google Scholar] [CrossRef]
- Deng, T.X.; Ma, X.Y.; Duan, A.Q.; Lu, X.R.; Abdel-Shafy, H. Genomic insights into selection signatures and candidate genes for milk production traits in buffalo population. Animal 2025, 19, 101427. [Google Scholar] [CrossRef]
- Yuan, J.; Li, S.; Sheng, Z.; Zhang, M.; Liu, X.; Yuan, Z.; Yang, N.; Jilan, C. Genome-wide run of homozygosity analysis reveals candidate genomic regions associated with environmental adaptations of Tibetan native chickens. BMC Genom. 2022, 23, 91. [Google Scholar] [CrossRef]
- Peripolli, E.; Munari, D.P.; Silva, M.V.G.B.; Lima, A.L.F.; Irgang, R.; Baldi, F. Runs of homozygosity: Current knowledge and applications in livestock. Anim. Genet. 2017, 48, 255–271. [Google Scholar] [CrossRef]
- Ma, R.; Liu, J.; Ma, X.; Yang, J. Genome-wide runs of homozygosity reveal inbreeding levels and trait-associated candidate genes in diverse sheep breeds. Genes 2025, 16, 316. [Google Scholar] [CrossRef]
- Szmatoła, T.; Gurgul, A.; Jasielczuk, I.; Oclon, E.; Ropka-Molik, K.; Stefaniuk-Szmukier, M.; Polak, G.; Tomczyk-Wrona, I.; Bugno-Poniewierska, M. Assessment and Distribution of Runs of Homozygosity in Horse Breeds Representing Different Utility Types. Animals 2022, 12, 3293. [Google Scholar] [CrossRef]
- Dlamini, N.M.; Visser, C.; Snyman, M.; Soma, P.; Muchadeyi, F.C. Genomic evaluation of resistance to haemonchus contortus in a south african dohne merino flock. Small Rumin. Res. 2019, 175, 117–125. [Google Scholar] [CrossRef]
- Abied, A.; Liu, X.; Sahlu, B.W.; Feng, X.; Ahbara, A.; Yun, P.; Ma, Y. Genome-wide analysis revealed homozygosity and demographic history of five chinese sheep breeds adapted to different environments. Genes 2020, 11, 1480. [Google Scholar] [CrossRef] [PubMed]
- Ghoreishifar, S.M.; Moradi-Shahrbabak, H.; Moradi-Shahrbabak, M.; Nicolazzi, E.L.; Stella, A.; van der Werf, J.H.J. Genomic measures of inbreeding and their association with production traits in Australian sheep breeds. Anim. Genet. 2019, 50, 655–658. [Google Scholar]
- Chen, Z.; Zhang, Z.; Wang, Z.; Zhang, Z.; Wang, Q.; Pan, Y. Heterozygosity and homozygosity regions affect reproductive success and the loss of reproduction: A case study with litter traits in pigs. Comput. Struct. Biotechnol. J. 2022, 20, 4060–4071. [Google Scholar] [CrossRef]
- Liu, S.; Xu, Y.; Chen, Z.; Li, H.; Zhang, Z.; Wang, Q.; Pan, Y. Genome-wide detection of runs of homozygosity and heterozygosity in tunchang pigs. Animal 2024, 18, 101236. [Google Scholar] [CrossRef] [PubMed]
- Gurgul, A.; Jasielczuk, I.; Miksza-Cybulska, A.; Kawecka, A.; Szmatoła, T.; Krupinski, J. Evaluation of genetic differentiation and genome-wide selection signatures in polish local sheep breeds. Livest. Sci. 2021, 251, 104635. [Google Scholar] [CrossRef]
- Jasielczuk, I.; Szmatoła, T.; Miksza-Cybulska, A.; Kawecka, A.; Gurgul, A. Linkage disequilibrium, historical effective population size and haplotype blocks in selected native sheep breeds. Small Rumin. Res. 2023, 228, 107109. [Google Scholar] [CrossRef]
- Biscarini, F.; Cozzi, P.; Gaspa, G.; Marras, G. Detect runs of homozygosity and runs of heterozygosity in diploid genomes using R. Genet. Sel. Evol. 2018, 50, 1–9. [Google Scholar]
- Santos, D.J.A.; Silva, F.F.; Braz, C.U.; Utsunomiya, A.T.H.; Bombasa, A.C.S.; Utsunomiya, Y.T.; Garcia, J.F. Unveiling ancestry, selection signature, and runs of heterozygosity in composite beef cattle through genomic analyses. Front. Genet. 2021, 12, 670034. [Google Scholar]
- Li, X.; Liu, J.; Zhang, W.; Wang, Z.; Li, C.; Li, J.; Zhang, Q. Genomic characterization of ROH and ROHet in Chinese indigenous sheep breeds. Genes 2022, 13, 1397. [Google Scholar] [CrossRef]
- McQuillan, R.; Leutenegger, A.L.; Abdel-Rahman, R.; Franklin, C.S.; Pericic, M.; Barac-Latas, V.; Wilson, J.F. Runs of homozygosity in European populations. Am. J. Hum. Genet. 2008, 83, 359–372. [Google Scholar] [CrossRef]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Zhao, Y. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef] [PubMed]
- Mastrangelo, S.; Tolone, M.; Di Gerlando, R.; Fontanesi, L.; Sardina, M.T.; Portolano, B. Genomic inbreeding estimation in small populations: Evaluation of runs of homozygosity in three local dairy sheep breeds. Animal 2017, 11, 738–745. [Google Scholar] [CrossRef] [PubMed]
- Granero, A.; Anaya, G.; Demyda-Peyras, S.; Alcalde, M.J.; Arrebola, F.; Molina, A. Genomic Population Structure of the Main Historical Genetic Lines of Spanish Merino Sheep. Animals 2022, 12, 1327. [Google Scholar] [CrossRef] [PubMed]
- Signer-Hasler, H.; Burren, A.; Ammann, P.; Drögemülle, C.; Flury, C. Runs of homozygosity and signatures of selection: A comparison among eight local swiss sheep breeds. Anim. Genet. 2019, 50, 512–525. [Google Scholar] [CrossRef]
- Megdiche, S.; Mastrangelo, S.; Hamouda, M.B.; Lenstra, J.A.; Ciani, E. A combined multi-cohort approach reveals novel and known genome-wide selection signatures for wool traits in merino and merino-derived sheep breeds. Front. Genet. 2019, 10, 1025. [Google Scholar] [CrossRef]
- Ceccobelli, S.; Landi, V.; Senczuk, G.; Mastrangelo, S.; Sardina, M.T.; Jemaa, S.B.; Pilla, F. A comprehensive analysis of the genetic diversity and environmental adaptability in worldwide merino and merino-derived sheep breeds. Genet. Sel. Evol. 2023, 55, 24. [Google Scholar] [CrossRef]
- Kerr, E.; Marr, M.M.; Collins, L.; Dubarry, K.; Salavati, M.; Scinto, A.; Woolley, S.; Clark, E.L. Analysis of genotyping data reveals the unique genetic diversity represented by the breeds of sheep native to the United Kingdom. BMC Genom. Data 2024, 25, 82. [Google Scholar] [CrossRef]
- Zsolnai, A.; Egerszegi, I.; Rózsa, L.; Anton, I. Genetic status of lowland-type Racka sheep colour variants. Animal 2021, 15, 100080. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Larranaga, O.; Langa, J.; Rendo, F.; Manzano, C.; Iriondo, M.; Estonba, A. Genomic selection signatures in sheep from the western pyrenees. Genet. Sel. Evol. 2018, 50, 9. [Google Scholar] [CrossRef]
- Hodge, M.J.; Rindfleish, S.J.; Heras-Saldana, S.; Stephen, C.; Pant, S.D. Heritability and genetic parameters for semen traits in australian sheep. Animals 2022, 12, 2946. [Google Scholar] [CrossRef]
- Rezvannejad, E.; Asadollahpour Nanaei, H.; Esmailizadeh, A. Detection of candidate genes affecting milk production traits in sheep using whole-genome sequencing analysis. Vet. Med. Sci. 2022, 8, 1197–1204. [Google Scholar] [CrossRef]
- Li, M.; Lu, Y.; Gao, Z.; Yue, D.; Hong, J.; Wu, J.; Xi, D.; Deng, W.; Chong, Y. Pan-Omics in Sheep: Unveiling Genetic Landscapes. Animals 2024, 14, 273. [Google Scholar] [CrossRef] [PubMed]
- Hudson, A.J.; Moore, A.N.; Elniski, D.; Joseph, J.; Yee, J.; Russell, A.G. Evolutionarily divergent spliceosomal snrnas and a conserved non-coding rna processing motif in giardia lamblia. Nucleic Acids Res. 2012, 40, 10995–11008. [Google Scholar] [CrossRef] [PubMed]
- Signer-Hasler, H.; Henkel, J.; Bangerter, E.; Bulut, Z.; The VarGoats Consortium; Drögemüller, C.; Leeb, T.; Flury, C. Runs of homozygosity in Swiss goats reveal genetic changes associated with domestication and modern selection. Genet. Sel. Evol. 2022, 54, 6. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Gil, B.; Arranz, J.J.; Pong-Wong, R.; García-Gámez, E.; Kijas, J.; Wiener, P. Application of Selection Mapping to Identify Genomic Regions Associated with Dairy Production in Sheep. PLoS ONE 2014, 9, e94623. [Google Scholar] [CrossRef]
- Pazzola, M.; Vacca, G.M.; Paschino, P.; Bittante, G.; Dettori, M.L. Novel Genes Associated with Dairy Traits in Sarda Sheep. Animals 2021, 11, 2207. [Google Scholar] [CrossRef]
- Dettori, M.L.; Pazzola, M.; Petretto, E.; Vacca, G.M. Association Analysis between SPP1, POFUT1 and PRLR Gene Variation and Milk Yield, Composition and Coagulation Traits in Sarda Sheep. Animals 2020, 10, 1216. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, J.; Zhao, F.; Ren, H.; Xu, L.; Lu, J.; Zhang, S.; Zhang, X.; Wei, C.; Lu, G.; et al. Genome-Wide Association Studies for Growth and Meat Production Traits in Sheep. PLoS ONE 2013, 8, e66569. [Google Scholar] [CrossRef]
- Smołucha, G.; Gurgul, A.; Jasielczuk, I.; KawĂŞcka, A.; Miksza-Cybulska, A. A genome-wide association study for prolificacy in three polish sheep breeds. J. Appl. Genet. 2021, 62, 323–326. [Google Scholar] [CrossRef]
- Keene, J.D.; Komisarow, J.M.; Friedersdorf, M.B. Splicing regulation by U4/U6.U5 tri-snRNP complex and associated factors. RNA 1998, 4, 816–829. [Google Scholar]
- Koseniuk, A.; Ropka-Molik, K.; Rubiś, D.; Smołucha, G. Genetic background of coat colour in sheep. Arch. Anim. Breed. 2018, 61, 173–178. [Google Scholar] [CrossRef]
- Li, W.; Guo, J.; Li, F.; Niu, C. Evaluation of crossbreeding of australian superfine merinos with gansu alpine finewool sheep to improve wool characteristics. PLoS ONE 2016, 11, e0166374. [Google Scholar] [CrossRef] [PubMed]
- Kawecka, A.; Krupinski, M. Sheep in the Polish Carpathians: Genetic resources conservation of the Podhale zackel and coloured mountain sheep. Geomat. Landmanagement Landsc. 2014, 1, 35–42. [Google Scholar] [CrossRef]
- Jakobsson, M.E.; Małecki, J.; Falnes, P.O. Regulation of eukaryotic elongation factor 1 alpha (eEF1A) by dynamic lysine methylation. RNA Biol. 2018, 15, 314–319. [Google Scholar] [CrossRef] [PubMed]





| Breed | Breed Abbreviation | Number of Individuals | Type of Use | Wool Type | Reproduction |
|---|---|---|---|---|---|
| Polish Mountain Sheep | PMS | 104 | Multi-purpose | Mixed | Low |
| Podhale Zackel | PZ | 100 | Multi-purpose | Mixed | Low |
| Colored Mountain Sheep | CMS | 97 | Multi-purpose | Mixed | Low |
| Swiniarka | SW | 99 | Multi-purpose | Mixed | Low |
| Uhruska | UHR | 69 | Wool and meat | Solid | Medium |
| Old-type Merino | MPOT | 50 | Wool and meat | Solid | Medium |
| Polish Merino of Colored Variety | MPC | 59 | Wool and meat | Solid | Medium |
| Black-headed | BH | 104 | Meat | Solid | Medium |
| Wrzosówka | WRZ | 90 | Sheepskin | Mixed | High |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szmatola, T.; Ropka-Molik, K.; Jasielczuk, I.; Kawęcka, A.; Gurgul, A. Exploring Runs of Homozygosity and Heterozygosity in Sheep Breeds Maintained in Poland. Genes 2025, 16, 709. https://doi.org/10.3390/genes16060709
Szmatola T, Ropka-Molik K, Jasielczuk I, Kawęcka A, Gurgul A. Exploring Runs of Homozygosity and Heterozygosity in Sheep Breeds Maintained in Poland. Genes. 2025; 16(6):709. https://doi.org/10.3390/genes16060709
Chicago/Turabian StyleSzmatola, Tomasz, Katarzyna Ropka-Molik, Igor Jasielczuk, Aldona Kawęcka, and Artur Gurgul. 2025. "Exploring Runs of Homozygosity and Heterozygosity in Sheep Breeds Maintained in Poland" Genes 16, no. 6: 709. https://doi.org/10.3390/genes16060709
APA StyleSzmatola, T., Ropka-Molik, K., Jasielczuk, I., Kawęcka, A., & Gurgul, A. (2025). Exploring Runs of Homozygosity and Heterozygosity in Sheep Breeds Maintained in Poland. Genes, 16(6), 709. https://doi.org/10.3390/genes16060709





