Evaluating Genetic Diversity and Regional Variation in Tswana Goats of Botswana
Abstract
1. Introduction
2. Materials and Methods
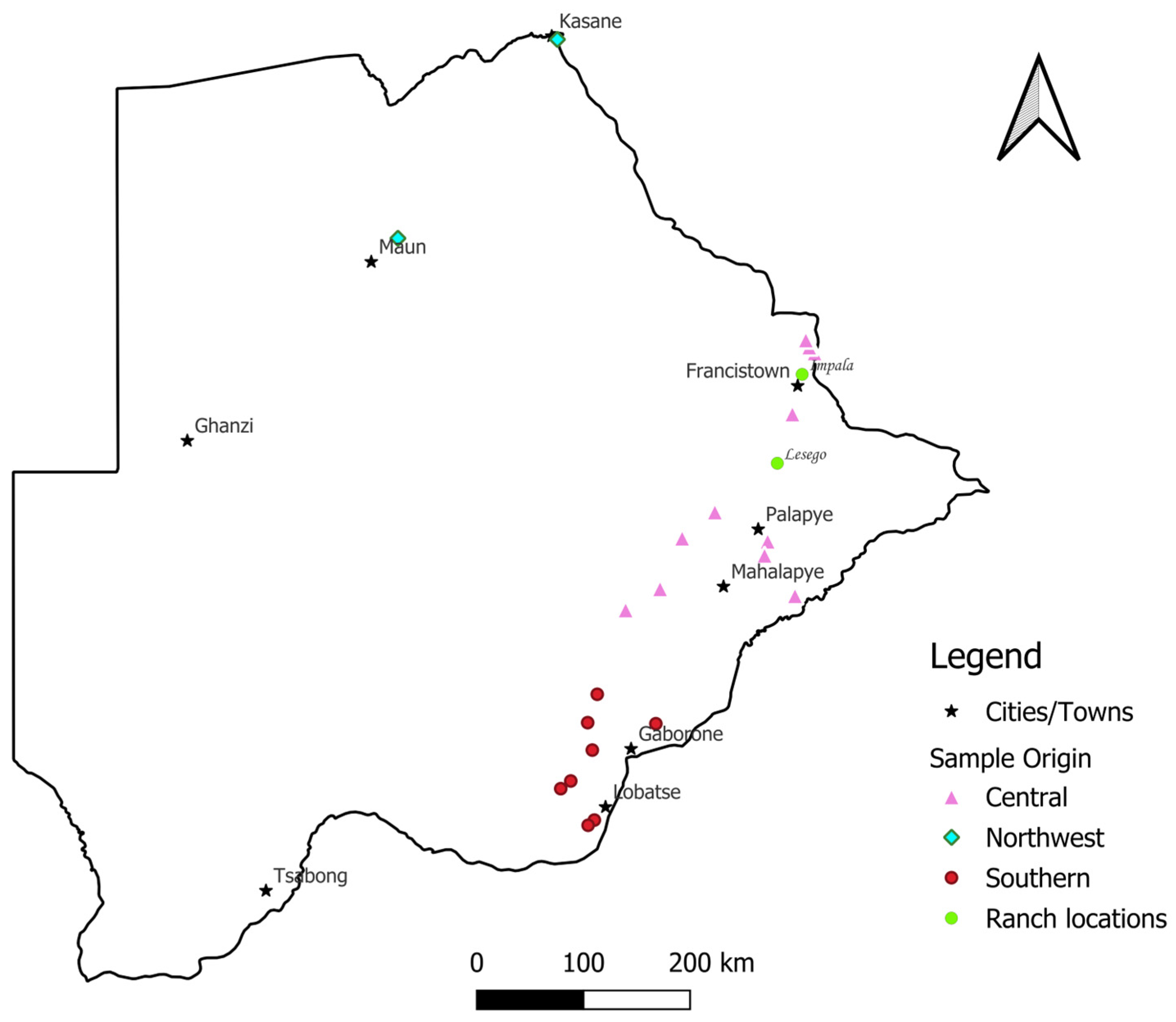
2.1. Study Area and Sample Collection
2.2. Genotyping and Quality Control (QC)
3. Results
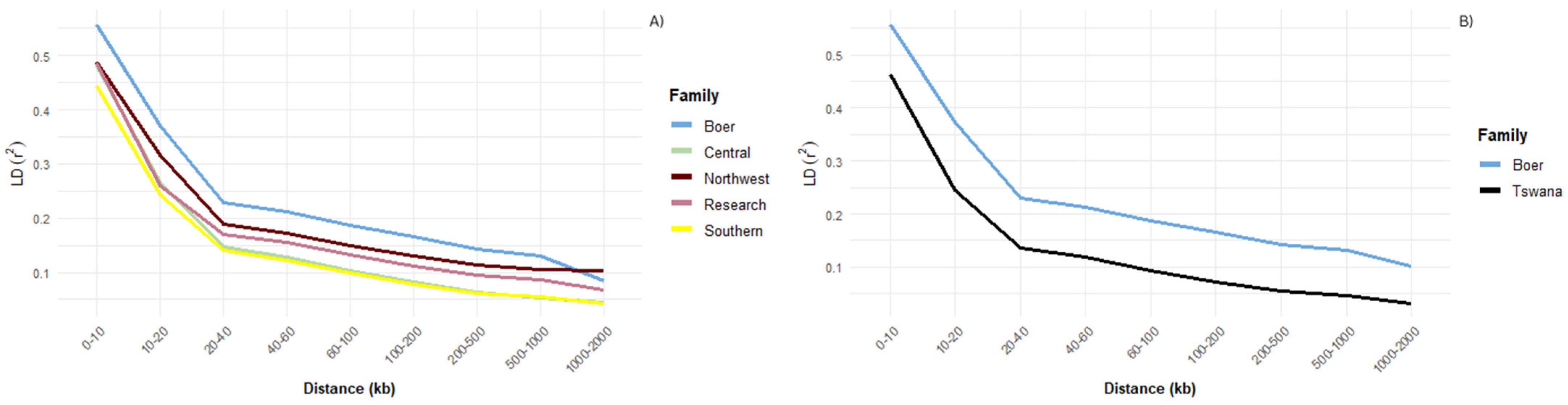
3.1. Linkage Disequilibrium
3.2. Effective Population Size
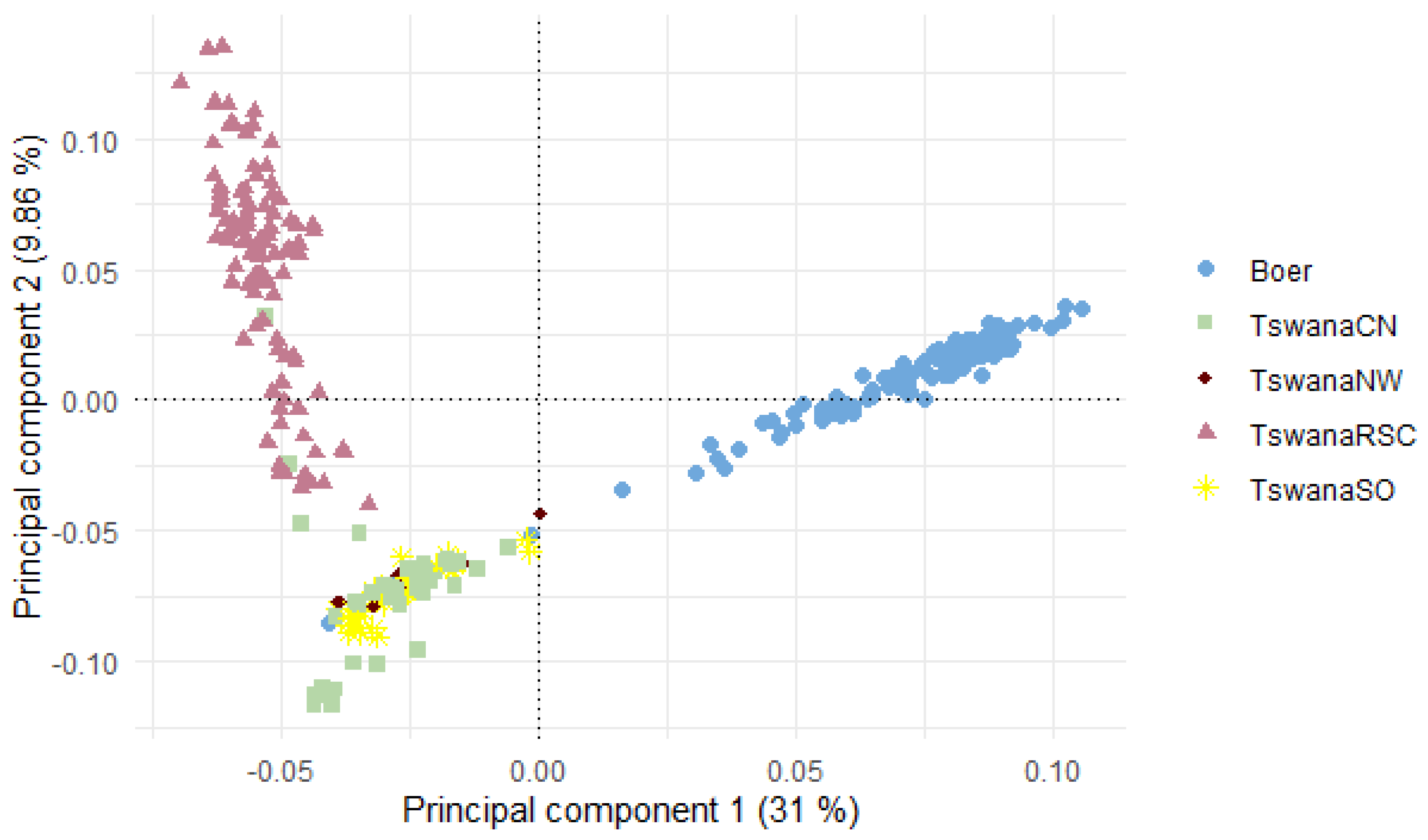
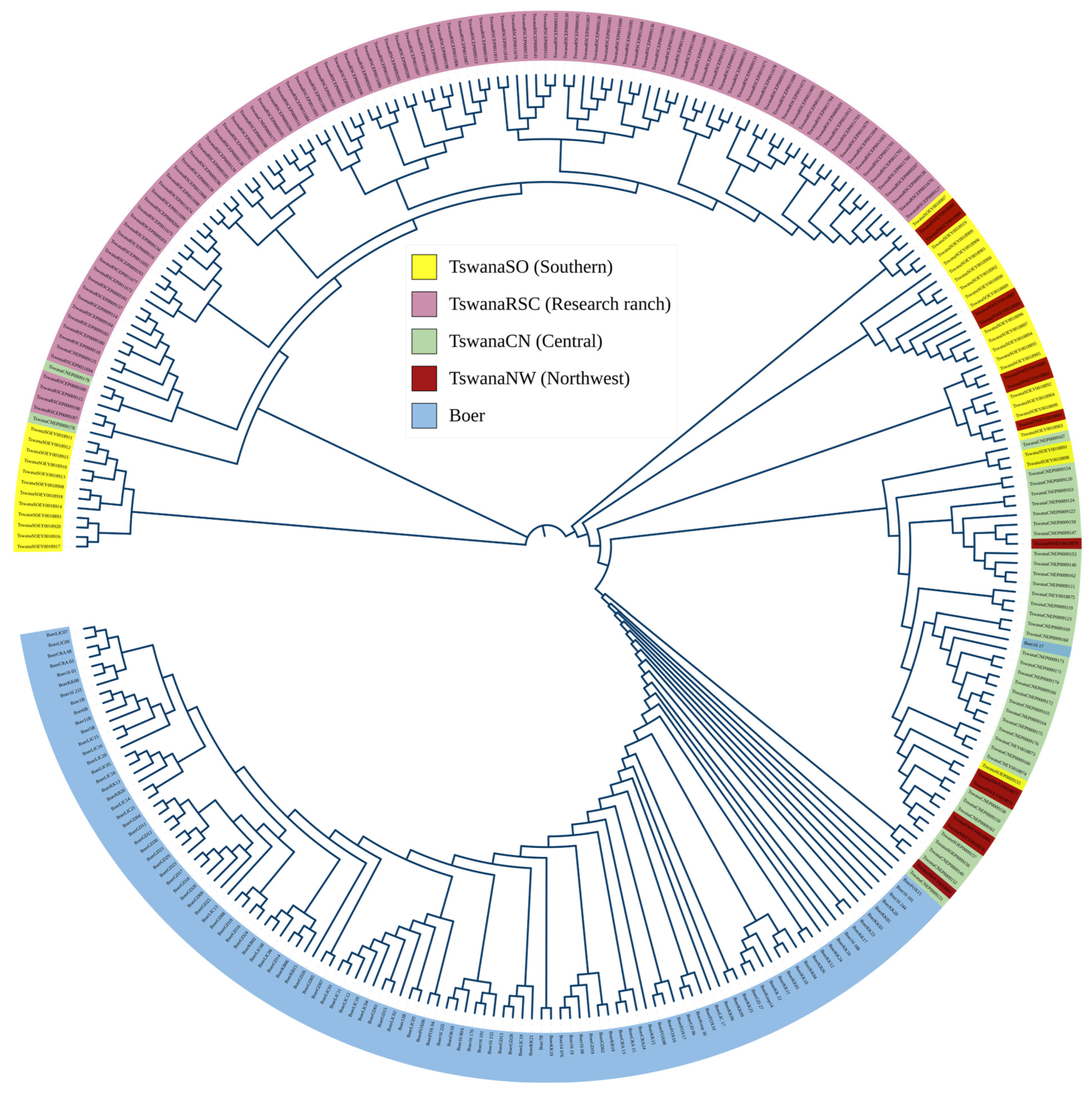
3.3. Population Structure Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| NARDI | National agricultural research and development institute |
| LD | Linkage disequilibrium |
| PCA | Principal component analysis |
| MAF | Minor allele frequency |
| HWE | Hardy–Weinberg Equilibrium |
| SNP | Single nucleotide polymorphism |
| n | Number of samples |
| Ho | Observed heterozygosity |
| He | Expected heterozygosity |
| FIS | Inbreeding coefficient |
| r2 | Squared correlation coefficient |
| Ne | Effective population size |
References
- Mekuriaw, G.; Gizaw, S.; Dessie, T.; Mwai, O.; Djikeng, A.; Tesfaye, K. A Review on Current Knowledge of Genetic Diversity of Domestic Goats (Capra hircus) Identified by Microsatellite Loci: How those Efforts are Strong to Support the Breeding Programs? J. Life Sci. Biomed. 2016, 6, 22–32. Available online: www.jlsb.science-line.com (accessed on 17 November 2023).
- Statistics Botswana. Annual Agricultural Survey Report 2019. 2020. Available online: https://statsbots.org.bw (accessed on 10 October 2022).
- Pule-Meulenberg, F.; Dakota, F.D. Assessing the symbiotic dependency of grain and tree legumes on N2 fixation for their N nutrition in five agro-ecological zones of Botswana. Symbiosis 2009, 48, 68–77. [Google Scholar] [CrossRef]
- FAO. AQUASTAT Country Profile-Botswana; FAO: Rome, Italy, 2005; Available online: https://fao.org/ (accessed on 24 January 2025).
- Adogla-Bessa, T.; Aganga, A.A. Responses of Tswana goats to various lengths of water deprivation. S. Afr. J. Anim. Sci. 2000, 30, 87–91. [Google Scholar] [CrossRef]
- Baleseng, L.; Kgosikoma, O.; Makgekgenene, A.; Coleman, M.; Morley, P.; Baker, D.O.; Bahta, S. Performance of goats and sheep under communal grazing in Botswana using various growth measures. In Proceedings of the Seventh International Conference on Agricultural Statistics, Rome, Italy, 24–26 October 2016. [Google Scholar]
- Monau, P.I.; Visser, C.; Nsoso, S.J.; Van Marle-Köster, E. Phenotypic and genetic characterization of indigenous Tswana goats. South Afr. J. Anim. Sci. 2018, 48, 925–934. [Google Scholar] [CrossRef]
- Nsoso, S.J.; Podisi, B.; Otsogile, E.; Mokhutshwane, B.S.; Ahmadu, B. Phenotypic Characterization of Indigenous Tswana Goats and Sheep Breeds in Botswana: Continuous Traits. Trop. Anim. Heal. Prod. 2004, 36, 789–800. [Google Scholar] [CrossRef] [PubMed]
- Animal Production Research Unit (APRU). Livestock and Range Research in Botswana; Animal Production Research Unit: Gaborone, Botswana, 2006. [Google Scholar]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- RStudio Team. RStudio: Integrated Development for R. RStudio; PBC: Boston, MA, USA, 2020; Available online: http://www.rstudio.com/ (accessed on 2 November 2023).
- Barbato, M.; Orozco-Terwengel, P.; Tapio, M.; Bruford, M.W. SNeP: A tool to estimate trends in recent effective population size trajectories using genome-wide SNP data. Front. Genet. 2015, 6, 109. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Lashmar, S.F.; Visser, C.; van Marle-Köster, E. SNP-based genetic diversity of South African commercial dairy and fiber goat breeds. Small Rumin. Res. 2016, 136, 65–71. [Google Scholar] [CrossRef]
- Mdladla, K.; Dzomba, E.F.; Huson, H.J.; Muchadeyi, F.C. Population genomic structure and linkage disequilibrium analysis of South African goat breeds using genome-wide SNP data. Anim. Genet. 2016, 47, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Onzima, R.B.; Upadhyay, M.R.; Doekes, H.P.; Brito, L.F.; Bosse, M.; Kanis, E.; Groenen, M.A.M.; Crooijmans, R.P.M.A. Genome-Wide Characterization of Selection Signatures and Runs of Homozygosity in Ugandan Goat Breeds. Front. Genet. 2018, 9, 318. [Google Scholar] [CrossRef] [PubMed]
- Chokoe, T.C.; Mdladla-Hadebe, K.; Muchadeyi, F.; Dzomba, E.; Matelele, T.; Mphahlele, T.; Mpofu, T.J.; Nephawe, K.; Mtileni, B. Genetic Diversity of South African Indigenous Goat Population from Four Provinces Using Genome-Wide SNP Data. Sustainability 2020, 12, 10361. [Google Scholar] [CrossRef]
- Tarekegn, G.M.; Wouobeng, P.; Jaures, K.S.; Mrode, R.; Edea, Z.; Liu, B.; Zhang, W.; Mwai, O.A.; Dessie, T.; Tesfaye, K.; et al. Genome-wide diversity and demographic dynamics of Cameroon goats and their divergence from east African, north African, and Asian conspecifics. PLoS ONE 2019, 14, e0214843. [Google Scholar] [CrossRef] [PubMed]
- Rahmatalla, S.A.; Arends, D.; Reissmann, M.; Ahmed, A.S.; Wimmers, K.; Reyer, H.; Brockmann, G.A. Whole genome population genetics analysis of Sudanese goats identifies regions harboring genes associated with major traits. BMC Genet. 2017, 18, 92. [Google Scholar] [CrossRef] [PubMed]
- Sallam, A.M.; Reyer, H.; Wimmers, K.; Bertolini, F.; Aboul-Naga, A.; Braz, C.U.; Rabee, A.E. Genome-wide landscape of runs of homozygosity and differentiation across Egyptian goat breeds. BMC Genom. 2023, 24, 573. [Google Scholar] [CrossRef] [PubMed]
- Brito, L.F.; Jafarikia, M.; Grossi, D.A.; Kijas, J.W.; Porto-Neto, L.R.; Ventura, R.V.; Salgorzaei, M.; Schenkel, F.S. Characterization of linkage disequilibrium, consistency of gametic phase and admixture in Australian and Canadian goats. BMC Genet. 2015, 16, 67. [Google Scholar] [CrossRef] [PubMed]
- Berihulay, H.; Islam, R.; Jiang, L.; Ma, Y. Genome-Wide Linkage Disequilibrium and the Extent of Effective Population Sizes in Six Chinese Goat Populations Using a 50K Single Nucleotide Polymorphism Panel. Animals 2019, 9, 350. [Google Scholar] [CrossRef] [PubMed]
- Deniskova, T.E.; Dotsev, A.V.; Fornara, M.S.; Sermyagin, A.A.; Reyer, H.; Wimmer, K.; Brem, G.; Zinovieva, N.A. The genomic architecture of the Russian population of Saanen goats in comparison with worldwide Saanen gene pool from five countries. Agric. Biol. 2020, 55, 285–294. Available online: https://www.agrobiology.ru/2-2020deniskova-eng.html (accessed on 2 July 2022). [CrossRef]
- Meuwissen, T. Genetic management of small populations: A review. Acta Agric. Scand. Sect. A Anim. Sci. 2009, 59, 71–79. [Google Scholar] [CrossRef]
- Monau, P.I.; Visser, C.; Muchadeyi, F.C.; Okpeku, M.; Nsoso, S.J.; van Marle-Köster, E. Population structure of indigenous southern African goats based on the Illumina Goat50K SNP panel. Trop. Anim. Health Prod. 2020, 52, 1795–1802. [Google Scholar] [CrossRef] [PubMed]

| Breed | n | Removed Animals | Excluded SNPs | ||||
|---|---|---|---|---|---|---|---|
| MAF < 5% | SNP (Call Rate < 95%) | HWE (0.00001) | Total SNPs Removed | Total SNPs Retained | |||
| Tswana | 192 | 2 | 1538 | 8684 | 76 | 10,298 | 47,037 |
| Boer | 112 | 1 | 2967 | 62,618 | 92 | 65,677 | 45,627 |
| Total | 304 | 3 | |||||
| Group | Region/Origin | n | Ho ± s.d. | He ± s.d. | MAF ± s.d. | FIS ± s.d. |
|---|---|---|---|---|---|---|
| TswanaCN | Central | 39 | 0.394 ± 0.000 | 0.401 ± 0.000 | 0.320 ± 0.122 | 0.019 ± 0.051 |
| TswanaSO | Southern | 32 | 0.397 ± 0.000 | 0.399 ± 0.000 | 0.318 ± 0.123 | 0.005 ± 0.038 |
| TswanaNW | Northwest | 12 | 0.364 ± 0.000 | 0.358 ± 0.000 | 0.289 ± 0.132 | −0.016 ± 0.064 |
| TswanaRSC | Ranches | 107 | 0.394 ± 0.000 | 0.380 ± 0.000 | 0.300 ± 0.132 | −0.038 ± 0.042 |
| Breed | Ho ± s.d. | He ± s.d | MAF ± s.d | FIS ± s.d |
|---|---|---|---|---|
| Tswana | 0.395 ± 0.019 | 0.400 ± 0.000 | 0.313 ± 0.127 | 0.013 ± 0.047 |
| Boer | 0.367 ± 0.022 | 0.375 ± 0.000 | 0.287 ±0.136 | 0.019 ± 0.060 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chalebgwa, A.B.; Monau, P.I.; Raphaka, K.; Hadebe, K.; Kgwatalala, P.; Nsoso, S.J. Evaluating Genetic Diversity and Regional Variation in Tswana Goats of Botswana. Genes 2025, 16, 678. https://doi.org/10.3390/genes16060678
Chalebgwa AB, Monau PI, Raphaka K, Hadebe K, Kgwatalala P, Nsoso SJ. Evaluating Genetic Diversity and Regional Variation in Tswana Goats of Botswana. Genes. 2025; 16(6):678. https://doi.org/10.3390/genes16060678
Chicago/Turabian StyleChalebgwa, Amantle Bonolo, Phetogo Ineeleng Monau, Kethusegile Raphaka, Khanyisile Hadebe, Patrick Kgwatalala, and Shalaulani James Nsoso. 2025. "Evaluating Genetic Diversity and Regional Variation in Tswana Goats of Botswana" Genes 16, no. 6: 678. https://doi.org/10.3390/genes16060678
APA StyleChalebgwa, A. B., Monau, P. I., Raphaka, K., Hadebe, K., Kgwatalala, P., & Nsoso, S. J. (2025). Evaluating Genetic Diversity and Regional Variation in Tswana Goats of Botswana. Genes, 16(6), 678. https://doi.org/10.3390/genes16060678






