Assessment of Genetic Diversity and Productive Traits in Crossbreed Cattle in the Caribbean Region, Colombia
Abstract
1. Introduction
2. Materials and Methods
2.1. Genomic Analysis
2.2. Commercially Important Traits
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ellegren, H.; Galtier, N. Determinants of Genetic Diversity. Nat. Rev. Genet. 2016, 17, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Brito, L.F.; Bédère, N.; Douhard, F.; Oliveira, H.R.; Arnal, M.; Peñagaricano, F.; Schinckel, A.P.; Baes, C.F.; Miglior, F. Review: Genetic Selection of High-Yielding Dairy Cattle Toward Sustainable Farming Systems in a Rapidly Changing World. Animal 2021, 15, 100292. [Google Scholar] [CrossRef] [PubMed]
- Herron, J.C.; Freeman, S. Mendelian Genetics in Populations II: Migration, Drift and Nonrandom Mating. In Evolutionary Analysis, 5th ed.; Pearson Education: London, UK, 2014; pp. 259–264. [Google Scholar]
- Groeneveld, L.F.; Lenstra, J.A.; Eding, H.; Toro, M.A.; Scherf, B.; Pilling, D.; Negrini, R.; Finlay, E.K.; Groeneveld, E.; Weigend, S. Genetic Diversity in Farm Animals—A Review. Anim. Genet. 2010, 41 (Suppl. S1), 6–31. [Google Scholar] [CrossRef]
- Makanjoula, B.O.; Maltecca, C.; Miglior, F.; Schenkel, F.S.; Baes, C.F. Effect of Recent and Ancient Inbreeding on Production and Fertility Traits in Canadian Holsteins. BMC Genom. 2020, 21, 605. [Google Scholar] [CrossRef]
- Meuwissen, T.H.E. Maximizing the Response of Selection with a Predefined Rate of Inbreeding. J. Anim. Sci. 1997, 75, 934–940. [Google Scholar] [CrossRef]
- González-Quintero, R.; Barahona-Rosales, R.; Bolívar-Vergara, D.M.; Chirinda, N.; Arango, J.; Pantévez, H.A.; Correa-Londoño, G.; Sánchez-Pinzón, M.S. Technical and Environmental Characterization of Dual-Purpose Cattle Farms and Ways of Improving Production: A Case Study in Colombia. Pastoralism 2020, 10, 19. [Google Scholar] [CrossRef]
- Cortés Mora, J.A.; Cotes Torres, A.; Cotes Torres, J.M. Structural Features of Dual-Purpose Cattle Production System in the Colombian Humid Tropic. Rev. Colomb. Cienc. Pecu. 2012, 25, 229–239. [Google Scholar] [CrossRef]
- González-Quintero, R.; Sánchez-Pinzón, M.S.; Bolívar-Vergara, D.M.; Chirinda, N.; Arango, J.; Pantévez, H.A.; Correa-Londoño, G.; Barahona-Rosales, R. Technical and Environmental Characterization of Very Small, Small, Medium, and Large Cow-Calf Operations in Colombia. Rev. Mex. Cienc. Pecu. 2020, 11, 183–200. [Google Scholar] [CrossRef]
- Rosero Alpala, J.A.; Rangel Garcia, W.D.; Rojas Barreto, A.; Burgos-Paz, W.O. Contribution of Genomic Data in Defining the Breed Composition of Dual-Purpose Cattle. Rev. Mex. Cienc. Pecu. 2021, 12, 1008–1024. [Google Scholar] [CrossRef]
- Schwarz, L.; Križanac, A.M.; Schneider, H.; Falker-Gieske, C.; Heise, J.; Liu, Z.; Bennewitz, J.; Thaller, G.; Tetens, J. Genetic and Genomic Analysis of Reproduction Traits in Holstein Cattle Using SNP Chip Data and Imputed Sequence Level Genotypes. BMC Genom. 2024, 25, 880. [Google Scholar] [CrossRef]
- Berry, D.P.; Spangler, M.L. Animal Board Invited Review: Practical Applications of Genomic Information in Livestock. Animal 2023, 17, 100996. [Google Scholar] [CrossRef] [PubMed]
- Bernini, F.; Punturiero, C.; Vevey, M.; Blanchet, V.; Milanesi, R.; Delledonne, A.; Bagnato, A.; Strillacci, M.G. Assessing Major Genes Allele Frequencies and the Genetic Diversity of the Native Aosta Cattle Female Population. Ital. J. Anim. Sci. 2023, 22, 1008–1022. [Google Scholar] [CrossRef]
- Igoshin, A.V.; Romashov, G.A.; Chernyaeva, E.N.; Elatkin, N.P.; Yudin, N.S.; Larkin, D.M. Comparative Analysis of Allele Frequencies for DNA Polymorphisms Associated with Disease and Economically Important Traits in the Genomes of Russian and Foreign Cattle Breeds. Russ. J. Genet. 2022, 26, 298–307. [Google Scholar] [CrossRef]
- Lopdell, T.J. Using QTL to Identify Genes and Pathways Underlying the Regulation and Production of Milk Components in Cattle. Animals 2023, 13, 911. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Biscarini, F.; Cozzi, P.; Gaspa, G.; Marras, G. detectRUNS: Detect Runs of Homozygosity and Runs of Heterozygosity in Diploid Genomes. R Package 2019; Institute for Statistics and Mathematics, Vienna University of Economics and Business: Vienna, Austria, 2019. [Google Scholar]
- McQuillan, R.; Leutenegger, A.-L.; Abdel-Rahman, R.; Franklin, C.S.; Pericic, M.; Barac-Lauc, L.; Smolej-Narancic, N.; Janicijevic, B.; Polasek, O.; Tenesa, A.; et al. Runs of Homozygosity in European Populations. Am. J. Hum. Genet. 2008, 83, 359–372. [Google Scholar] [CrossRef]
- Cesarani, A.; Sorbolini, S.; Criscione, A.; Bordonaro, S.; Pulina, G.; Battacone, G.; Marletta, D.; Gaspa, G.; Macciotta, N.P.P. Genome-wide Variability and Selection Signatures in Italian Island Cattle Breeds. Anim. Genet. 2018, 49, 371–383. [Google Scholar] [CrossRef]
- Toro-Ospina, A.M.; Herrera Rios, A.C.; Pimenta Schettini, G.; Vallejo Aristizabal, V.H.; Bizarria dos Santos, W.; Zapata, C.A.; Ortiz Morea, E.G. Identification of Runs of Homozygosity Islands and Genomic Estimated Inbreeding Values in Caqueteño Creole Cattle (Colombia). Genes 2022, 13, 1232. [Google Scholar] [CrossRef]
- Naranjo Guerrero, L.F.; Rogberg-Muñoz, A.; Rodríguez, N.; González Herrera, L.G. Genomic Diversity Study of Highly Crossbred Cattle Population in a Low and Middle Tropical Environment. Trop. Anim. Health Prod. 2024, 56, 258. [Google Scholar] [CrossRef]
- Galina, C.S.; Geffroy, M. Dual-Purpose Cattle Raised in Tropical Conditions: What Are Their Shortcomings in Sound Productive and Reproductive Function? Animals 2023, 13, 2224. [Google Scholar] [CrossRef]
- Soares, B.D.; Vargas, G.; Duffield, T.; Schenkel, F.; Squires, E.J. Genome-wide Association Study and Functional Analyses for Clinical and Subclinical Ketosis in Holstein Cattle. J. Dairy Sci. 2021, 104, 6789–6802. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shao, S.; Wang, Y.; Du, Z.; Yu, H.; Li, F.; Qin, Y. Ahdc1 is a Potent Regulator of Obesity and Energy Metabolism. Am. J. Physiol. Endocrinol. Metab. 2023, 325, E638–E648. [Google Scholar] [CrossRef] [PubMed]
- Liudkovska, V.; Krawczyk, P.S.; Brouze, A.; Gumińska, N.; Wegierski, T.; Cysewski, D.; Mackiewicz, Z.; Ewbank, J.J.; Drabikowski, K.; Mroczek, S.; et al. TENT5 Cytoplasmic Noncanonical Poly(A) Polymerases Regulate the Innate Immune Response in Animals. Sci. Adv. 2023, 9, eadd9468. [Google Scholar] [CrossRef]
- Li, J.; Huang, X.; Chen, Q.; Lei, X. Non-Additive Association of the Rs41257413 SNP in the GLYCAM1 Gene with Milk Protein Percentage in Chinese Holstein Cattle. SSRN 2025, 20, 5129267. [Google Scholar] [CrossRef]
- Shin, S.; Heo, J.; Yeo, J.; Lee, C.; Chung, E. Genetic Association of Phosphodiesterase 1B (PDE1B) with Carcass Traits in Korean Cattle. Mol. Biol. Rep. 2012, 39, 4869–4874. [Google Scholar] [CrossRef]
- Arif, A.; Bailey, S.; Izumi, N.; Anzelon, T.A.; Ozata, D.M.; Andersson, C.; Gainetdinov, I.; MacRae, I.J.; Tomari, Y.; Zamore, P.D. GTSF1 Accelerates Target RNA Cleavage by PIWI-Clade Argonaute Proteins. Nature 2022, 608, 618–625. [Google Scholar] [CrossRef]
- Mateescu, R.G.; Garrick, D.J.; Reecy, J.M. Network Analysis Reveals Putative Genes Affecting Meat Quality in Angus Cattle. Front. Genet. 2017, 8, 171. [Google Scholar] [CrossRef]
- Wu, J.; Wu, T.; Xie, X.; Niu, Q.; Zhao, Z.; Zhu, B.; Chen, Y.; Zhang, L.; Gao, X.; Niu, X.; et al. Genetic Association Analysis of Copy Number Variations for Meat Quality in Beef Cattle. Foods 2023, 12, 3986. [Google Scholar] [CrossRef]
- Jiang, W.; Mooney, M.H.; Shirali, M. Unveiling the Genetic Landscape of Feed Efficiency in Holstein Dairy Heifers: Insights into Heritability, Genetic Markers, and Pathways via Meta-Analysis. J. Anim. Sci. 2024, 102, skae040. [Google Scholar] [CrossRef]
- Carvalheiro, R.; Costilla, R.; Neves, H.H.R.; Albuquerque, L.G.; Moore, S.; Hayes, B.J. Unraveling Genetic Sensitivity of Beef Cattle to Environmental Variation Under Tropical Conditions. Genet. Sel. Evol. 2019, 51, 29. [Google Scholar] [CrossRef]
- Li, Y.; Yang, M.; Lou, A.; Yun, J.; Ren, C.; Li, X.; Xia, G.; Nam, K.; Yoon, D.; Jin, H.; et al. Integrated Analysis of Expression Profiles with Meat Quality Traits in Cattle. Sci. Rep. 2022, 12, 5926. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, J.; Guo, Y.; Zhang, L.; Xu, L.; Gao, X.; Zhu, B.; Gao, H.; Ni, H.; Chen, Y. Multi-Strategy Genome-Wide Association Studies Identify the DCAF16-NCAPG Region as a Susceptibility Locus for Average Daily Gain in Cattle. Sci. Rep. 2016, 6, 38073. [Google Scholar] [CrossRef]
- Yang, T.; Luo, H.; Lou, W.; Chang, Y.; Brito, L.F.; Zhang, H.; Ma, L.; Hu, L.; Wang, A.; Li, S. Genetic Background of Hematological Parameters in Holstein Cattle Based on Genome-Wide Association and RNA Sequencing Analyses. J. Dairy Sci. 2024, 107, 24345. [Google Scholar] [CrossRef]
- Martínez-Rocha, R.; Hidalgo, J.; Cesarani, A.; Ramírez-Valverde, R.; Núñez-Domínguez, R.; García-Muñiz, J.G.; Domínguez-Viveros, J. Assessment of Genetic Diversity, Runs of Homozygosity, and Signatures of Selection in Tropical Milking Criollo Cattle Using Pedigree and Genomic Data. Genes 2022, 13, 1896. [Google Scholar] [CrossRef]
- Peripolli, E.; Stafuzza, N.B.; Munari, D.P.; Lima, A.L.F.; Irgang, R.; Machado, M.A.; Panetto, J.C.D.C.; Ventura, R.V.; Baldi, F.; da Silva, M.V.G.B. Assessment of Runs of Homozygosity Islands and Estimates of Genomic Inbreeding in Gyr (Bos indicus) Dairy Cattle. BMC Genom. 2018, 19, 34. [Google Scholar] [CrossRef]
- Lozada-Soto, E.A.; Tiezzi, F.; Jiang, J.; Cole, J.B.; VanRaden, P.M.; Maltecca, C. Genomic Characterization of Autozygosity and Recent Inbreeding Trends in All Major Breeds of US Dairy Cattle. J. Dairy Sci. 2022, 105, 8956–8971. [Google Scholar] [CrossRef]
- Betancur Zambrano, M.F.; Rincón Flórez, J.C.; Herrera Rios, A.C.; Solarte Portilla, C.E.; Bedoya Berrio, G. de J. Evaluation of Runs of Homozygosity and Genomic Inbreeding in Holstein Cattle from Colombia. Semin. Ciências Agrárias 2020, 41 (Suppl. S2), 3397. [Google Scholar] [CrossRef]
- Rosero, J.A.; Álvarez, L.A.; Muñoz, J.E.; Durán, C.V.; Rodas, Á.G. Allelic Frequency of the Kappa-Casein Gene in Colombian and Creole Cattle Breeds. Rev. Colomb. Cienc. Pecu. 2012, 25, 97–108. [Google Scholar] [CrossRef]
- de Vitte, K.; Kerziene, S.; Klementavičiūtė, J.; de Vitte, M.; Mišeikienė, R.; Kudlinskienė, I.; Čepaitė, J.; Dilbiene, V.; Stankevičius, R. Relationship of β-casein Genotypes (A1A1, A1A2, and A2A2) to the Physicochemical Composition and Sensory Characteristics of Heifers’ Milk. J. Appl. Anim. Res. 2022, 50, 161–166. [Google Scholar] [CrossRef]
- Bisutti, V.; Pegolo, S.; Giannuzzi, D.; Mota, L.F.M.; Vanzin, A.; Toscano, A.; Trevisi, E.; Ajmone Marsan, P.; Brasca, M.; Cecchinato, A. The β-casein (CSN2) A2 Allelic Variant Alters Milk Protein Profile and Slightly Worsens Coagulation Properties in Holstein Heifers. J. Dairy Sci. 2022, 105, 3794–3809. [Google Scholar] [CrossRef]
- Andrade Ortega, S.G.; López Flores, J.O.; Sánchez Gómez, I.E. Allelic and Genotypic Frequencies of the Kappa Casein and βeta Lactoglobulin Gene in Brown Swiss Cattle. Calera 2021, 21, 42–48. [Google Scholar] [CrossRef]
- Bobbo, T.; Cipolat-Gotet, C.; Bittante, G.; Cecchinato, A. Short Communication: Association Analysis of Diacylglycerol Acyltransferase (DGAT1) Mutation on Chromosome 14 for Milk Yield and Composition Traits, Somatic Cell Score, and Coagulation Properties in Holstein Bulls. J. Dairy Sci. 2018, 101, 7075–7081. [Google Scholar] [CrossRef] [PubMed]
- Echeverri Zuluaga, J.J.; Vázquez Araque, N.A.; Gallo Garcia, Y.M. Polimorfismo del gen de la Somatotropina Bovina y su asociación con características de importancia en la producción lechera. Rev. Lasallista Investig. 2010, 7, 67–76. [Google Scholar]
- Bonilla, C.A.; Rubio, M.S.; Sifuentes, A.M.; Parra-Bracamonte, G.M.; Arellano, V.W.; Méndez, M.R.D.; Berruecos, J.M.; Ortiz, R. Association of CAPN1 316, CAPN1 4751 and TG5 Markers with Bovine Meat Quality Traits in Mexico. Genet. Mol. Res. 2010, 9, 2395–2405. [Google Scholar] [CrossRef]
- Desgarennes Alcalá, C.M.; Del Moral Ventura, S.; Meza Villalvazo, V.M.; Peña Castro, J.M.; Zárate Martínez, J.P.; Zavaleta, J.A. Allelic and Genotypic Frequencies of CAPN1 and CAST, Two Genes Associated to Meat Quality, in Cattle from the Papaloapan Basin. Nova Sci. 2017, 9, 211–228. [Google Scholar] [CrossRef]
- Casas, E.; White, S.N.; Wheeler, T.L.; Shackelford, S.D.; Koohmaraie, M.; Riley, D.G.; Chase, C.C., Jr.; Johnson, D.D.; Smith, T.P.L. Effects of Calpastatin and μ-Calpain Markers in Beef Cattle on Tenderness Traits. J. Anim. Sci. 2006, 84, 520–525. [Google Scholar] [CrossRef]

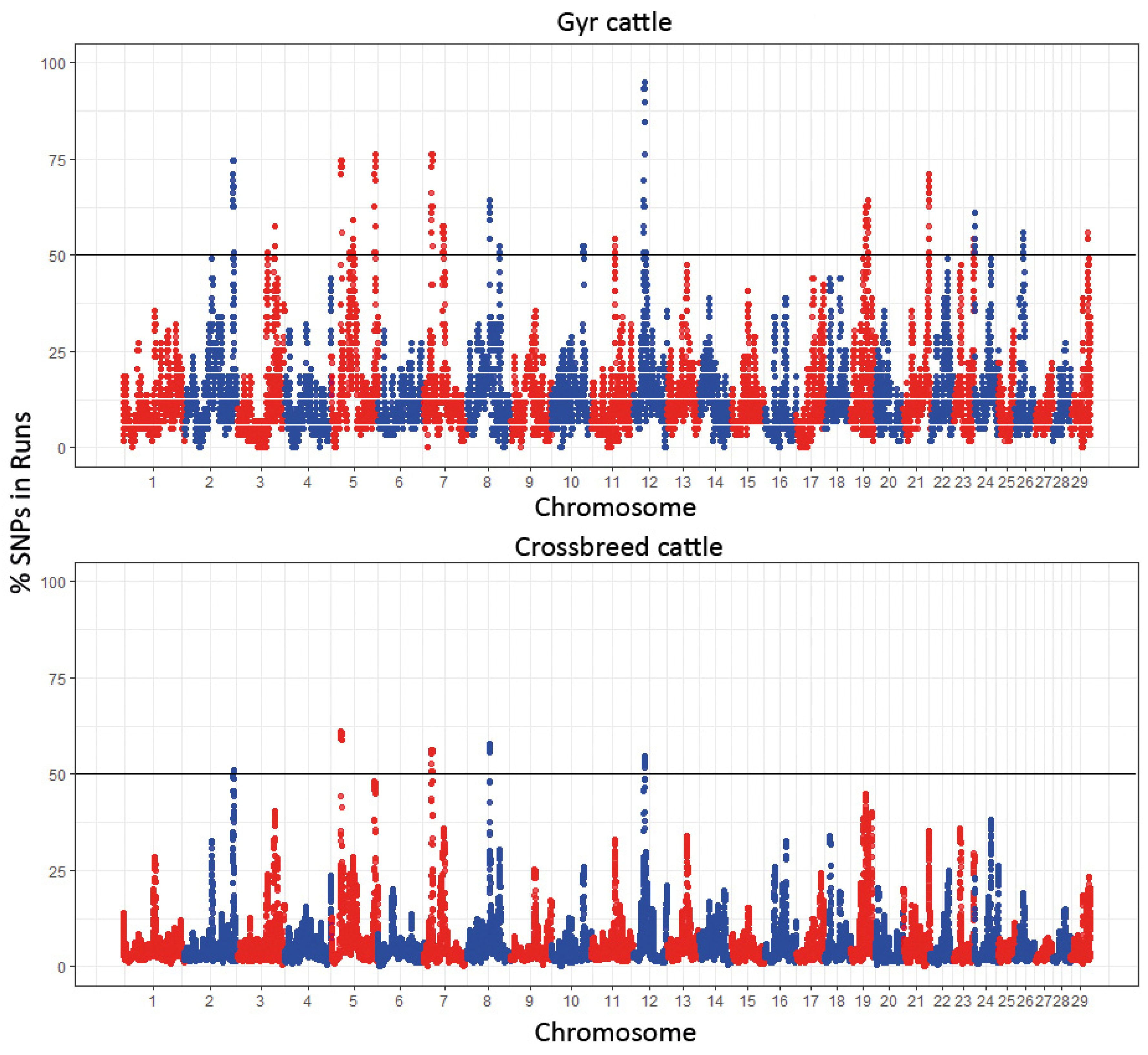

| BTA | nSNP | From | To | Associated Gene |
|---|---|---|---|---|
| 2 | 22 | 125,691,492 | 126,383,381 | FGR, AHDC1, WASF2, MAP3K6, TMEM222, SYYL1, GPN2, KDF1, TENT5B |
| 5 | 52 | 25,259,925 | 26,964,045 | GLYCAM1, PDE1B, GTSF1, COPZ1, MAP3K12, TF7, CALCOCO1, HOXC4-8-10-11-13, SMUG1, ATP5MC2 |
| 7 | 51 | 19,764,739 | 21,612,020 | FSD1, NMRK2, ATCAY, DAPK3, TEKTIP1, GIPC3, SHF ANKRD24, SHD, TMIGD2, MATK, CELF5, NCLN, TLE6, MOB3A, S1PR4, APBA3 |
| 8 | 51 | 58,010,694 | 59,902,102 | RHF32, SPATA31G1, PIGO, STOML2, ATOSB, RUSC2, CIMIP2B, TESK1, CD72, SIT1 |
| 12 | 48 | 27,933,503 | 29,563,846 | STARD13, KL, PDS5B, N4BP2L2, ZAR1L, FRY, RXFP2 |
| Group | <2 | 2–4 | 4–8 | 8–16 | >16 | |
|---|---|---|---|---|---|---|
| Crossbreed | n ROH | 14,290 | 9399 | 2028 | 663 | 181 |
| Mean length | 1.45 | 2.64 | 5.4 | 10.85 | 23.5 | |
| Gyr | n ROH | 2493 | 1860 | 529 | 236 | 142 |
| Mean length | 1.44 | 2.68 | 5.42 | 11.19 | 26.11 |
| GEN | SNP ID | Description | Genotypic Frequencies | Allelic Frequencies | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gyr | Cross | Gyr | Cross | Gyr | Cross | Gyr | Cross | Gyr | Cross | |||
| A1A1 | A1A2 | A2A2 | A1 | A2 | ||||||||
| CSN2 | rs43703013 | Beta Casein | 0.00 | 0.00 | 0.17 | 0.08 | 0.83 | 0.92 | 0.08 | 0.04 | 0.92 | 0.96 |
| rs43703011 | 0.02 | 0.01 | 0.19 | 0.17 | 0.8 | 0.82 | 0.11 | 0.09 | 0.89 | 0.91 | ||
| AA | AB | BB | A | B | ||||||||
| CSN3 | rs43706475 | Kappa Casein | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 1.00 | 0.00 | 0.00 |
| rs110014544 | 0.78 | 0.80 | 0.22 | 0.19 | 0.00 | 0.01 | 0.89 | 0.11 | 0.89 | 0.11 | ||
| LGB | rs110180463 | Beta lactoglobulin | 0.00 | 0.01 | 0.02 | 0.17 | 0.98 | 0.9 | 0.01 | 0.10 | 0.99 | 0.82 |
| rs110066229 | 0.19 | 0.11 | 0.46 | 0.48 | 0.36 | 0.41 | 0.42 | 0.35 | 0.58 | 0.65 | ||
| rs110641366 | 0.00 | 0.06 | 0.10 | 0.42 | 0.90 | 0.51 | 0.05 | 0.27 | 0.95 | 0.73 | ||
| rs109625649 | 0.17 | 0.10 | 0.47 | 0.48 | 0.36 | 0.42 | 0.41 | 0.34 | 0.59 | 0.66 | ||
| CoA:diacylglycerol acyltransferase 1 | AA | AG | GG | A | G | |||||||
| DGAT1 | rs109234250 | 0.84 | 0.65 | 0.14 | 0.31 | 0.02 | 0.04 | 0.91 | 0.81 | 0.09 | 0.19 | |
| Growth hormone | TT | TG | GG | T | G | |||||||
| GH1 | rs109191047 | 0.00 | 0.50 | 0.20 | 0.35 | 0.98 | 0.60 | 0.01 | 0.22 | 0.99 | 0.78 | |
| CAPN1_316 | rs17872000 | Calpain | CC | CG | GG | C | G | |||||
| 0.00 | 0.00 | 0.00 | 0.10 | 1.00 | 0.90 | 0.00 | 0.05 | 1.00 | 0.95 | |||
| CAPN1_530 | rs17871051 | AA | AG | GG | A | G | ||||||
| 0.00 | 0.00 | 0.07 | 0.12 | 0.93 | 0.88 | 0.03 | 0.06 | 0.97 | 0.94 | |||
| CAPN1_4571 | rs17872050 | TT | TC | CC | T | G | ||||||
| 0.73 | 0.69 | 0.27 | 0.27 | 0.00 | 0.03 | 0.86 | 0.83 | 0.14 | 0.17 | |||
| CAST_282 | rs110955059 | Calpastatin | CC | CG | GG | C | G | |||||
| 0.29 | 0.26 | 0.42 | 0.53 | 0.29 | 0.22 | 0.50 | 0.52 | 0.50 | 0.48 | |||
| AA | AG | GG | A | G | ||||||||
| CAST_22870 | rs41255587 | 0.05 | 0.19 | 0.27 | 0.51 | 0.58 | 0.30 | 0.19 | 0.45 | 0.81 | 0.55 | |
| CAST_22959 | rs109221039 | 0.69 | 0.39 | 0.29 | 0.48 | 0.02 | 0.13 | 0.84 | 0.63 | 0.16 | 0.37 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Serrano, A.; Ahumada-Velasco, M.; Cárdenas Beltrán, J.M. Assessment of Genetic Diversity and Productive Traits in Crossbreed Cattle in the Caribbean Region, Colombia. Genes 2025, 16, 677. https://doi.org/10.3390/genes16060677
Rodríguez-Serrano A, Ahumada-Velasco M, Cárdenas Beltrán JM. Assessment of Genetic Diversity and Productive Traits in Crossbreed Cattle in the Caribbean Region, Colombia. Genes. 2025; 16(6):677. https://doi.org/10.3390/genes16060677
Chicago/Turabian StyleRodríguez-Serrano, Andrés, Marcos Ahumada-Velasco, and Jesús María Cárdenas Beltrán. 2025. "Assessment of Genetic Diversity and Productive Traits in Crossbreed Cattle in the Caribbean Region, Colombia" Genes 16, no. 6: 677. https://doi.org/10.3390/genes16060677
APA StyleRodríguez-Serrano, A., Ahumada-Velasco, M., & Cárdenas Beltrán, J. M. (2025). Assessment of Genetic Diversity and Productive Traits in Crossbreed Cattle in the Caribbean Region, Colombia. Genes, 16(6), 677. https://doi.org/10.3390/genes16060677





