GhA01EP1 of Upland Cotton Stimulates Precocity, Improved Water Deficit Tolerance, and High Seed Yield in Transgenic Arabidopsis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatment
2.2. Phenotype, Seed Yield, and Protein and Starch Contents Comparison Between Transgenic Lines and Col-0
2.3. Root Growth Assay of Transgenic Arabidopsis Under Water Deficit Stress
2.4. Water Deficit Resistance Evaluation of Transgenic Arabidopsis via Drought-Rehydration Method
2.5. Transcriptome Sequencing Analysis
2.6. Biochemical Indicators Determination
2.7. qRT-PCR Validation of Transcriptome Data
2.8. Screening of Protein Interaction with GhA01EP1 by Glutathione-S-Transferase (GST) Pull-Down Assay and Liquid Chromatography Tandem Mass Spectrometry (LC-MS/MS) Analysis
2.9. Validation of Protein Interactions Using GST Pull-Down Assay
2.10. Validation of Protein Interactions Using Co-Immunoprecipitation (Co-IP) Assay
3. Results
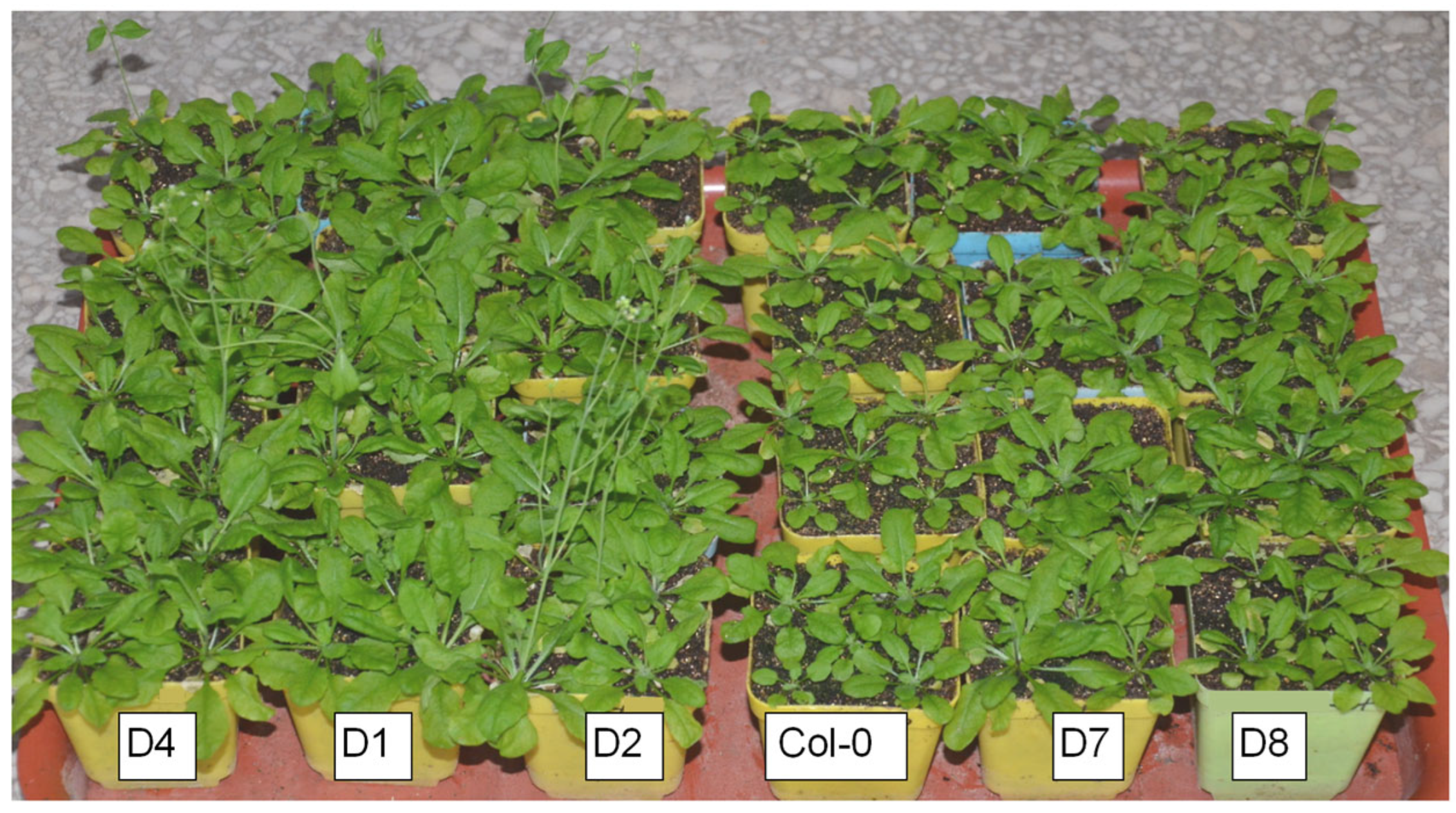
3.1. GhA01EP1 Promotes the Development of GhA01EP1-Transgenic Arabidopsis and Results in Earlier Flowering Time
3.2. GhA01EP1 Affects GhA01EP1-Transgenic Arabidopsis Morphology and Seed Yield
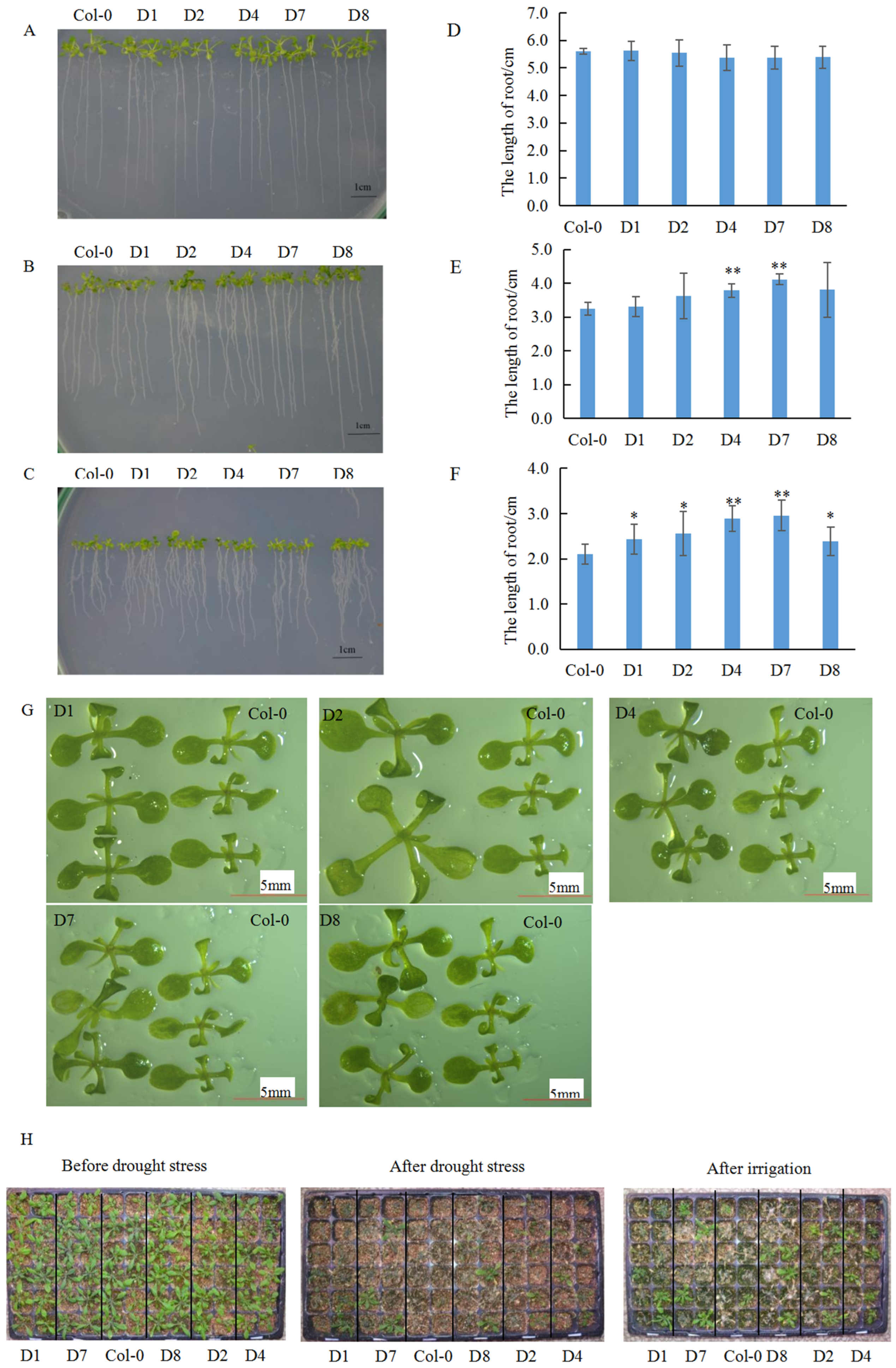
3.3. The GhA01EP1 Enhances Water Deficit Tolerance in GhA01EP1-Transgenic Arabidopsis
3.4. Transcriptome Analysis of Col-0 and GhA01EP1-Transgenic Arabidopsis
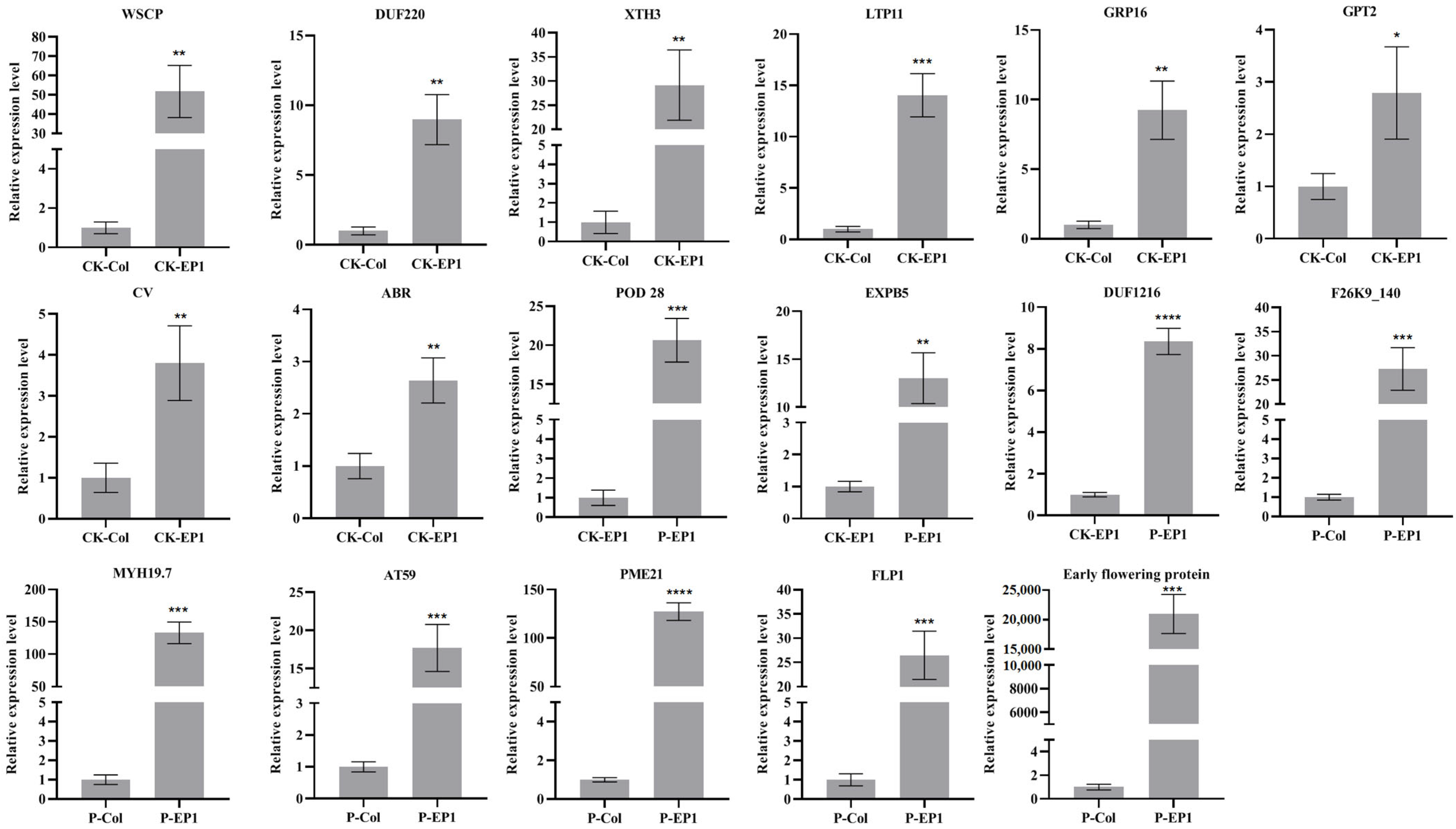
3.5. Validation of RNA-Seq Results by qRT-PCR
3.6. Comparison of Biochemical Indicators Between Col-0 and GhA01EP1-Transgenic Arabidopsis
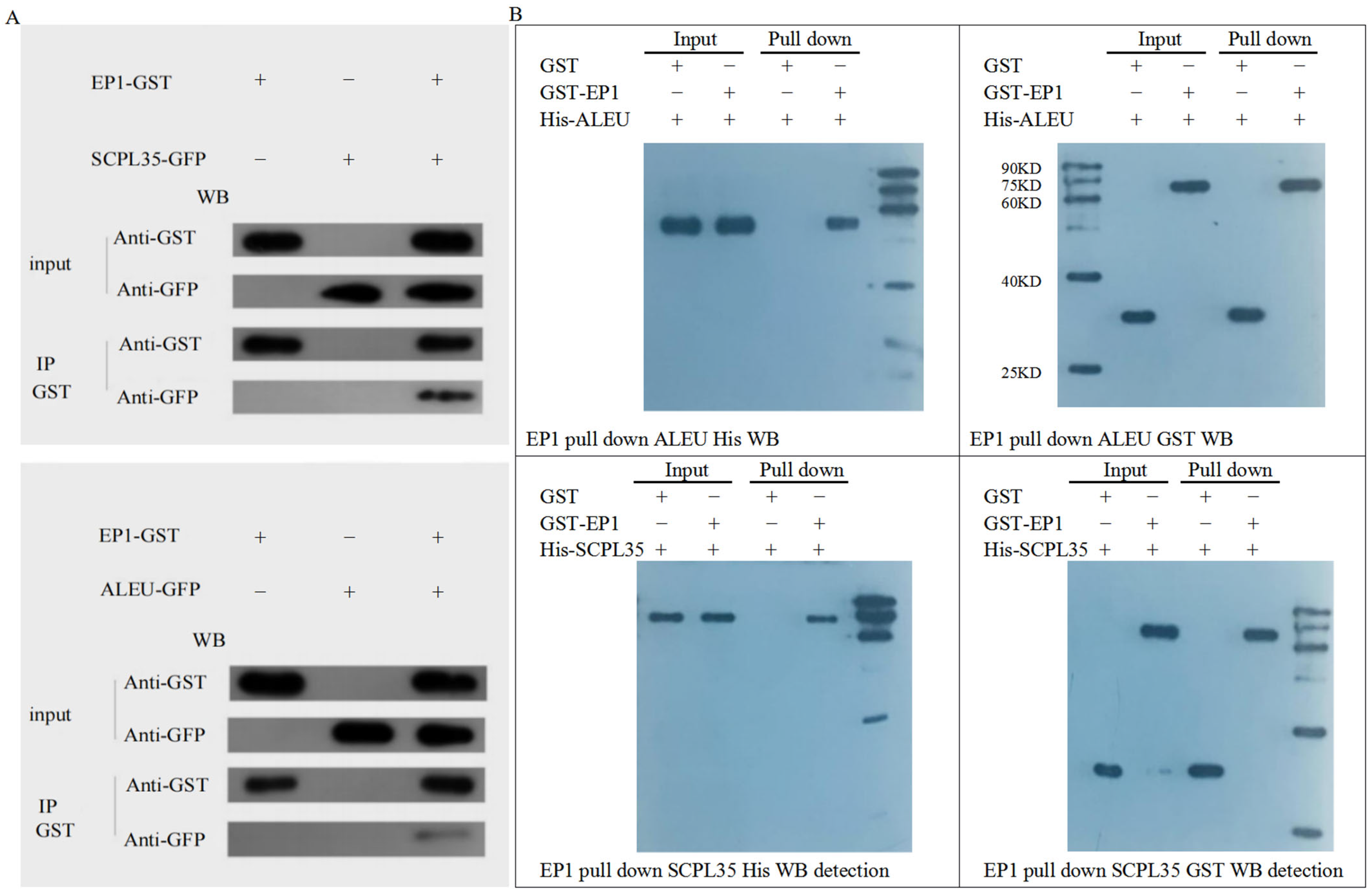
3.7. Screening of Proteins Interacting with GhA01EP1 in GhA01EP1-Transgenic Arabidopsis
4. Discussion
4.1. Mechanism of GhA01EP1 in Enhancing Water Deficit Tolerance
4.2. Mechanism of GhA01EP1 Regulation of Precocity and Rapid Growth of Plants
4.3. Mechanism of GhA01EP1 Influence on Yield Traits
4.4. Analysis of Interacting Proteins of GhA01EP1
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Co-IP | Co-immunoprecipitation |
| Col | Columbia |
| DEGs | Differentially expressed genes |
| Gly | Glyoxalase |
| GO | Gene ontology |
| GST | Glutathione S-transferase |
| JA-Ile | Jasmonoyl isoleucine |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LC-MS/MS | Liquid chromatography tandem mass spectrometry |
| MDA | Malondialdehyde |
| MG | Methylglyoxal |
| POD | Peroxidase |
| QQS | Qua-quine starch |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| ROS | Reactive oxygen species |
References
- Kamoun, S. The secretome of plant-associated fungi and oomycetes. In Plant Relationships, 2nd ed.; Deising, H.B., Barry, S., Carl, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; Volume 5, pp. 173–180. [Google Scholar] [CrossRef]
- Li, R.; Ma, X.Y.; Zhang, Y.J.; Zhang, Y.J.; Zhu, H.; Shao, S.N.; Zhang, D.D.; Klosterman, S.J.; Dai, X.F.; Subbarao, K.V.; et al. Genome-wide identification and analysis of a cotton secretome reveals its role in resistance against Verticillium dahliae. BMC Biol. 2023, 21, 166. [Google Scholar] [CrossRef] [PubMed]
- Li, S.X.; Showalter, A.M. Cloning and developmental/stress-regulated expression of a gene encoding a tomato arabinogalactan protein. Plant Mol. Biol. 1996, 32, 641–652. [Google Scholar] [CrossRef] [PubMed]
- Nothnagel, E.A. Proteoglycans and related components in plant cells. Int. Rev. Cytol. 1997, 174, 195–291. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Chen, M.; Showalter, A.M. Developmental expression and perturbation of arabinogalactan-proteins during seed germination and seedling growth in tomato. Physiol. Plant 2001, 112, 442–450. [Google Scholar] [CrossRef]
- Langan, K.J.; Nothnagel, E.A. Cell surface arabinogalactan-proteins and their relation to cell proliferation and viability. Protoplasma 1997, 196, 87–98. [Google Scholar] [CrossRef]
- Fragkostefanakis, S.; Dandachi, F.; Kalaitzis, P. Expression of arabinogalactan proteins during tomato fruit ripening and in response to mechanical wounding, hypoxia and anoxia. Plant Physiol. Bioch. 2012, 52, 112–118. [Google Scholar] [CrossRef]
- Gao, M.G.; Showalter, A.M. Yariv reagent treatment induces programmed cell death in Arabidopsis cell cultures and implicates arabinogalactan protein involvement. Plant J. 1999, 19, 321–331. [Google Scholar] [CrossRef]
- Lamport, D.T.A.; Kieliszewski, M.J.; Showalter, A.M. Salt stress upregulates periplasmic arabinogalactan proteins: Using salt stress to analyse AGP function. New Phytol. 2006, 169, 479–492. [Google Scholar] [CrossRef]
- Breen, S.; Williams, S.J.; Outram, M.; Kobe, B.; Solomon, P.S. Emerging insights into the functions of pathogenesis-related protein 1. Trends Plant Sci. 2017, 22, 871–879. [Google Scholar] [CrossRef]
- Yang, Y.L.; Fan, P.; Liu, J.X.; Xie, W.J.; Liu, N.; Niu, Z.B.; Li, Q.Q.; Song, J.; Tian, Q.J.; Bao, Y.G.; et al. Thinopyrum intermedium TiAP1 interacts with a chitin deacetylase from Blumeria graminis f. sp. tritici and increases the resistance to Bgt in wheat. Plant Biotechnol. J. 2022, 20, 454–467. [Google Scholar] [CrossRef]
- Lei, Y.; Gao, J.S.; Li, Y.Y.; Song, C.W.; Guo, Q.; Guo, L.L.; Hou, X.G. Functional characterization of PoEP1 in regulating the flowering stage of tree peony. Plants 2024, 13, 1642. [Google Scholar] [CrossRef] [PubMed]
- Engelen, F.A.V.; Hartog, M.V.; Thomas, T.L.; Thomas, T.T.; Taylor, B.; Sturm, A.; Kammen, A.V.; Vries, S.C.D. The carrot secreted glycoprotein gene EP1 is expressed in the epidermis and has sequence homology to Brassica S-locus glycoproteins: For cell and molecular biology. Plant J. 1993, 4, 855–862. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhao, C.P.; Zhao, L.Y.; Liu, X.; Liu, S.E.; Wang, K.H.; Wang, Z.X.; Geng, J.Y.; Guo, B.S. Cloning and functional analysis of epidermis-specific secreted glycoprotein gene GhA01EP1 in cotton. J. Cotton Sci. 2021, 33, 448–458. [Google Scholar] [CrossRef]
- Ji, H.T.; Wang, Y.N.; Cloix, C.; Li, K.X.; Jenkins, G.I.; Wang, S.F.; Shang, Z.L.; Shi, Y.T.; Yang, S.H.; Li, X. The Arabidopsis RCC1 family protein TCF1 regulates freezing tolerance and cold acclimation through modulating lignin biosynthesis. PLoS Genet. 2015, 11, e1005471. [Google Scholar] [CrossRef]
- Wu, A.H.; Allu, A.D.; Garapati, P.; Siddiqui, H.; Dortay, H.; Zanor, M.I.; Asensi-Fabado, M.A.; Munne’-Bosch, S.; Antonio, C.; Tohge, T.; et al. JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 2012, 24, 482–506. [Google Scholar] [CrossRef]
- Kaur, C.; Sharma, S.; Singla-Pareek, S.L.; Sopory, S.K. Methylglyoxal, triose phosphateisomerase and glyoxalase pathway: Implications in abiotic stress and signaling in plants. In Elucidation of Abiotic Stress Signaling in Plants; Pandey, G.K., Ed.; Springer: New York, NY, USA, 2015; pp. 347–366. [Google Scholar] [CrossRef]
- Hoque, T.S.; Hossain, M.A.; Mostofa, M.G.; Burritt, D.j.; Fujita, M.; Tran, L.S.P. Methylglyoxal: An emerging signaling molecule in plant abiotic stress responses and tolerance. Front. Plant Sci. 2016, 7, 1341. [Google Scholar] [CrossRef]
- Ghosh, A.; Kushwaha, H.R.; Hasan, M.R.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. Presence of unique glyoxalase III proteins in plants indicates the existence of shorter route for methylglyoxal detoxification. Sci. Rep. 2016, 6, 18358. [Google Scholar] [CrossRef]
- Kaur, C.; Ghosh, A.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. Glyoxalases and stress tolerance in plants. Biochem. Soc. T 2014, 42, 485–490. [Google Scholar] [CrossRef]
- Mustafiz, A.; Ghosh, A.; Tripathi, A.K.; Kaur, C.; Ganguly, A.K.; Bhavesh, N.S.; Tripathi, J.K.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. A unique Ni2+-dependent and methylglyoxal-inducible rice glyoxalase I possesses a single active site and functions in abiotic stress response. Plant J. 2014, 78, 951–963. [Google Scholar] [CrossRef]
- Wu, C.; Ma, C.Q.; Pan, Y.; Gong, S.L.; Zhao, C.X.; Chen, S.X.; Li, H.Y. Sugar beet M14 glyoxalase I gene can enhance plant tolerance to abiotic stresses. J. Plant Res. 2013, 126, 415–425. [Google Scholar] [CrossRef]
- Xu, M.L.; Zuo, D.Y.; Wang, Q.L.; Lv, L.M.; Zhang, Y.P.; Jiao, H.X.; Zhang, X.; Yang, Y.; Song, G.L.; Cheng, H.L. Identification and molecular evolution of the GLX genes in 21 plant species: A focus on the Gossypium hirsutum. BMC Genom. 2023, 24, 474. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.A.; Hossain, M.Z.; Fujita, M. Stress-induced changes of methylglyoxal level and glyoxalase I activity in pumpkin seedlings and cDNA cloning of glyoxalase I gene. Aust. J. Crop Sci. 2009, 3, 53–64. [Google Scholar]
- Rajwanshi, R.; Kumar, D.; Yusuf, M.A.; DebRoy, S.; Sarin, N.B. Stress-inducible overexpression of glyoxalase I is preferable to its constitutive overexpression for abiotic stress tolerance in transgenic Brassica juncea. Mol. Breed. 2016, 36, 76. [Google Scholar] [CrossRef]
- Zeng, Z.M. Functional Characterization of Glyoxalasei and Xyloglucan Endotransglycosylase in Rice (Oryza sativa L.). Ph.D. Dissertation, Chongqing University, Chongqing, China, 2016. [Google Scholar]
- Singla-Pareek, S.L.; Yadav, S.K.; Pareek, A.; Reddy, M.K.; Sopory, S.K. Transgenic tobacco overexpressing GLYoxalase pathway enzymes grow and set viable seeds in zinc-piked soils. Plant Physiol. 2006, 140, 613–623. [Google Scholar] [CrossRef]
- Sankaranarayanan, S.; Jamshed, M.; Kumar, A.; Skori, L.; Scandola, S.; Wang, T.; Spiege, D.; Samuel, M.A. Glyoxalase goes green: The expanding roles of glyoxalase in plants. Int. J. Mol. Sci. 2017, 18, 898. [Google Scholar] [CrossRef]
- Rugnone, M.L.; Soverna, A.F.; Sanchez, S.E.; Schlaen, R.G.; Carlos, E.H.; Seymour, D.K.; Mancini, E.; Chernomoretz, A.; Weigel, D.; Más, P.; et al. LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc. Natl. Acad. Sci. USA 2013, 110, 12120–12125. [Google Scholar] [CrossRef]
- Xie, Q.G.; Wang, P.; Liu, X.; Yuan, L.; Wang, L.B.; Zhang, C.G.; Li, Y.; Xing, H.Y.; Zhi, L.Y.; Yue, Z.L.; et al. LNK1 and LNK2 are transcriptional coactivators in the Arabidopsis circadian oscillator. Plant Cell 2014, 26, 2843–2857. [Google Scholar] [CrossRef]
- Fornara, F.; Panigrahi, K.C.S.; Gissot, L.; Sauerbrunn, N.; Rühl, M.; Jarillo, J.A.; Coupland, G. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 2009, 17, 75–86. [Google Scholar] [CrossRef]
- Huq, E.; Tepperman, J.M.; Quail, P.H. GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2000, 97, 9789–9794. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kiba, T.; Henriques, R.; Mizuno, T.; Chua, N.H.; Sakakibara, H. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 2010, 22, 594–605. [Google Scholar] [CrossRef]
- Liu, T.; Carlsson, J.; Takeuchi, T.; Newton, L.; Farre, E.M. Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. Plant J. 2013, 76, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Castillejo, C.; Pelaz, S. The balance between Constans and Tempranillo activities determines FT expression to trigger flowering. Curr. Biol. 2008, 18, 1338–1343. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.T.; Lin, S.M.; Deng, J.L.; Keasling, J.D.; Luo, X.Z. Engineering yeast for the de novo synthesis of jasmonates. Nat. Synth. 2024, 3, 224–235. [Google Scholar] [CrossRef]
- Lshiguro, S.; Kawai-Oda, A.; Ueda, J.; Nishida, L.; Okada, K. The DEFECTIVE IN ANTHER DEHISCIENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 2001, 13, 2191–2209. [Google Scholar] [CrossRef]
- Koo, A.J.K.; Cookea, T.F.; Howea, G.A. Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-L-isoleucine. Proc. Natl. Acad. Sci. USA 2011, 108, 9298–9303. [Google Scholar] [CrossRef]
- Woodward, C.; Bemis, S.M.; Hill, E.J.; Sawa, S.; Koshiba, T.; Torii, K.U. Interaction of auxin and ERECTA in elaborating Arabidopsis inflorescence architecture revealed by the activation tagging of a new member of the YUCCA family putative flavin monooxygenases. Plant Physiol. 2005, 139, 192–203. [Google Scholar] [CrossRef]
- Zhao, Y.; Xing, L.; Wang, X.G.; Hou, Y.J.; Gao, J.H.; Wang, P.C.; Duan, C.G.; Zhu, X.H.; Zhu, J.K. The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci. Signal. 2014, 7, ra53. [Google Scholar] [CrossRef]
- Ordoñez-Herrera, N.; Trimborn, L.; Menje, M.; Henschel, M.; Robers, L.; Kaufholdt, D.; Hänsch, R.; Adrian, J.; Ponnu, J.; Hoecker, U. The transcription factor COL12 is a substrate of the COP1/SPA E3 ligase and regulates flowering time and plant architecture. Plant Physiol. 2018, 176, 1327–1340. [Google Scholar] [CrossRef]
- Auldridge, M.E.; Block, A.; Vogel, J.T.; Dabney-Smith, C.; Mila, I.; Bouzayen, M.; Magallanes-Lundback, M.; DellaPenna, D.; McCarty, D.R.; Klee, H.J. Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J. 2006, 45, 982–993. [Google Scholar] [CrossRef]
- Brewer, P.B.; Yoneyama, K.; Filardo, F.; Meyers, E.; Scaffidi, A.; Frickey, T.; Akiyama, K.; Seto, Y.; Dun, E.A.; Cremer, J.E.; et al. Lateral branching oxidoreductase acts in the final stages of strigolactone biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 6301–6306. [Google Scholar] [CrossRef]
- Li, L.; Zheng, W.G.; Zhu, Y.B.; Ye, H.X.; Tang, B.Y.; Arendsee, Z.W.; Jones, D.; Li, R.R.; Ortiz, D.; Zhao, X.F.; et al. QQS orphan gene regulates carbon and nitrogen partitioning across species via NF-YC interactions. Proc. Natl. Acad. Sci. USA 2015, 112, 14734–14739. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.D.; Liu, D.; Lv, X.M.; Wang, Y.; Xun, Z.L.; Liu, Z.X.; Li, F.L.; Lu, H. The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen development in Arabidopsis. Plant Cell 2014, 26, 2939–2961. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, D.; Zhao, C.; Zhang, X.; Zhang, H.; Yuan, C.; Wang, K.; Liu, S.; Geng, J.; Guo, B. GhA01EP1 of Upland Cotton Stimulates Precocity, Improved Water Deficit Tolerance, and High Seed Yield in Transgenic Arabidopsis. Genes 2025, 16, 669. https://doi.org/10.3390/genes16060669
Li D, Zhao C, Zhang X, Zhang H, Yuan C, Wang K, Liu S, Geng J, Guo B. GhA01EP1 of Upland Cotton Stimulates Precocity, Improved Water Deficit Tolerance, and High Seed Yield in Transgenic Arabidopsis. Genes. 2025; 16(6):669. https://doi.org/10.3390/genes16060669
Chicago/Turabian StyleLi, Dan, Cunpeng Zhao, Xiaohui Zhang, Haina Zhang, Chen Yuan, Kaihui Wang, Suen Liu, Junyi Geng, and Baosheng Guo. 2025. "GhA01EP1 of Upland Cotton Stimulates Precocity, Improved Water Deficit Tolerance, and High Seed Yield in Transgenic Arabidopsis" Genes 16, no. 6: 669. https://doi.org/10.3390/genes16060669
APA StyleLi, D., Zhao, C., Zhang, X., Zhang, H., Yuan, C., Wang, K., Liu, S., Geng, J., & Guo, B. (2025). GhA01EP1 of Upland Cotton Stimulates Precocity, Improved Water Deficit Tolerance, and High Seed Yield in Transgenic Arabidopsis. Genes, 16(6), 669. https://doi.org/10.3390/genes16060669





