Abstract
Background/Objectives: Whole-genome sequencing (WGS) data provide valuable information about the genetic architecture of local livestock but have not yet been applied to Russian native goats, in particular, the Orenburg and Karachay breeds. A preliminary search for selection signatures based on single nucleotide polymorphism (SNP) genotype data in these breeds was not informative. Therefore, in this study, we aimed to address runs of homozygosity (ROHs) patterns and find the respective signatures of selection overlapping candidate genes in Orenburg and Karachay goats using the WGS approach. Methods: Paired-end libraries (150 bp reads) were constructed for each animal. Next-generation sequencing was performed using a NovaSeq 6000 sequencer (Illumina, Inc., San Diego, CA, USA), with ~20X genome coverage. ROHs were identified in sliding windows, and ROH segments shared by at least 50% of the samples were considered as ROH islands. Results: ROH islands were identified on chromosomes CHI3, CHI5, CHI7, CHI12, CHI13, and CHI15 in Karachay goats; and CHI3, CHI11, CHI12, CHI15, and CHI16 in Orenburg goats. Shared ROH islands were found on CHI12 (containing the PARP4 and MPHOSPH8 candidate genes) and on CHI15 (harboring STIM1 and RRM1). The Karachay breed had greater ROH length and higher ROH number compared to the Orenburg breed (134.13 Mb and 695 vs. 78.43 Mb and 438, respectively). The genomic inbreeding coefficient (FROH) varied from 0.032 in the Orenburg breed to 0.054 in the Karachay breed. Candidate genes associated with reproduction, milk production, immunity-related traits, embryogenesis, growth, and development were identified in ROH islands in the studied breeds. Conclusions: Here, we present the first attempt of elucidating the ROH landscape and signatures of selection in Russian local goat breeds using WGS analysis. Our findings will pave the way for further insights into the genetic mechanisms underlying adaption and economically important traits in native goats.
1. Introduction
The utilization of large-scale genomic data to search for selective sweeps and genome-wide associations in the genetic make-up of domestic goats (Capra hircus Linnaeus, 1758; CHI) is key to pinpointing changes pertaining to their domestication, breed formation, and recent selection [1]. For instance, based on whole-genome sequencing (WGS) data in Pakistani Teddy goats, Saif et al. [2] found genes in selective sweep regions that were associated with body weight, reproduction, milk production, litter size, wool production, and coat color. In another investigation using WGS data [3], strong selective signatures were detected that harbored novel genes associated with cashmere traits and environmental adaptation. Gudra et al. [4] identified 26 genetic variants associated with somatic cell count in Latvian local goats. GWAS performed in Leizhou goats revealed a significant variant (g.53666634T>C) affecting leg length traits [5]. In dairy goats, Xiong et al. [6] identified genes associated with milk production traits and reproduction traits. Sheriff et al. [7] detected nine strong regions spanning 163 genes that may affect adaptation to arid and semi-arid environments and immune response in Ethiopian indigenous goats. Wang et al. [8] proposed a number of genes as potential candidates for fecundity traits in highly prolific Jining gray goats.
Searching for runs of homozygosity (ROHs) is a widespread genomic and bioinformatic approach used to detect selection signatures in livestock species [9,10,11,12,13]. Many reports have involved the identification of candidate genes overlapping with ROH islands and related to economically important and adaption traits in farm animals [14,15,16,17,18]. For example, using whole-genome resequencing data, Li et al. [19] and Zhao et al. [20] applied this approach to find candidate genes that are responsible for reproduction traits, hair follicle development, fat tail formation, growth, and feed intake in Chinese Hu sheep. Analyzing the whole genomes of 100 American minks, Davoudi et al. [21] detected 63 ROHs embracing genes associated with fur quality, body size, and reproduction. Ayalew et al. [22] analyzed whole-genome sequences of African zebu cattle to address genetic variants associated with heat tolerance and high immune resistance. In the case of domestic goats, Sun et al. [23] identified the IL2, IL7, and KIT genes that overlapped with ROH islands and were involved in immune processes in Jianchang Black goats. Signer-Hasler et al. [1] performed an analysis of 226 caprine genomes from 11 populations and identified ROH islands possessing 2 candidate causative genetic variants that might be related to domestication.
The Orenburg goat breed (Figure 1a) is an endemic and landmark breed that produces soft cashmere fibers [24,25,26]. This breed, created in the Orenburg province near the Urals Mountain, is adapted to harsh continental climate conditions and not able to manifest high-quality down performance in other regions. The breed gained world fame when a down-knitted shawl made from Orenburg goat down won golden medals at the World Fair [27]. In addition, among the variety of local goat breeds, the Orenburg breed is one of the most prolific. The fertility of goats of this breed averages 140 kids per 100 goats. About 52.6% of females give birth to twins, and less frequently to triplets (about 2%) [28].

Figure 1.
The representative studied goat breeds of Russian origin: (a) a family of Orenburg goats (a black male and two white females); and (b) a herd of Karachay goats.
The reducing demand for down-knitted pieces has led to a dramatic decrease in the population size of this breed. In the crisis state of the down knitting industry, Orenburg goats are also used to obtain, in addition to down, other products, including meat [29,30]. Indeed, Orenburg goats can be raised under intensive breeding conditions, demonstrating excellent meat quality [29].
The surveillance of modern remaining populations and genetic monitoring using microsatellite markers [31] and single nucleotide polymorphism (SNP) arrays [32] have revealed that the original genomic components remain in the Orenburg breed. Currently, the initial steps to conserve the gene pool of this endemic breed are fortunately taking place by implementing its gamete preservation [32].
The Karachay breed (less often Karachai [33,34,35,36], sometimes Karachaevskaya [33,37] and Karachaev [37,38,39], the latter name being not entirely correct) originates from North Caucasus regions and is characterized by stamina, unpretentiousness, and high resilience to the local conditions [40] (Figure 1b). The birth rate of Karachay goats is within the range of 1.19–1.26 [34,41]. This is a dual-purpose breed (i.e., used for meat and milk), and the goats may be raised under conditions of ecologically friendly maintenance on mountain and foothill pastures [35]. Importantly, a few genomic studies were previously conducted to address their production traits. In particular, a GWAS revealed several genes that were significantly associated with the dry matter content and fatty acids in the milk of Karachay goats [36]. In addition, Selionova et al. [35] reported that a GG genotype at a single SNP locus, rs268269710, was associated with a heavier live weight and a greater carcass yield in young Karachay goats. Despite a complex of beneficial traits, indigenous Karachay goats are not as highly productive as commercial breeds [42]. Because of that, this breed is used as a maternal breed to develop the first Russian meat goat breed by crossing Karachay dams with Kalahari Red sires [39].
In our previous studies [38,43], we addressed the genetic diversity and population structure of the Orenburg and Karachay goat breeds based on SNP genotype data. However, the detection of selection signatures in the genomes of Orenburg and Karachay goats underlying adaptability and other economically important traits is relevant and will be instrumental for their further conservation, sustainable maintenance, and performance improvement. In this respect, the aim of the present study was to identify the breed-specific ROH landscape, potential selective sweeps, and candidate genes located within ROH islands in the Orenburg and Karachay goat breeds based on WGS analysis.
2. Materials and Methods
2.1. Animal Sampling
The samples utilized in this study were represented by tissue specimens of goats from the Orenburg (n = 19) and Karachay (n = 20) breeds, two Russian local breeds that originated in different environments under different selection pressures and characterized by different production performances. The sample sizes of the two breeds assigned to whole-genome sequencing were limited by the funding policy annual restrictions, while being compensated by the high depth of sequencing coverage (see Section 2.2). For ethical animal handling and sampling, see the Institutional Review Board Statement section. Prior to collecting samples, the following procedures were performed. The 1.7 mL Eppendorf tubes were numbered and filled in with 1 mL of 95% ethyl alcohol. To collect auricular samples from the goats, tagging forceps for livestock were used. The skin sampling sites were aseptically cleaned to avoid contamination. All procedures were performed by trained personnel wearing sterile medical gloves. The size of the auricle samples was not less than 0.5 by 0.5 cm. After collection, each ear biopsy was immediately placed in an Eppendorf tube, with ethyl alcohol completely covering a sample. The tubes were closed tightly and transported to the Center for Collective Use of Research Equipment “Bioresources and Bioengineering of Agricultural Animals” at the L.K. Ernst Federal Research Center for Animal Husbandry (LKEFRCAH).
For this study, we selected only typical, healthy, unrelated individuals that met the breed standards. The kinship and breed assignment of the sampled goats were preliminarily checked by microsatellite analysis. Sample collection of the Karachay goats (females) was performed on one flock at the Ladozhsky breeding farm, an LKEFRCAH branch situated in Krasnodar Krai, South Russia (45.304996, 39.891568), on 24 March 2020. The local climate is dry subtropical, transitional from moderate continental with long hot summers and warm winters (the average annual temperature is about +13.3 °C). The average age of the goats varied from 1.5 to 2.2 years. Their fiber color was brown and red-brown. Sampling from the Orenburg breed was performed on three flocks in three locations in Orenburg Oblast (formerly the Orenburg province) (51.135854, 57.945351, 51.289560, 58.178482, and 51.355471, 56). The climate is continental, with warm summers (along with frequent droughts) and cold winters. Ten Orenburg samples were collected at the Guberlinsky nucleus farm on 16 March 2017, and included three sires and four dams aged four years and three young females aged one year. Nine females (aged 2–3 years) were collected from two small holders’ farms on 19 March 2024. The fiber color of all studied goats was gray.
Figure 2 shows the sampling locations for the Orenburg (marked in red color) and Karachay breeds (marked in blue color), along with silhouettes of the representatives of the studied breeds. The map of the sampling sites (Figure 2) was created using R packages maps (Version 3.3.0) [44] and ggplot2 (Version 3.5.2) [45].
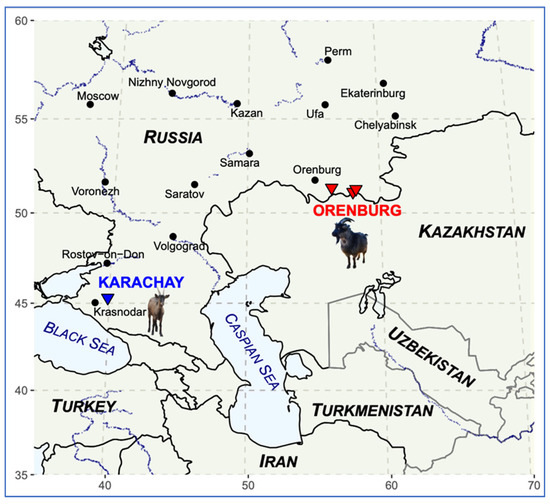
Figure 2.
The map with the corresponding sampling sites of goats for the Orenburg (▼) and Karachay (▼) breeds.
2.2. DNA Extraction and Whole-Genome Sequencing
Genomic DNA was isolated from tissue samples using the DNA Extran 2 kit (Syntol, Moscow, Russia). In order to assess the quantity and quality of the double-stranded DNA, its concentration was determined using a Qubit™ fluorometer (Invitrogen/Life Technologies, Waltham, MA, USA), while the ratio of absorption at 260 and 280 nm (OD 260/280) was measured using a NanoDrop 8000 device (Thermo Fisher Scientific Inc., Waltham, MA, USA). The minimum amount of DNA used to prepare sequencing libraries was 3 μg; therefore, the threshold DNA concentration was 30 ng/μL in at least 100 μL volume. When the concentration was lower, the DNA was subject to re-isolation in order to enhance the quantity of the initial material. In addition, to check the integrity of the DNA, the electrophoresis was run in 1% agarose gels.
To make the sequencing libraries, TruSeq DNA Nano Library Prep kits (Illumina, Inc., San Diego, CA, USA) and the Accel-NGS® 2S Plus DNA Library Kit (Integrated DNA Technologies, Inc., Coralville, IA, USA) for Illumina® Platforms (Swift Biosciences, Inc., Ann Arbor, MI, USA) were utilized. Paired-end libraries with a fragment size of 150 bp were constructed for each goat. Next-generation sequencing was performed using a NovaSeq 6000 sequencer (Illumina, Inc., USA). Genome coverage was 19×–20×. Alignment to the reference goat genome assembly ARS1.2 (https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_001704415.2/, accessed on 25 April 2025, https://ftp.ensembl.org/pub/release-113/fasta/capra_hircus/dna/, accessed on 25 April 2025) was executed using bwa-mem2 tools (Version 2.2.1) [46] and SAMtools (Version 1.21) [47].
2.3. Data Processing and in Silico Analyses
A total of 20,554,866 SNPs were used for ROH identification. Prior to further constructing Neighbor-Net graphs and performing principal component analysis (PCA), the SNPs were pruned using their linkage disequilibrium criterion. After pruning, a total of 4,588,309 SNPs were left for the subsequent analyses. Pairwise identity-by-state (IBS) distances were computed using PLINK v1.9 [48] (with the software setting --distance 1-ibs). The Neighbor-Net graphs based on the matrix of IBS distances were visualized using SplitsTree 4.14.5 software [49]. PCA was performed using PLINK v1.9 [48] and visualized using the R package ggplot2 [45].
The ROH regions were identified using sliding windows in the R package detectRUNS [50] using the following program settings: runs <- slidingRUNS.run(‘outdata_roh.ped’, ‘outdata_roh.map’, windowSize = 50, threshold = 0.05, minSNP = 50, ROHet = FALSE, maxOppWindow = 1, maxMissWindow = 5, maxGap = 100,000, minLengthBps = 100,000, minDensity = 1/10,000. Herewith, the following parameters were used to identify the ROHs:
- (1)
- windowSize is the size of the sliding window (i.e., number of SNP loci; default = 15);
- (2)
- threshold is the threshold of overlapping windows of the same state (homozygous/heterozygous) to call an SNP in a RUN (default = 0.05);
- (3)
- minSNP is the minimum number of SNPs in a RUN (default = 3);
- (4)
- ROHet is a heterozygosity/homozygosity parameter for whether runs of heterozygosity (ROHet) or homozygosity (ROHom) are detected (default = FALSE);
- (5)
- maxOppWindow is the maximum number of homozygous/heterozygous SNPs in the sliding window (default = 1);
- (6)
- maxMissWindow is the maximum number of missing SNPs in the sliding window (default = 1);
- (7)
- maxGap is the maximum distance between consecutive SNPs to be still considered a potential run (default = 106 bp);
- (8)
- minLengthBps is the minimum length of a run in bp (defaults to 1000 bp = 1 Kb);
- (9)
- minDensity is the minimum number of SNPs per Kb (defaults to 0.1 = 1 SNP every 10 Kb);
- (10)
- maxOppRun is the maximum number of opposite genotype SNPs in the run (optional);
- (11)
- maxMissRun is the maximum number of missing SNPs in the run (optional).
ROH segments shared by at least 50% of the samples were considered as ROH islands.
We calculated ROHs for each goat and then categorized ROHs according to the following four length bins (classes): 0.1–0.2 Mb, 0.2–0.4 Mb, 0.4–0.8 Mb, and >0.8 Mb. For each breed and length class, the total number of detected ROHs was computed for each individual. To determine the mean sum of ROHs, the total ROH length for each goat in the populations was calculated, and the results were averaged by breed group.
The genomic inbreeding coefficient based on ROHs (FROH) was computed as the sum of the length of all ROHs per goat proportioned to the total autosomal SNP coverage.
3. Results
3.1. Between-Breed Genetic Differentiation
As an initial step, we investigated a pattern of genetic relations and differentiation between the two studied goat breeds. Figure 3 shows the Neighbor-Net plot for the Orenburg and Karachay breeds. The breeds were clearly differentiated by forming their own clusters.
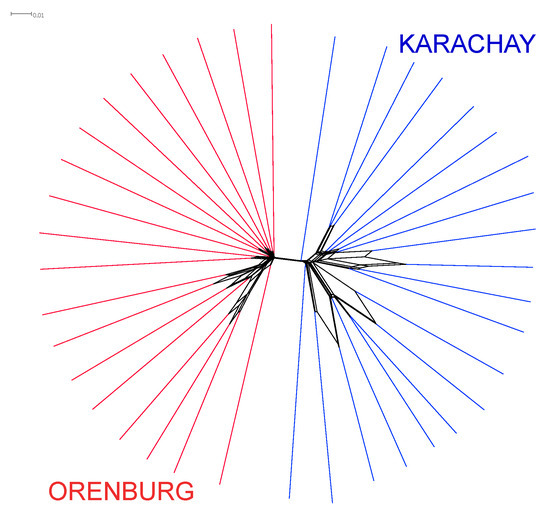
Figure 3.
Individual Neighbor-Net graph using IBS distances plotted for the Orenburg and Karachay breeds based on the WGS data analysis.
PCA revealed that PC1 apparently differentiated the Orenburg goats from the Karachay breed (Figure 4), and this distinct differentiation pattern for the two breeds was consistent with the Neighbor-Net analysis plot (Figure 3).
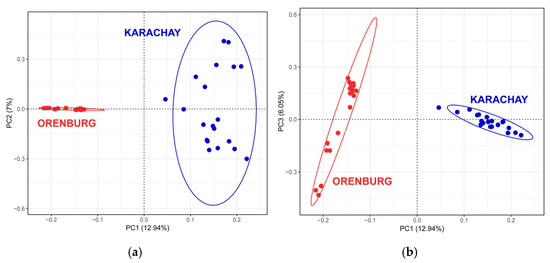
Figure 4.
Principal component analysis performed for Orenburg and Karachay goats based on the WGS data analysis: (a) for the first two principal components (PC1 and PC2); and (b) for the first and third principal components (PC1 and PC3).
3.2. Runs of Homozogosity Patterns
As a result of the subsequent in silico analysis, we found out that the Karachay breed had a greater ROH length and higher ROH number in comparison with the Orenburg breed (Table 1). Regarding the individual values of these indicators, the maximum values of the ROH number and ROH length varied from 540 to 2075 and from 102.96 to 404.4 Kb in the Orenburg and Karachay breeds, respectively.

Table 1.
Runs of homozygosity (ROH) distribution values 1 in the Orenburg and Karachay breeds based on the WGS data analysis.
The values of FROH varied from 0.032 in the Orenburg breed to 0.054 in the Karachay breed. The maximum value of FROH detected in the Karachay breed was four times higher than that computed for the Orenburg breed (0.16 vs. 0.04, respectively).
When dividing the ROHs into length classes (Figure 5), only short ROHs were identified. The ROHs accounted for class 0.1–0.2 Mb were the most frequent in the studied breeds (Figure 5a). The mean number of ROHs for class 0.1–0.2 Mb was 329.95 ± 10.79 and 480.85 ± 63.46 in the Orenburg and Karachay breeds, respectively. In class 0.2–0.4 Mb, the mean number of ROHs was 90.47 ± 5.42 for the Orenburg breed and 175.15 ± 26.23 for the Karachay breed. The ROHs from class 0.4–0.8 Mb were less frequent and varied from 17.79 ± 1.81 in the Orenburg breed to 36.70 ± 6.09 in the Karachay breed. The ROHs included in class >0.8 Mb were the rarest in both breeds, and accounted for 0.58 ± 0.18 in the Orenburg breed and 2.55 ± 0.48 in the Karachay breed.
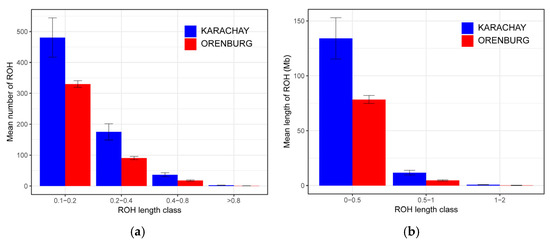
Figure 5.
Distribution of runs of homozygosity (ROH) according to length classes in Orenburg and Karachay goats based on the WGS data analysis: (a) mean number of ROHs per breed; and (b) mean length of ROHs per breed.
The mean length of ROHs per breed was the largest in class 0–0.5 Mb, corresponding to 78.43 ± 3.66 and 134.13 ± 18.79 in the Orenburg and Karachay breeds, respectively (Figure 5b). The mean length of ROHs included into class 0.5–1 Mb varied from 4.71 ± 0.57 in the Orenburg breed to 11.73 ± 2.08 in the Karachay breed. The lowest mean lengths of ROHs per breed were detected for class 1–2, as follows: 0.66 ± 0.20 and 0.13 ± 0.09 in the Orenburg and Karachay breeds, respectively.
3.3. Distribution of ROH Islands and Candidate Genes
We found ROH islands in the genome of the Orenburg breed on chromosomes CHI3, CHI11, CHI12, CHI15, and CHI16. In Karachay goats, ROH islands were identified on CHI3, CHI5, CHI7, CHI12, CHI13, and CHI15 (Figure 6).
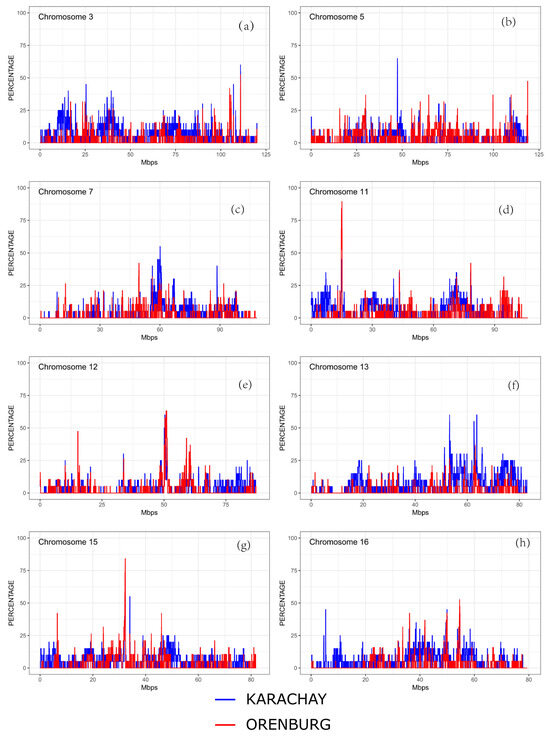
Figure 6.
ROH islands identified in the Orenburg and Karachay breeds on the following chromosomes: (a) CHI3, (b) CHI5, (c) CHI7, (d) CHI11, (e) CHI12, (f) CHI13, (g) CHI15, and (h) CHI16.
For the most frequently occurring (in more than 50% of animals) islands of homozygosity, with an indication of the beginning and end of the regions, the number of SNPs in ROHs and the genes located in them are presented in Table 2.

Table 2.
Genes located in the ROH islands (common in ≥50% in each breed) in the Orenburg and Karachay breeds based on the WGS analysis.
The shared ROH islands were found on CHI12 (50,342,201–51,155,729 and 50,286,354–53,239,419 bp in the Orenburg and Karachay breeds, respectively) and on CHI15 (32,124,217–32,379,590 and 32,188,813–32,383,957 bp in the Orenburg and Karachay breeds, respectively). The former contained the genes encoding poly (ADP-ribose) polymerase family member 4 (PARP4) and M-phase phosphoprotein 8 (MPHOSPH8), whereas the latter included the genes for stromal interaction molecule 1 (STIM1) and ribonucleotide reductase catalytic subunit M1 (RRM1). The ROH segments on CHI11 spanning from 14,885,086 to 14,956,388 bp, 14,957,644 to 14,990,520 bp, 14,991,874 to 15,026,370 bp, and 15,064,670 to 15,155,143 bp and a ROH segment on CHI15 (32,269,497 to 32,363,003 bp) were common in ≥80% of the Orenburg breed.
4. Discussion
Traditional and autochthonous livestock breeds are an important reservoir of genetic diversity [51,52,53,54,55]. The Orenburg and Karachay breeds represent local goats that are well adapted for exploiting in a specific environment. The Orenburg breed is raised in steppes in continental climate conditions with strong winter winds and hot summers. The Karachay breed inhabits mountains and foothills in the North Caucasus region and may be successfully raised in the southern regions of Russia with higher summer temperatures. However, the genetic potential of these breeds has been consistently underestimated. Moreover, the Orenburg breed gene pool was threatened by the shutting down of gene pool farms [26,32].
In the present study, we examined the patterns of ROH distribution in these two Russian goat breeds based on WGS. Interestingly, Karachay goats exhibited larger genome coverage in ROHs compared to the Orenburg breed. This pattern is in accordance with our previous results obtained in a different sampling experiment of Karachay goats using DNA arrays [38]. According to Bertolini et al. [56], sometimes such a pattern may be observed in local breeds due to geographic isolation. In the case of the Orenburg goats examined in this study, we used samples from three geographically distant populations, which may affect the ROH coverage.
In general, the patterns of ROH distribution in Russian goats were compatible with the estimates in other goat breeds. For instance, the ROH length varied from 101.5 to 14,801.1 Kb in Mongolian cashmere goats [57]. In Swiss local breeds, the mean number of ROHs varied from 1174 to 1436, and the average length of ROHs ranged between 246 and 433 Kb [1]. In one study on Chinese goats [58], the average ROH length was 0.184 ± 0.102 Mb. The genomic coefficient (FROH) values calculated in our study were in agreement with those obtained in Mongolian cashmere goats (0.026) [57], Swiss local goats (0.137–0.263) [1], Hainan black goats (0.107–0.186) [58], Latvian local goats (0.249) [4], and Ethiopian indigenous goats (0.016–0.261) [7]. Peng et al. [59] reported that the mean FROH values for African, European, and Bezoar goats were 0.073, 0.079 and 0.182, respectively.
We did not identify any long ROHs that reflect the recent inbreeding events [60]; moreover, our results are compatible with those obtained in Hu sheep, in the genome of which ROH segments with lengths from 300 Kb to 1 Mb were predominant [20]. Bian et al. [61] reported that ROHs were categorized into length classes of 0.5–1, 1–2, and 2–4 Mb in the Xiangxi white buffalo. A comparative genomic analysis of African zebu and taurine breeds revealed that the Holstein breed showed a significant accumulation of ROHs in all ROH classes, including 0.5–1 Mb, 1–2 Mb, and >2 Mb [22]. In Mongolian cashmere goats, the highest number of ROHs, representing 38.39% of the total ROH count, had the lengths of 0–0.5 Mb [57]. In Swiss local goats, the ROH segments longer than 1 Mb exceeded 50% of all detected ROHs [1]. ROHs with lengths of 0.1–0.2 Mb accounted for 72.41% of all ROH fragments, and ROHs with lengths over 1 Mb made up only 0.08% in Chinese goats [58]. Short ROHs (≤500 Kb) dominated in Latvian local goats [4]. Zhao et al. [20] assumed that short ROHs detected using WGS may point out the absence of inbreeding events in recent generations in livestock populations.
Based on the WGS data, we found ROH islands in both Russian breeds that harbored the potentially relevant candidate genes. The ROH island on CHI17 was common for both breeds and contained the STIM1 and RRM1 genes. STIM1 was found to be under strong selection in many goat groups, including Swiss [1], Chinese [8,23,62,63], and Pakistani breeds [2,64]. The STIM1 gene was related to body mass and weight in Teddy goats [2] and can be associated with horn/poll phenotypes in goats [63]. Sun et al. [23] suggested that STIM1 was involved in genetic adaptations to local environmental conditions in Jintang Black goats. Wang et al. [8] identified a region harboring the STIM1 and RRM1 genes potentially involved in reproduction in Jining gray goats. Furthermore, the region including RRM1 and STIM1 might be connected with gain in beef steers [65,66]. In addition, Sun et al. [23] reported that eight SNPs within the STIM1 gene were fixed in several black goat breeds, while the Bezoars carry the mutant alleles. Also, Zheng et al. [67] found that the STIM1–RRM1 haplotype in domestic goats was putatively introgressed from an ibex-like species and may be linked to neural function or behavior.
In both breeds, we found a ROH island on CHI12 that was wider in the Orenburg breed (~50.3–51.1 Mb) and contained the common PARP4 and MPHOSPH8 genes. This region overlapped, or was close to, the signatures of selection reported by Kim et al. [68], Bertolini et al. [56], Dadousis et al. [69], and Pegolo et al. [70] based on the identification of ROH hot spots. Kim et al. [68] suggested that this genomic region may be associated with adaptations to natural and climatic conditions. The ROH island identified in the Karachay breed, along with PARP4 and MPHOSPH8, included the zinc finger MYM-type containing 2 (ZMYM2) gene. This gene was involved in DNA methylation patterning in the early embryonic development of mammals [71] and in the regulation of spermatogenesis and cell cycles in goats [72]. In addition, ZMYM2 might be related to adaptive functions in Mediterranean sheep and goats [73]. It was also found in ROH islands in other Russian goat breeds [43], as well as in several sheep breeds [74].
A large group of genes, including those for gap junction protein β 2 (GJB2), gap junction protein β 6 (GJB6), crystallin lambda 1 (CRYL1), intraflagellar transport 88 (IFT88), interleukin 17 (IL17D), eukaryotic translation elongation factor 1 α lysine methyltransferase 1 (EEF1AKMT1), exp ortin 4 (XPO4), large tumor suppressor kinase 2 (LATS2), and Sin3A-associated protein 18 (SAP18), were identified in a ROH island in the Orenburg breed on CHI12. GJB2 and GJB6 regulate the formation of gap junction in the cochlea and play key roles in hearing [75]. Pegolo et al. [70] assumed that this region, found in ROH hot spots in diverse goat breeds, could have undergone selection even prior to the domestication of goats because enhanced hearing is beneficial to detect potential threats in natural habitats. Additionally, the GJB2 gene, associated with body size and growth, may be subject to positive selection in the Boer [76] and Chinese goat breeds [77]. It was also found in the ROH islands in Chinese sheep breeds [20,78,79]. GJB6 and SAP18 may be involved in the regulation of reproductive performance in sheep by influencing ovarian and embryonic development, as well as spermatogenesis [19]. Quan et al. [80] proposed specific variants of the GJB6 gene related to larger litter size as molecular markers for selective breeding.
The CRYL1 gene is involved in the development of the nervous system and kidneys during embryogenesis in sheep [81] and in low mating behavior in Rasa Aragonesa rams [82]. Several studies have suggested a role of the IFT88 gene in cashmere and mohair fineness in goats [83,84,85]. In addition, this gene plays an important role in spermatogenesis processes [86,87] and is under selection pressure in several Russian sheep breeds [88]. The IL17D gene is involved in the inflammatory response to respiratory infections [89]. Chen et al. [90] reported that the LATS2 gene affected milk fat secretion and activation of lactogenesis in goats. In addition, this gene is involved in the regulation of reproduction functions in sheep [91] and cattle [92].
The genome regions under selection pressure were found in the Orenburg breed on CHI11 and CHI16. These contain the genes encoding Yip1 domain family member 4 (YIPF4), baculoviral IAP repeat containing 6 (BIRC6), tetratricopeptide repeat domain 27 (TTC27), and RAB GTPase activating protein 1-like (RABGAP1L). YIPF4 might be associated with litter size in Dazu black goats [93]. In addition, the YIP4 gene was proposed as a candidate gene that helps them potentially adapt to nutrient stress conditions [94]. The TTC27 gene, associated with milk production traits, was identified to be under selection pressure in the Pakistani Teddy [2] and Dera-Din-Panah [64] goat breeds. RABGAP1L is involved in endocytosis and endosome maturation [95]. The BIRC6 gene was identified in a ROH island in Chinese Cashmere goats [96] and was found to be under selection in Pakistani Teddy goats [2]. This gene was reported to play a role in the regulation of fertility traits and early embryonic development in different livestock species [97,98], including sheep [99]. In addition, the BIRC6 gene was linked with body weight at 6 months in Bos indicus [100] and affected average daily gain in beef cattle [101].
Finally, a number of genes were identified in ROH islands in Karachay goats. These included signaling lymphocytic activation molecule family member 1 (SLAMF1), SIL1 nucleotide exchange factor (SIL1), pre-mRNA processing factor 6 (PRPF6), sterile α motif domain containing 10 (SAMD10), zinc finger protein 512B (ZNF512B), uridine–cytidine kinase 1 like 1 (UCKL1), tumor protein p53 inducible nuclear protein 2 (TP53INP2), nuclear receptor coactivator 6 (NCOA6), and γ-glutamyltransferase 7 (GGT7). SLAMF1 is an immune-related gene in goats and other ruminant species [102,103]. The SIL1 gene, related to immunity traits, was found in ROH islands in three indigenous Chinese cattle populations (Leiqiong, Lufeng, and Hainan) [104]. The PRPF6 gene was associated with milk production traits in dairy goats [105]. ZNF512B may play a role in the regulation of feather morphogenesis in ducks [106]. The TP53INP2 gene was involved in the positive regulation of differentiation in bovine adipocytes [107] and brown adipose tissue energy metabolism in humans [108]. Additionally, it was shown that the knockout of this gene increased skeletal muscle mass in mice [109,110]. NCOA6 and GGT7 were reported as candidates for fiber color in alpaca [111]. NCOA6 was involved in the regulation of milk fat synthesis in dairy cattle and located within a quantitative trait locus for milk production traits [112,113].
5. Conclusions
In this study, we performed WGS in the Orenburg and Karachay goat breeds to find ROH islands under selective pressure and estimate an inbreeding level in their genomes. The observed FROH values in this study suggested low to moderate inbreeding in the studied breeds. The ROH islands were found on CHI3, CHI5, CHI7, CHI12, CHI13, and CHI15 in the Karachay goats; and on CHI3, CHI11, CHI12, CHI15, and CHI16 in the Orenburg breed. The shared ROH islands were found on CHI12 and on CHI15. Furthermore, we identified the candidate genes within the ROH islands. The results suggest that the majority of genes within one ROH island in the Karachay goats were associated with traits related to immunity, body weight, and milk production. Genes harboring in the ROH islands in the Orenburg goats were relevant to reproduction, increased litter size, growth, and development. In addition, a region on CHI11 that was common for 80% of the individuals and includes the YIPF4, BIRC6, and TTC27 genes may be potentially considered as a target region for selection in the Orenburg breed. We thus identified candidate genes that reflect the essential biological specifics of the studied goat breeds. Our findings may be used for developing conservation strategies and for future genetic improvements in native Russian goats.
Author Contributions
Conceptualization, T.E.D., A.V.D., and N.A.Z.; methodology, A.V.D., T.E.D., and A.S.A.; software, A.V.D.; validation, T.E.D., A.V.D., M.N.R., and N.A.Z.; formal analysis, A.V.D., A.S.A., T.E.D., and O.A.K.; investigation, S.N.P., A.N.F., S.A.P., E.A.G., I.V.G., M.A.V., N.A.C., and A.D.S.; resources, T.E.D., S.V.L., and N.A.Z.; data curation, A.V.D., T.E.D., and N.A.Z.; writing—original draft preparation, T.E.D., A.V.D., and N.A.Z.; writing—review and editing, T.E.D., A.V.D., O.A.K., D.K.G., M.N.R., and N.A.Z.; visualization, T.E.D., A.V.D., and M.N.R.; supervision, D.K.G. and N.A.Z.; project administration, T.E.D. and N.A.Z.; funding acquisition, T.E.D. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Russian Science Foundation (RSF), Grant No. 24-46-02012 (https://rscf.ru/project/24-46-02012/, accessed on 25 April 2025).
Institutional Review Board Statement
The animals used in this study were treated in accordance with, and all relevant procedures were conducted according to, the ethical guidelines approved by the LKEFRCAH Commission on the Ethics of Animal Experiments (Protocol No. 2 dated 24 February 2025). The tissue samples (fragments of the auricle) of the goats from the populations of the Orenburg and Karachay breeds were collected by trained personnel by using tagging forceps for livestock under strict veterinary rules in accordance with those in place for executing laboratory research (tests) in the implementation of the veterinary control (supervision), as approved by the Eurasian Economic Commission Council Decision No. 80 (dated 10 November 2017), in the course of the work with the goat herd in 2024.
Informed Consent Statement
Not applicable.
Data Availability Statement
The WGS data for Orenburg and Karachay goats used in this study are available upon request from the corresponding author.
Acknowledgments
We are grateful to Vladimir N. Frolov and Almas Zh. Sapinov for providing access to the Orenburg breed herds and for technical support at the time of animal sampling.
Conflicts of Interest
The authors declare no conflicts of interest. The RSF had no role in the design of this study; in the collection, analyses, or interpretation of the data; in the writing of the manuscript; or in the decision to publish the results.
References
- Signer-Hasler, H.; Henkel, J.; Bangerter, E.; Bulut, Z.; Drögemüller, C.; Leeb, T.; Flury, C. Runs of homozygosity in Swiss goats reveal genetic changes associated with domestication and modern selection. Genet. Sel. Evol. 2022, 54, 6. [Google Scholar] [CrossRef] [PubMed]
- Saif, R.; Henkel, J.; Mahmood, T.; Ejaz, A.; Ahmad, F.; Zia, S. Detection of whole genome selection signatures of Pakistani Teddy goat. Mol. Biol. Rep. 2021, 48, 7273–7280. [Google Scholar] [CrossRef] [PubMed]
- Dan, H.; Zhong, H.; Akhatayeva, Z.; Lin, K.; Xu, S. Whole-genome selective scans detect genes associated with cashmere traits and climatic adaptation in cashmere goats (Capra hircus) in China. Genes 2025, 16, 292. [Google Scholar] [CrossRef]
- Gudra, D.; Valdovska, A.; Jonkus, D.; Kairisa, D.; Galina, D.; Ustinova, M.; Viksne, K.; Fridmanis, D.; Kalnina, I. Genetic characterization of the Latvian local goat breed and genetic traits associated with somatic cell count. Animal 2024, 18, 101154. [Google Scholar] [CrossRef]
- Liu, J.; Dong, S.; Lv, J.; Li, Y.; Sun, B.; Guo, Y.; Deng, M.; Liu, D.; Liu, G. Screening of SNP loci related to leg length trait in Leizhou goats based on whole-genome resequencing. Int. J. Mol. Sci. 2024, 25, 12450. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Bao, J.; Hu, W.; Shang, M.; Zhang, L. Whole-genome resequencing reveals genetic diversity and selection characteristics of dairy goat. Front. Genet. 2023, 13, 1044017. [Google Scholar] [CrossRef]
- Sheriff, O.; Ahbara, A.M.; Haile, A.; Alemayehu, K.; Han, J.L.; Mwacharo, J.M. Whole-genome resequencing reveals genomic variation and dynamics in Ethiopian indigenous goats. Front. Genet. 2024, 15, 1353026. [Google Scholar] [CrossRef]
- Wang, J.J.; Li, Z.D.; Zheng, L.Q.; Zhang, T.; Shen, W.; Lei, C.Z. Genome-wide detection of selective signals for fecundity traits in goats (Capra hircus). Gene 2022, 818, 146221. [Google Scholar] [CrossRef]
- He, S.; Di, J.; Han, B.; Chen, L.; Liu, M.; Li, W. Genome-wide scan for runs of homozygosity identifies candidate genes related to economically important traits in Chinese Merino. Animals 2020, 10, 524. [Google Scholar] [CrossRef]
- Zinovieva, N.A.; Dotsev, A.V.; Sermyagin, A.A.; Deniskova, T.E.; Abdelmanova, A.S.; Kharzinova, V.R.; Sölkner, J.; Reyer, H.; Wimmers, K.; Brem, G. Selection signatures in two oldest Russian native cattle breeds revealed using high-density single nucleotide polymorphism analysis. PLoS ONE 2020, 15, e0242200. [Google Scholar] [CrossRef]
- Abdelmanova, A.S.; Dotsev, A.V.; Romanov, M.N.; Stanishevskaya, O.I.; Gladyr, E.A.; Rodionov, A.N.; Vetokh, A.N.; Volkova, N.A.; Fedorova, E.S.; Gusev, I.V.; et al. Unveiling comparative genomic trajectories of selection and key candidate genes in egg-type Russian White and meat-type White Cornish chickens. Biology 2021, 10, 876. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Liu, J.; Ma, X.; Yang, J. Genome-wide runs of homozygosity reveal inbreeding levels and trait-associated candidate genes in diverse sheep breeds. Genes 2025, 16, 316. [Google Scholar] [CrossRef] [PubMed]
- Romanov, M.N.; Shakhin, A.V.; Abdelmanova, A.S.; Volkova, N.A.; Efimov, D.N.; Fisinin, V.I.; Korshunova, L.G.; Anshakov, D.V.; Dotsev, A.V.; Griffin, D.K.; et al. Dissecting selective signatures and candidate genes in grandparent lines subject to high selection pressure for broiler production and in a local Russian chicken breed of Ushanka. Genes 2024, 15, 524. [Google Scholar] [CrossRef]
- Liu, D.; Chen, Z.; Zhao, W.; Guo, L.; Sun, H.; Zhu, K.; Liu, G.; Shen, X.; Zhao, X.; Wang, Q.; et al. Genome-wide selection signatures detection in Shanghai Holstein cattle population identified genes related to adaption, health and reproduction traits. BMC Genom. 2021, 22, 747. [Google Scholar] [CrossRef]
- Fedorova, E.S.; Dementieva, N.V.; Shcherbakov, Y.S.; Stanishevskaya, O.I. Identification of key candidate genes in runs of homozygosity of the genome of two chicken breeds, associated with cold adaptation. Biology 2022, 11, 547. [Google Scholar] [CrossRef]
- Liu, X.; Peng, Y.; Zhang, X.; Chen, W.; Chen, Y.; Wei, L.; Zhu, Q.; Khan, M.Z.; Wang, C. Potential genetic markers associated with environmental adaptability in herbivorous livestock. Animals 2025, 15, 748. [Google Scholar] [CrossRef]
- Dementieva, N.V.; Kudinov, A.A.; Larkina, T.A.; Mitrofanova, O.V.; Dysin, A.P.; Terletsky, V.P.; Tyshchenko, V.I.; Griffin, D.K.; Romanov, M.N. Genetic variability in local and imported germplasm chicken populations as revealed by analyzing runs of homozygosity. Animals 2020, 10, 1887. [Google Scholar] [CrossRef] [PubMed]
- Romanov, M.N.; Abdelmanova, A.S.; Fisinin, V.I.; Gladyr, E.A.; Volkova, N.A.; Koshkina, O.A.; Rodionov, A.N.; Vetokh, A.N.; Gusev, I.V.; Anshakov, D.V.; et al. Selective footprints and genes relevant to cold adaptation and other phenotypic traits are unscrambled in the genomes of divergently selected chicken breeds. J. Anim. Sci. Biotechnol. 2023, 14, 35. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Z.; Fang, Y.; Cao, C.; Zhang, Z.; Pan, Y.; Wang, Q. Runs of homozygosity revealed reproductive traits of Hu sheep. Genes 2022, 13, 1848. [Google Scholar] [CrossRef]
- Zhao, F.; Xie, R.; Fang, L.; Xiang, R.; Yuan, Z.; Liu, Y.; Wang, L. Analysis of 206 whole-genome resequencing reveals selection signatures associated with breed-specific traits in Hu sheep. Evol. Appl. 2024, 17, e13697. [Google Scholar] [CrossRef]
- Davoudi, P.; Do, D.N.; Rathgeber, B.; Colombo, S.; Sargolzaei, M.; Plastow, G.; Wang, Z.; Miar, Y. Characterization of runs of homozygosity islands in American mink using whole-genome sequencing data. J. Anim. Breed. Genet. 2024, 141, 507–520. [Google Scholar] [CrossRef] [PubMed]
- Ayalew, W.; Wu, X.; Tarekegn, G.M.; Sisay Tessema, T.; Naboulsi, R.; Van Damme, R.; Bongcam-Rudloff, E.; Edea, Z.; Enquahone, S.; Yan, P. Whole-genome resequencing reveals selection signatures of Abigar cattle for local adaptation. Animals 2023, 13, 3269. [Google Scholar] [CrossRef]
- Sun, X.; Guo, J.; Li, L.; Zhong, T.; Wang, L.; Zhan, S.; Lu, J.; Wang, D.; Dai, D.; Liu, G.E.; et al. Genetic diversity and selection signatures in Jianchang Black goats revealed by whole-genome sequencing data. Animals 2022, 12, 2365. [Google Scholar] [CrossRef] [PubMed]
- Orekhov, A.A. Goats. In Animal Genetic Resources of the USSR; FAO Animal Production and Health Paper; Dmitriev, N.G., Ernst, L.K., Eds.; Food and Agriculture Organization of the United Nations: Rome, Italy, 1989; Volume 65, pp. 344–365. Available online: https://www.fao.org/4/ah759e/AH759E14.htm (accessed on 25 April 2025).
- Tarasova, E.I.; Frolov, A.N.; Lebedev, S.V.; Romanov, M.N. Istoriya, razvedeniye, selektsiya i genetika orenburgskoy porody koz [History, Breeding, Selection and Genetics of the Orenburg Goat Breed]. In Molecular Genetic Technologies for Analysis of Gene Expression Related to Animal Productivity and Disease Resistance, Materials of the 3rd International Scientific and Practical Conference, Moscow, Russia, 30 September 2021; Pozyabin, S.V., Kochish, I.I., Romanov, M.N., Eds.; Ministry of Agriculture of the Russian Federation, Federal State Budgetary Educational Institution of Higher Education “Moscow State Academy of Veterinary Medicine and Biotechnology–MVA named after K.I. Scriabin”; Sel’skokhozyaistvennye tekhnologii: Moscow, Russia, 2021; pp. 450–454. [Google Scholar] [CrossRef]
- Tarasova, E.I.; Frolov, A.N.; Lebedev, S.V.; Romanov, M.N. Landmark native breed of the Orenburg goats: Progress in its breeding and genetics and future prospects. Anim. Biotechnol. 2023, 34, 5139–5154. [Google Scholar] [CrossRef] [PubMed]
- Royal Commission. Official Descriptive and Illustrated Catalogue of the Great Exhibition of the Works of Industry of All Nations; Spicer Brothers: London, UK, 1851; Volume 3, p. 1383. [Google Scholar]
- Petrov, N.I. Produktivnost’ i nasledovanie masti potomstvom orenburgskikh koz [Productivity and inheritance of color type by the progeny of Orenburg goats]. Vestn. Myasn. Skotovod. [Her. Beef Cattle Breed.] 2015, 4, 47–50. Available online: https://www.elibrary.ru/item.asp?id=25286682 (accessed on 25 April 2025). (In Russian with English summary).
- Belkov, G.I.; Panin, V.A. Kachestvennyye i kolichestvennyye pokazateli myasnoy produktivnosti koz orenburgskoy porody [Qualitative and quantitative indicators of meat productivity of Orenburg goats]. Izv. Orenburg. Gos. Agrar. Un-Ta [Izv. Orenburg State Agrar. Univ.] 2021, 3, 304–307, (In Russian with English summary). [Google Scholar]
- Kharlamov, A.V.; Panin, V.A. Kompleksnaya otsenka pokazateley produktivnosti koz orenburgskoy porody v usloviyakh Orenburgskogo regiona [Comprehensive assessment of productivity indicators of Orenburg goats in the conditions of the Orenburg region]. Nauka i obraz. [Sci. Educ.] 2023, 1–2, 123–132, (In Russian with English summary). [Google Scholar]
- Abdelmanova, A.S.; Deniskova, T.E.; Petrov, S.N.; Frolov, A.N.; Platonov, S.A.; Gladyr, E.A.; Gusev, I.V.; Lebedev, S.V.; Zinovieva, N.A. Otsenka dinamiki geneticheskogo raznoobraziya populyatsiy orenburgskoy porody koz s ispol’zovaniyem mikrosatellitnykh markerov [Assessment of the dynamics of genetic diversity of Orenburg goat breed populations by microsatellite markers]. Dostizheniya Nauki I Tekhniki APK [Achiev. Sci. Technol. AIC] 2024, 38, 50–56. [Google Scholar]
- Deniskova, T.E.; Dotsev, A.V.; Abdelmanova, A.S.; Petrov, S.N.; Frolov, A.N.; Platonov, S.A.; Gladyr, E.A.; Gusev, I.V.; Selionova, M.I.; Rodionov, A.N.; et al. Genetic diversity in the Orenburg goat breed revealed by single-nucleotide polymorphism (SNP) analysis: Initial steps in saving a threatened population. Genes 2024, 15, 1375. [Google Scholar] [CrossRef]
- Scherf, B.D. (Ed.) World Watch List for Domestic Animal Diversity, 2nd ed.; FAO, UNEP: Rome, Italy, 1995; Available online: https://www.fao.org/faostat/en/ (accessed on 26 November 2015).
- Aybazov, M.M.; Selionova, M.I.; Mamontova, T.V. Ekster’yernyye i nekotoryye biologicheskiye pokazateli karachayevskikh koz [Exterior and some biological indices of Karachai goats]. Zootehniâ [Zootech] 2019, 12, 5–9, (In Russian with English summary). [Google Scholar]
- Selionova, M.; Aibazov, M.; Sermyagin, A.; Belous, A.; Deniskova, T.; Mamontova, T.; Zharkova, E.; Zinovieva, N. Genome-wide association and pathway analysis of carcass and meat quality traits in Karachai young goats. Animals 2023, 13, 3237. [Google Scholar] [CrossRef] [PubMed]
- Selionova, M.; Trukhachev, V.; Aibazov, M.; Sermyagin, A.; Belous, A.; Gladkikh, M.; Zinovieva, N. Genome-wide association study of milk composition in Karachai goats. Animals 2024, 14, 327. [Google Scholar] [CrossRef]
- Selionova, M.I.; Trukhachev, V.I.; Aybazov, A.-M.M.; Stolpovsky, Y.A.; Zinovieva, N.A. Genetic markers of goats (review). Sel’skokhozyaistvennaya Biol. [Agric. Biol.] 2021, 56, 1031–1048. [Google Scholar] [CrossRef]
- Deniskova, T.E.; Dotsev, A.V.; Selionova, M.I.; Reyer, H.; Sölkner, J.; Fornara, M.S.; Aybazov, A.M.; Wimmers, K.; Brem, G.; Zinovieva, N.A. SNP-based genotyping provides insight into the West Asian origin of Russian local goats. Front. Genet. 2021, 12, 708740. [Google Scholar] [CrossRef]
- Sermyagin, A.A.; Deniskova, T.E.; Gusev, I.V.; Petrov, S.N.; Rodionov, A.N.; Dotsev, A.V.; Zinovieva, N.A. Identification of SNPs associated with growth and development traits of goats (Capra hircus Linnaeus, 1758) from the resource population in age dynamics. Sel’skokhozyaistvennaya Biol. [Agric. Biol.] 2024, 59, 633–648. [Google Scholar] [CrossRef]
- Selionova, M.I.; Mamontova, T.V.; Aibazov, A.-M.M. Osobennosti reproduktivnoy funktsii karachayevskikh koz v zavisimosti ot raznykh geograficheskikh rayonov razvedeniya [Features of the reproductive function of Karachay goats depending on different geographical areas of breeding]. Izv. Timiryazevsk. S-Kh. Akad. [Izv. Timiryazev Agric. Acad.] 2021, 2, 114–122, (In Russian with English summary). [Google Scholar]
- Mamontova, T.V.; Gadzhiev, Z.K.; Aibazov, A.-M.M. Produktivnyye i vosproizvoditel’nyye osobennosti mestnykh karachayevskikh koz [Productive and reproductive features of local Karachay goats]. Sb. Nauch. Tr. Stavropol’skogo Nauch.-Issled. In-Ta Zhivotnovodstva I Kormoproizvodstva [Collect. Sci. Pap. Stavropol Res. Inst. Anim. Husb. Forage Prod.] 2011, 1, 15–17. Available online: https://www.elibrary.ru/item.asp?id=17294528 (accessed on 25 April 2025).
- Aybazov, M.M.; Selionova, M.I.; Seitov, M.S.; Bikteev, S.M. Avtokhtonnaya karachayevskaya poroda koz: Genotipy i nekotoryye ekster’yernyye i inter’yernyye pokazateli [Autochthonous Karachay breed of goats: Genotypes and some exterior and interior indicators]. Izv. Orenburg. Gos. Agrar. Un-Ta [Izv. Orenburg State Agrar. Univ.]. 2022, 5, 300–306, (In Russian with English summary). [Google Scholar]
- Deniskova, T.E.; Dotsev, A.V.; Selionova, M.I.; Aibazov, A.-M.M.; Zinovieva, N.A. Search for signatures of selection in the genomes of domestic goats (Capra hircus L.) raised in Russia using detection of ROH islands. Sel’skokhozyaistvennaya Biol. [Agric. Biol.] 2024, 59, 620–632. [Google Scholar] [CrossRef]
- Becker, R.A.; Wilks, A.R.; Brownrigg, R.; Minka, T.P.; Deckmyn, A. Maps: Draw Geographical Maps. R Package Version 3.3.0. 2018. Available online: https://CRAN.R-project.org/package=maps (accessed on 25 April 2025).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009; ISBN 978-0-387-98141-3. [Google Scholar] [CrossRef]
- Vasimuddin, M.; Misra, S.; Li, H.; Aluru, S. Efficient Architecture-aware Acceleration of BWAMEM for Multicore Systems. In Proceedings of the 2019 IEEE International Parallel and Distributed Processing Symposium (IPDPS), Rio de Janeiro, Brazil, 20–24 May 2019; IEEE Xplore: Piscataway, NJ, USA, 2019; pp. 314–324. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Biscarini, F.; Paolo Cozzi, P.; Gaspa, G.; Marras, G. detectRUNS: Detect Runs of Homozygosity and Runs of Heterozygosity in Diploid Genomes; R Package Version 0.9.6; The Comprehensive R Archive Network (CRAN); Institute for Statistics and Mathematics, Vienna University of Economics and Business: Vienna, Austria, 2019; Available online: https://cran.r-project.org/web/packages/detectRUNS/index.html (accessed on 25 April 2025).
- Romanov, M.N.; Sölkner, J.; Zinovieva, N.A.; Wimmers, K.; Weigend, S. Editorial: Traditional and up-to-date genomic insights into domestic animal diversity. Front. Genet. 2023, 13, 1117708. [Google Scholar] [CrossRef] [PubMed]
- Moiseyeva, I.G. Otechestvennyye porody kur [Native breeds of domestic fowl]. In Geneticheskie Nesursy Sel’skokhozyajstvennykh Zhivothykh: Redkie I Ischezayushchie Otechestvennye Porody [Farm Animal Genetic Resources: Rare and Endangered Native Breeds]; Moiseyeva, I.G., Zakharov, I.A., Mitichashvili, R.S., Eds.; Nauka: Moscow, Russia, 1992; pp. 11–112. Available online: https://agris.fao.org/search/en/providers/122621/records/647396783ed73003714cc15a (accessed on 25 April 2025). (In Russian)
- Dehghanzadeh, H.; Mirhosseini, S.Z.; Romanov, M.N.; Ghorbani, A. Evaluation of genetic variability and distances among five Iranian native chicken populations using RAPD markers. Pak. J. Biol. Sci. 2009, 12, 866–871. [Google Scholar] [CrossRef]
- Larkina, T.A.; Barkova, O.Y.; Peglivanyan, G.K.; Mitrofanova, O.V.; Dementieva, N.V.; Stanishevskaya, O.I.; Vakhrameev, A.B.; Makarova, A.V.; Shcherbakov, Y.S.; Pozovnikova, M.V.; et al. Evolutionary subdivision of domestic chickens: Implications for local breeds as assessed by phenotype and genotype in comparison to commercial and fancy breeds. Agriculture 2021, 11, 914. [Google Scholar] [CrossRef]
- Kulibaba, R.O.; Sakhatskyi, M.I.; Griffin, D.K.; Romanov, M.N. Molecular diversity of Ukrainian native chicken breeds: A review. Worlds Poult. Sci. J. 2024, 80, 1265–1292. [Google Scholar] [CrossRef]
- Bertolini, F.; Cardoso, T.F.; Marras, G.; Nicolazzi, E.L.; Rothschild, M.F.; Amills, M.; AdaptMap consortium. Genome-wide patterns of homozygosity provide clues about the population history and adaptation of goats. Genet. Sel. Evol. 2018, 50, 59. [Google Scholar] [CrossRef]
- Wang, R.; Wang, X.; Qi, Y.; Li, Y.; Na, Q.; Yuan, H.; Rong, Y.; Ao, X.; Guo, F.; Zhang, L.; et al. Genetic diversity analysis of Inner Mongolia cashmere goats (Erlangshan subtype) based on whole genome re-sequencing. BMC Genom. 2024, 25, 698. [Google Scholar] [CrossRef]
- An, Z.X.; Shi, L.G.; Hou, G.Y.; Zhou, H.L.; Xun, W.J. Genetic diversity and selection signatures in Hainan black goats revealed by whole-genome sequencing data. Animal 2024, 18, 101147. [Google Scholar] [CrossRef]
- Peng, W.; Zhang, Y.; Gao, L.; Wang, S.; Liu, M.; Sun, E.; Lu, K.; Zhang, Y.; Li, B.; Li, G.; et al. Investigation of selection signatures of dairy goats using whole-genome sequencing data. BMC Genom. 2025, 26, 234. [Google Scholar] [CrossRef]
- Curik, I.; Ferencakovic, M.; Sölkner, J. Inbreeding and runs of homozygosity: A possible solution to an old problem. Livest. Sci. 2014, 166, 26–34. [Google Scholar] [CrossRef]
- Bian, C.; Luo, Y.; Li, J.; Cheng, H.; He, F.; Duan, H.; Ahmed, Z.; Lei, C.; Yi, K. Inference of genetic diversity, population structure, and selection signatures in Xiangxi white buffalo of China through whole-genome resequencing. Genes 2024, 15, 1450. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zhong, J.; Li, L.; Zhong, T.; Wang, L.; Song, T.; Zhang, H. Comparative genome analyses reveal the unique genetic composition and selection signals underlying the phenotypic characteristics of three Chinese domestic goat breeds. Genet. Sel. Evol. 2019, 51, 70. [Google Scholar] [CrossRef]
- Wan, X.; Jing, J.N.; Wang, D.F.; Lv, F.H. Whole-genome selective scans detect genes associated with important phenotypic traits in goat (Capra hircus). Front. Genet. 2023, 14, 1173017. [Google Scholar] [CrossRef]
- Saif, R.; Mahmood, T.; Zia, S.; Henkel, J.; Ejaz, A. Genomic selection pressure discovery using site-frequency spectrum and reduced local variability statistics in Pakistani Dera-Din-Panah goat. Trop. Anim. Health Prod. 2023, 55, 331. [Google Scholar] [CrossRef]
- Snelling, W.M.; Allan, M.F.; Keele, J.W.; Kuehn, L.A.; Thallman, R.M.; Bennett, G.L.; Ferrell, C.L.; Jenkins, T.G.; Freetly, H.C.; Nielsen, M.K.; et al. Partial-genome evaluation of postweaning feed intake and efficiency of crossbred beef cattle. J. Anim. Sci. 2011, 89, 1731–1741. [Google Scholar] [CrossRef]
- Lindholm-Perry, A.K.; Kern, R.J.; Kuehn, L.A.; Snelling, W.M.; Miles, J.R.; Oliver, W.T.; Freetly, H.C. Differences in transcript abundance of genes on BTA15 located within a region associated with gain in beef steers. Gene 2015, 572, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Wang, X.; Li, M.; Li, Y.; Yang, Z.; Wang, X.; Pan, X.; Gong, M.; Zhang, Y.; Guo, Y.; et al. The origin of domestication genes in goats. Sci. Adv. 2020, 6, eaaz5216. [Google Scholar] [CrossRef]
- Kim, E.S.; Elbeltagy, A.R.; Aboul-Naga, A.M.; Rischkowsky, B.; Sayre, B.; Mwacharo, J.M.; Rothschild, M.F. Multiple genomic signatures of selection in goats and sheep indigenous to a hot arid environment. Heredity 2016, 116, 255–264. [Google Scholar] [CrossRef]
- Dadousis, C.; Cecchi, F.; Ablondi, M.; Fabbri, M.C.; Stella, A.; Bozzi, R. Keep Garfagnina alive. An integrated study on patterns of homozygosity, genomic inbreeding, admixture and breed traceability of the Italian Garfagnina goat breed. PLoS ONE 2021, 16, e0232436. [Google Scholar] [CrossRef]
- Pegolo, S.; Bisutti, V.; Mota, L.F.M.; Cecchinato, A.; Amalfitano, N.; Dettori, M.L.; Pazzola, M.; Vacca, G.M.; Bittante, G. Genome-wide landscape of genetic diversity, runs of homozygosity, and runs of heterozygosity in five Alpine and Mediterranean goat breeds. J. Anim. Sci. Biotechnol. 2025, 16, 33. [Google Scholar] [CrossRef] [PubMed]
- Graham-Paquin, A.L.; Saini, D.; Sirois, J.; Hossain, I.; Katz, M.S.; Zhuang, Q.K.; Kwon, S.Y.; Yamanaka, Y.; Bourque, G.; Bouchard, M.; et al. ZMYM2 is essential for methylation of germline genes and active transposons in embryonic development. Nucleic Acids Res. 2023, 51, 7314–7329. [Google Scholar] [CrossRef] [PubMed]
- Bo, D.; Jiang, X.; Liu, G.; Xu, F.; Hu, R.; Wassie, T.; Chong, Y.; Ahmed, S.; Liu, C.; Girmay, S. Multipathway synergy promotes testicular transition from growth to spermatogenesis in early-puberty goats. BMC Genom. 2020, 21, 372. [Google Scholar] [CrossRef]
- Serranito, B.; Cavalazzi, M.; Vidal, P.; Taurisson-Mouret, D.; Ciani, E.; Bal, M.; Rouvellac, E.; Servin, B.; Moreno-Romieux, C.; Tosser-Klopp, G.; et al. Local adaptations of Mediterranean sheep and goats through an integrative approach. Sci. Rep. 2021, 11, 21363. [Google Scholar] [CrossRef]
- Deniskova, T.E.; Dotsev, A.V.; Zinovieva, N.A. [Identification of candidate genes associated with economically significant traits based on the analysis of homozygosity islands in the genome of sheep breeds bred in Russia]. Dostizheniya Nauki I Tekhniki APK [Achiev. Sci. Technol. AIC] 2023, 37, 80–86, (In Russian with English summary). [Google Scholar]
- Pandya, A.; Arnos, K.S.; Xia, X.J.; Welch, K.O.; Blanton, S.H.; Friedman, T.B.; Garcia Sanchez, G.; Liu, X.Z.; Morell, R.; Nance, W.E. Frequency and distribution of GJB2 (connexin 26) and GJB6 (connexin 30) mutations in a large North American repository of deaf probands. Genet. Med. 2003, 5, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Onzima, R.B.; Upadhyay, M.R.; Doekes, H.P.; Brito, L.F.; Bosse, M.; Kanis, E.; Groenen, M.A.M.; Crooijmans, R.P.M.A. Genome-wide characterization of selection signatures and runs of homozygosity in Ugandan goat breeds. Front. Genet. 2018, 9, 318. [Google Scholar] [CrossRef]
- Li, G.; Tang, J.; Huang, J.; Jiang, Y.; Fan, Y.; Wang, X.; Ren, J. Genome-wide estimates of runs of homozygosity, heterozygosity, and genetic load in two Chinese indigenous goat breeds. Front. Genet. 2022, 13, 774196. [Google Scholar] [CrossRef]
- Abied, A.; Xu, L.; Sahlu, B.W.; Xing, F.; Ahbara, A.; Pu, Y.; Lin, J.; Berihulay, H.; Islam, R.; He, X.; et al. Genome-wide analysis revealed homozygosity and demographic history of five Chinese sheep breeds adapted to different environments. Genes 2020, 11, 1480. [Google Scholar] [CrossRef]
- Liu, J.; Shi, L.; Li, Y.; Chen, L.; Garrick, D.; Wang, L.; Zhao, F. Estimates of genomic inbreeding and identification of candidate regions that differ between Chinese indigenous sheep breeds. J. Anim. Sci. Biotechnol. 2021, 12, 95. [Google Scholar] [CrossRef]
- Quan, K.; Shi, H.; Wei, C.; Li, J.; Liu, K.; Wang, H.; Sun, W.; Han, H. Genetic diversity, reproductive performance, and genetic enhancement strategies in Huang-Huai goats. Front. Genet. 2025, 16, 1549051. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Chen, S.; Qiao, L.; Zhang, S.; Liu, Q.; Yang, K.; Pan, Y.; Liu, J.; Liu, W. Four markers useful for the distinction of intrauterine growth restriction in sheep. Animals 2023, 13, 3305. [Google Scholar] [CrossRef] [PubMed]
- Lakhssassi, K.; Sarto, M.P.; Lahoz, B.; Alabart, J.L.; Folch, J.; Serrano, M.; Calvo, J.H. Blood transcriptome of Rasa Aragonesa rams with different sexual behavior phenotype reveals CRYL1 and SORCS2 as genes associated with this trait. J. Anim. Sci. 2023, 101, skad098. [Google Scholar] [CrossRef]
- Fu, X.; Zhao, B.; Tian, K.; Wu, Y.; Suo, L.; Ba, G.; Ciren, D.; De, J.; Awang, C.; Gun, S.; et al. Integrated analysis of lncRNA and mRNA reveals novel insights into cashmere fineness in Tibetan cash-mere goats. PeerJ 2020, 8, e10217. [Google Scholar] [CrossRef]
- Bai, Z.X.; Xu, Y.N.; Gu, M.; Cai, W.; Zhang, Y.; Qin, Y.; Chen, R.; Sun, Y.; Wu, Y.; Wang, Z. Proteomic analysis of coarse and fine skin tissues of Liaoning cashmere goat. Funct. Integr. Genom. 2022, 22, 503–513. [Google Scholar] [CrossRef]
- Liu, Z.; Li, H.; Luo, Y.; Li, J.; Sun, A.; Ahmed, Z.; Zhang, B.; Lei, C.; Yi, K. Comprehensive whole-genome resequencing unveils genetic diversity and selective signatures of the Xiangdong black goat. Front. Genet. 2024, 15, 1326828. [Google Scholar] [CrossRef] [PubMed]
- San Agustin, J.T.; Pazour, G.J.; Witman, G.B. Intraflagellar transport is essential for mammalian spermiogenesis but is absent in mature sperm. Mol. Biol. Cell 2015, 26, 4358–4372. [Google Scholar] [CrossRef]
- Wei, X.; Wang, X.; Yang, C.; Gao, Y.; Zhang, Y.; Xiao, Y.; Ju, Z.; Jiang, Q.; Wang, J.; Liu, W.; et al. CFAP58 is involved in the sperm head shaping and flagellogenesis of cattle and mice. Development 2024, 151, dev202608. [Google Scholar] [CrossRef]
- Yurchenko, A.A.; Deniskova, T.E.; Yudin, N.S.; Dotsev, A.V.; Khamiruev, T.N.; Selionova, M.I.; Egorov, S.V.; Reyer, H.; Wimmers, K.; Brem, G.; et al. High-density genotyping reveals signatures of selection related to acclimation and economically important traits in 15 local sheep breeds from Russia. BMC Genom. 2019, 20 (Suppl. 3), 294. [Google Scholar] [CrossRef]
- Elder, L.A.; Hinnant, H.R.; Mandella, C.M.; Claus-Walker, R.A.; Parrish, L.M.; Slanzon, G.S.; McConnel, C.S. Differential gene expression in peripheral leukocytes of pre-weaned Holstein heifer calves with respiratory disease. PLoS ONE 2023, 18, e0285876. [Google Scholar] [CrossRef]
- Chen, Z.; Luo, J.; Zhang, C.; Ma, Y.; Sun, S.; Zhang, T.; Loo, J.J. Mechanism of prolactin inhibition of miR-135b via methylation in goat mammary epithelial cells. J. Cell. Physiol. 2018, 233, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.M.; Zhang, T.T.; An, S.Y.; El-Samahy, M.A.; Yang, H.; Wan, Y.J.; Meng, F.X.; Xiao, S.H.; Wang, F.; Lei, Z.H. Expression of Hippo signaling pathway components in Hu sheep male reproductive tract and spermatozoa. Theriogenology 2019, 126, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Plewes, M.R.; Hou, X.; Zhang, P.; Liang, A.; Hua, G.; Wood, J.R.; Cupp, A.S.; Lv, X.; Wang, C.; Davis, J.S. Yes-associated protein 1 is required for proliferation and function of bovine granulosa cells in vitro. Biol. Reprod. 2019, 101, 1001–1017. [Google Scholar] [CrossRef] [PubMed]
- E, G.X.; Zhu, Y.B.; Basang, W.D.; Na, R.S.; Han, Y.G.; Zeng, Y. Comparative and selection sweep analysis of CNV was associated to litter size in Dazu black goats. Anim. Biotechnol. 2021, 32, 792–797. [Google Scholar] [CrossRef]
- Mahar, K.; Gurao, A.; Kumar, A.; Pratap Singh, L.; Chitkara, M.; Gowane, G.R.; Ahlawat, S.; Niranjan, S.K.; Pundir, R.K.; Kataria, R.S.; et al. Genomic inbreeding analysis reveals resilience and genetic diversity in Indian yak populations. Gene 2024, 928, 148787. [Google Scholar] [CrossRef]
- Workman, A.M.; Heaton, M.P.; Webster, D.A.; Harhay, G.P.; Kalbfleisch, T.S.; Smith, T.P.L.; Falkenberg, S.M.; Carlson, D.F.; Sonstegard, T.S. Evaluating large spontaneous deletions in a bovine cell line selected for bovine viral diarrhea virus resistance. Viruses 2021, 13, 2147. [Google Scholar] [CrossRef]
- Zhao, Q.; Huang, C.; Chen, Q.; Su, Y.; Zhang, Y.; Wang, R.; Su, R.; Xu, H.; Liu, S.; Ma, Y.; et al. Genomic inbreeding and runs of homozygosity analysis of cashmere goat. Animals 2024, 14, 1246. [Google Scholar] [CrossRef]
- Salilew-Wondim, D.; Hölker, M.; Rings, F.; Phatsara, C.; Mohammadi-Sangcheshmeh, A.; Tholen, E.; Schellander, K.; Tesfaye, D. Depletion of BIRC6 leads to retarded bovine early embryonic development and blastocyst formation in vitro. Reprod. Fertil. Dev. 2010, 22, 564–579. [Google Scholar] [CrossRef]
- Guan, D.; Luo, N.; Tan, X.; Zhao, Z.; Huang, Y.; Na, R.; Zhang, J.; Zhao, Y. Scanning of selection signature provides a glimpse into important economic traits in goats (Capra hircus). Sci. Rep. 2016, 6, 36372. [Google Scholar] [CrossRef]
- El-Halawany, N.; Zhou, X.; Al-Tohamy, A.F.; El-Sayd, Y.A.; Shawky, A.E.; Michal, J.J.; Jiang, Z. Genome-wide screening of candidate genes for improving fertility in Egyptian native Rahmani sheep. Anim. Genet. 2016, 47, 513. [Google Scholar] [CrossRef]
- Kour, A.; Deb, S.M.; Nayee, N.; Niranjan, S.K.; Raina, V.S.; Mukherjee, A.; Patil, C.S. Novel insights into genome-wide associations in Bos indicus reveal genetic linkages between fertility and growth. Anim. Biotechnol. 2021, 34, 39–55. [Google Scholar] [CrossRef] [PubMed]
- Serão, N.V.; González-Peña, D.; Beever, J.E.; Bollero, G.A.; Southey, B.R.; Faulkner, D.B.; Rodriguez-Zas, S.L. Bivariate genome-wide association analysis of the growth and intake components of feed efficiency. PLoS ONE 2013, 8, e78530. [Google Scholar] [CrossRef]
- Sarkar, J.; Balamurugan, V.; Sen, A.; Saravanan, P.; Sahay, B.; Rajak, K.K.; Rasool, T.J.; Bhanuprakash, V.; Singh, R.K. Sequence analysis of morbillivirus CD150 receptor-signaling lymphocyte activation molecule (SLAM) of different animal species. Virus Genes 2009, 39, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Wang, T.; Li, Z.; Wan, Y.; Yang, B.; Zeng, W.; Zhang, Y.; Wang, J. MicroRNA-218 Regulates signaling lymphocyte activation molecular (SLAM) mediated peste des petits ruminants virus infectivity in goat peripheral blood mononuclear cells. Front. Immunol. 2019, 10, 2201. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhao, G.; Lin, X.; Zhang, J.; Hou, G.; Zhang, L.; Liu, D.; Li, Y.; Li, J.; Xu, L. Genomic inbreeding and runs of homozygosity analysis of indigenous cattle populations in southern China. PLoS ONE 2022, 17, e0271718. [Google Scholar] [CrossRef]
- Zhao, J.; Shi, C.; Kamalibieke, J.; Gong, P.; Mu, Y.; Zhu, L.; Lv, X.; Wang, W.; Luo, J. Whole genome and transcriptome analyses in dairy goats identify genetic markers associated with high milk yield. Int. J. Biol. Macromol. 2025, 292, 139192. [Google Scholar] [CrossRef]
- Xu, Q.; Xi, Y.; Ma, S.; Wang, J.; Li, J.; Han, C.; Li, L.; Wang, J.; Liu, H. Transcriptome profiling of morphogenetic differences between contour and flight feathers in duck. Br. Poult. Sci. 2022, 63, 597–604. [Google Scholar] [CrossRef]
- Zhang, W.; Li, P.; Wang, S.; Cheng, G.; Wang, L.; Mi, X.; Su, X.; Wang, Y.; Zan, L. TP53INP2 promotes bovine adipocytes differentiation through autophagy activation. Animals 2019, 9, 1060. [Google Scholar] [CrossRef]
- Sabaté-Pérez, A.; Romero, M.; Sànchez-Fernàndez-de-Landa, P.; Carobbio, S.; Mouratidis, M.; Sala, D.; Engel, P.; Martínez-Cristóbal, P.; Villena, J.A.; Virtue, S.; et al. Autophagy-mediated NCOR1 degradation is required for brown fat maturation and thermogenesis. Autophagy 2023, 19, 904–925. [Google Scholar] [CrossRef]
- Sala, D.; Ivanova, S.; Plana, N.; Ribas, V.; Duran, J.; Bach, D.; Turkseven, S.; Laville, M.; Vidal, H.; Karczewska-Kupczewska, M.; et al. Autophagy-regulating TP53INP2 mediates muscle wasting and is repressed in diabetes. J. Clin. Investig. 2014, 124, 1914–1927. [Google Scholar] [CrossRef]
- Verbrugge, S.A.J.; Schönfelder, M.; Becker, L.; Yaghoob Nezhad, F.; Hrabě de Angelis, M.; Wackerhage, H. Genes whose gain or loss-of-function increases skeletal muscle mass in mice: A systematic literature review. Front. Physiol. 2018, 9, 553. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, M.N.; Raudsepp, T.; Alshanbari, F.; Gutiérrez, G.; Ponce de León, F.A. Chromosomal localization of candidate genes for fiber growth and color in alpaca (Vicugna pacos). Front. Genet. 2019, 10, 583. [Google Scholar] [CrossRef] [PubMed]
- Olsen, H.G.; Knutsen, T.M.; Kohler, A.; Svendsen, M.; Gidskehaug, L.; Grove, H.; Nome, T.; Sodeland, M.; Sundsaasen, K.K.; Kent, M.P.; et al. Genome-wide association mapping for milk fat composition and fine mapping of a QTL for de novo synthesis of milk fatty acids on bovine chromosome 13. Genet. Sel. Evol. 2017, 49, 20. [Google Scholar] [CrossRef] [PubMed]
- Knutsen, T.M.; Olsen, H.G.; Tafintseva, V.; Svendsen, M.; Kohler, A.; Kent, M.P.; Lien, S. Unravelling genetic variation underlying de novo-synthesis of bovine milk fatty acids. Sci. Rep. 2018, 8, 2179. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).