Comparative Genomic Analysis Across Multiple Species to Identify Candidate Genes Associated with Important Traits in Chickens
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome Data Acquisition
2.2. Gene Family Clustering Analysis
2.3. Phylogenetic Tree Construction and Differentiation Time Calculation
2.4. Analysis of Gene Family Expansion and Contraction
2.5. Positive Choice Analysis
2.6. Genome-Wide Replication Event Analysis
2.7. Genomic Covariance Analysis
2.8. LTR-RT Insertion Time Analysis
2.9. Annotation and Pathway Analysis
3. Results
3.1. Identification of Candidate Genes Associated with Key Chicken Traits via Gene Family Clustering Analysis
3.1.1. Distribution of Gene Family Copy Number by Species
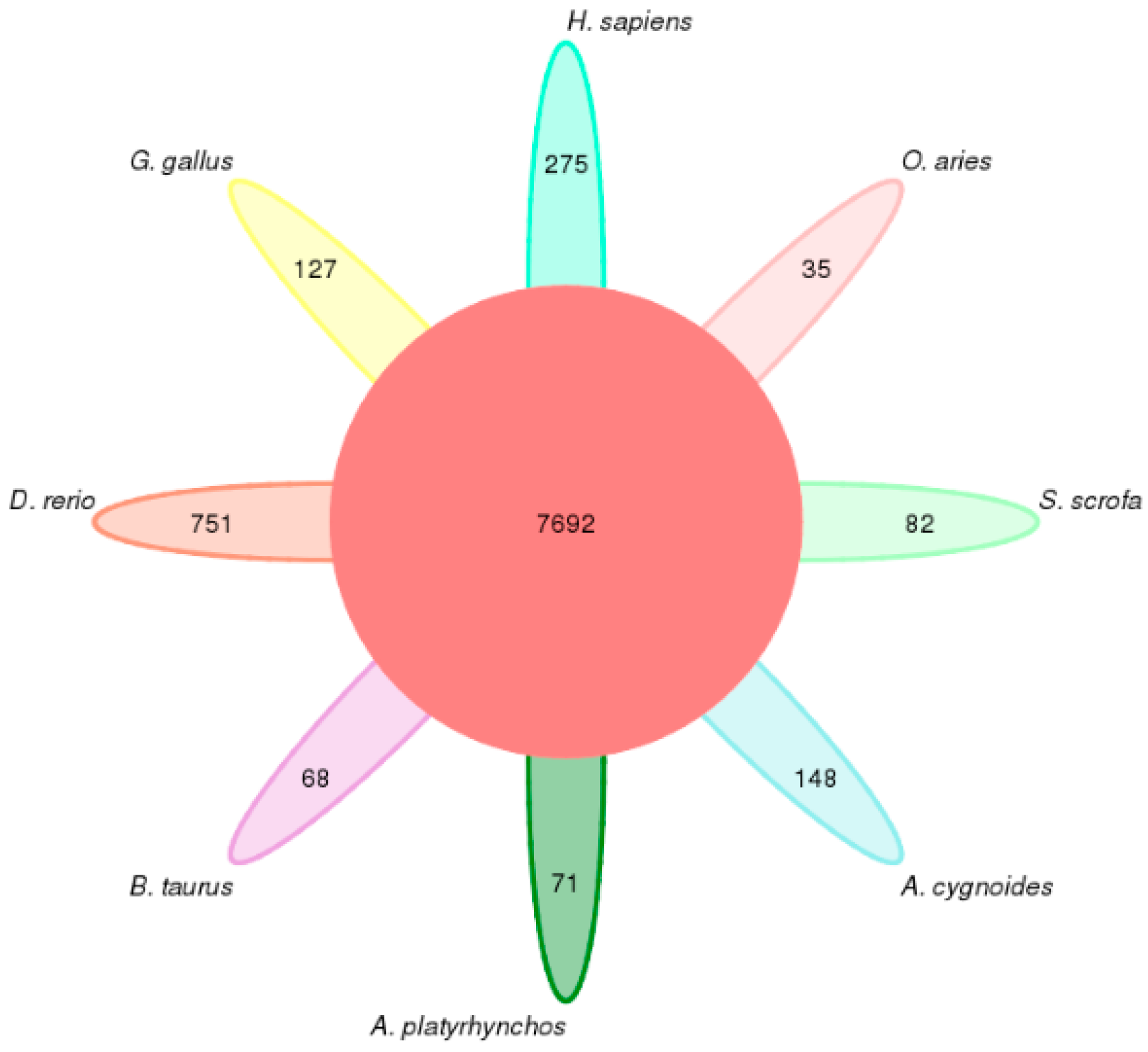
3.1.2. Identification and Statistics of Species-Specific Gene Families
3.1.3. Statistics on the Number of Gene Families Specific to Each Species
3.2. Screening Candidate Genes Related to Important Chicken Traits Based on Gene Family Expansion and Contraction Analysis
3.3. Screening Candidate Genes Related to Important Chicken Traits Based on Positive Selection Analysis
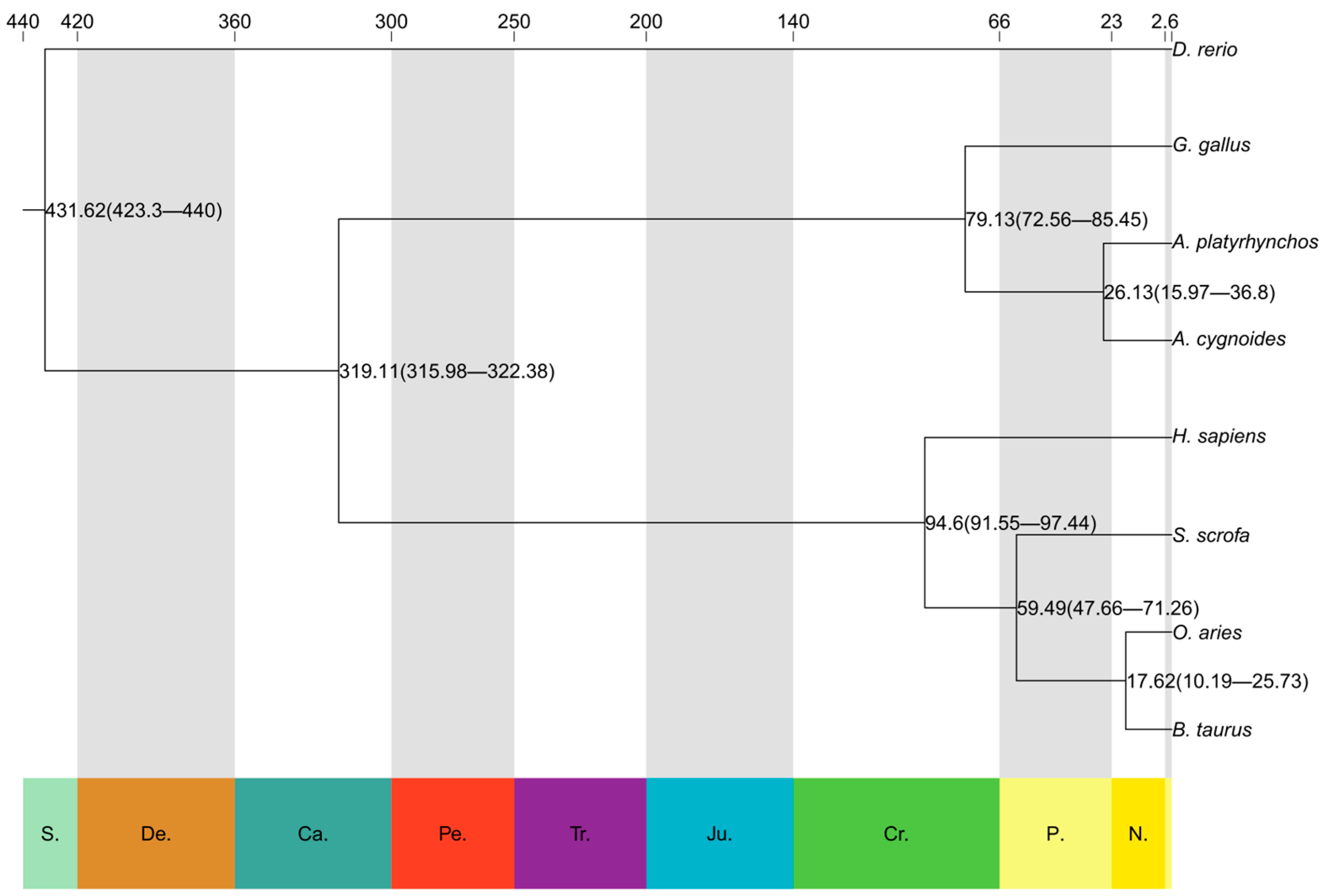
3.4. Phylogenetic Tree Construction and Differentiation Time Calculation
3.5. Genome-Wide Duplication Events Between Chickens and Other Species Analyzed Based on the Ks Approach
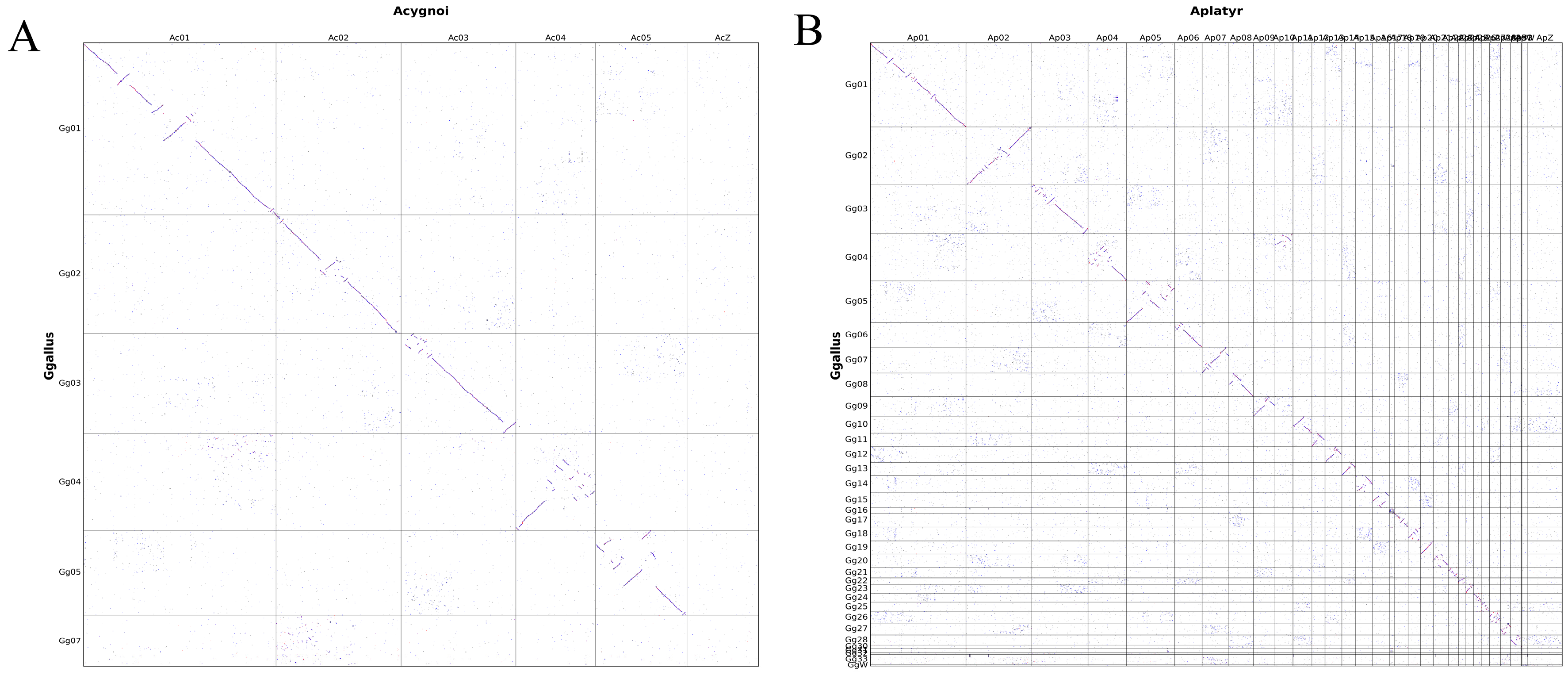
3.6. Comparison of Genomic Structural Differences Between Chickens, Ducks, and Geese Based on Genomic Collinearity Analysis
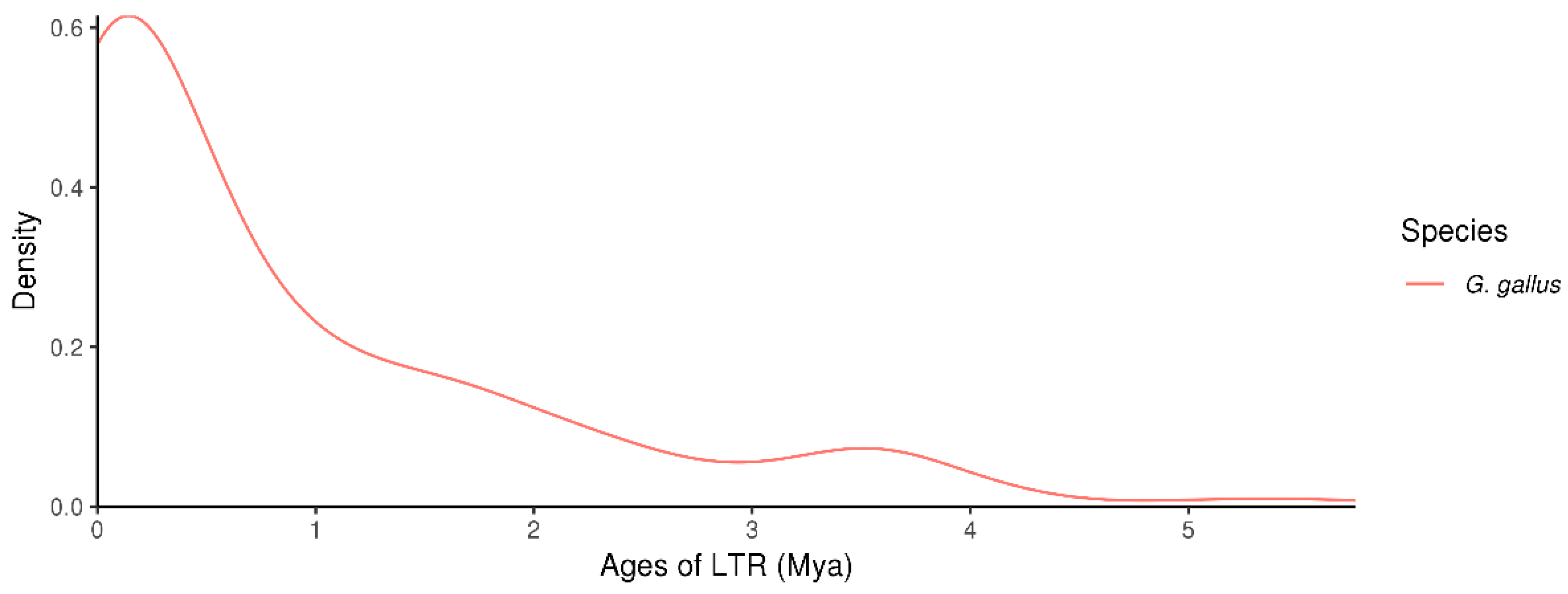
3.7. Distribution of LTR-RT Insertion Ages in the Chicken Genome
3.8. Association Analysis Between Candidate Genes and Important Chicken Traits
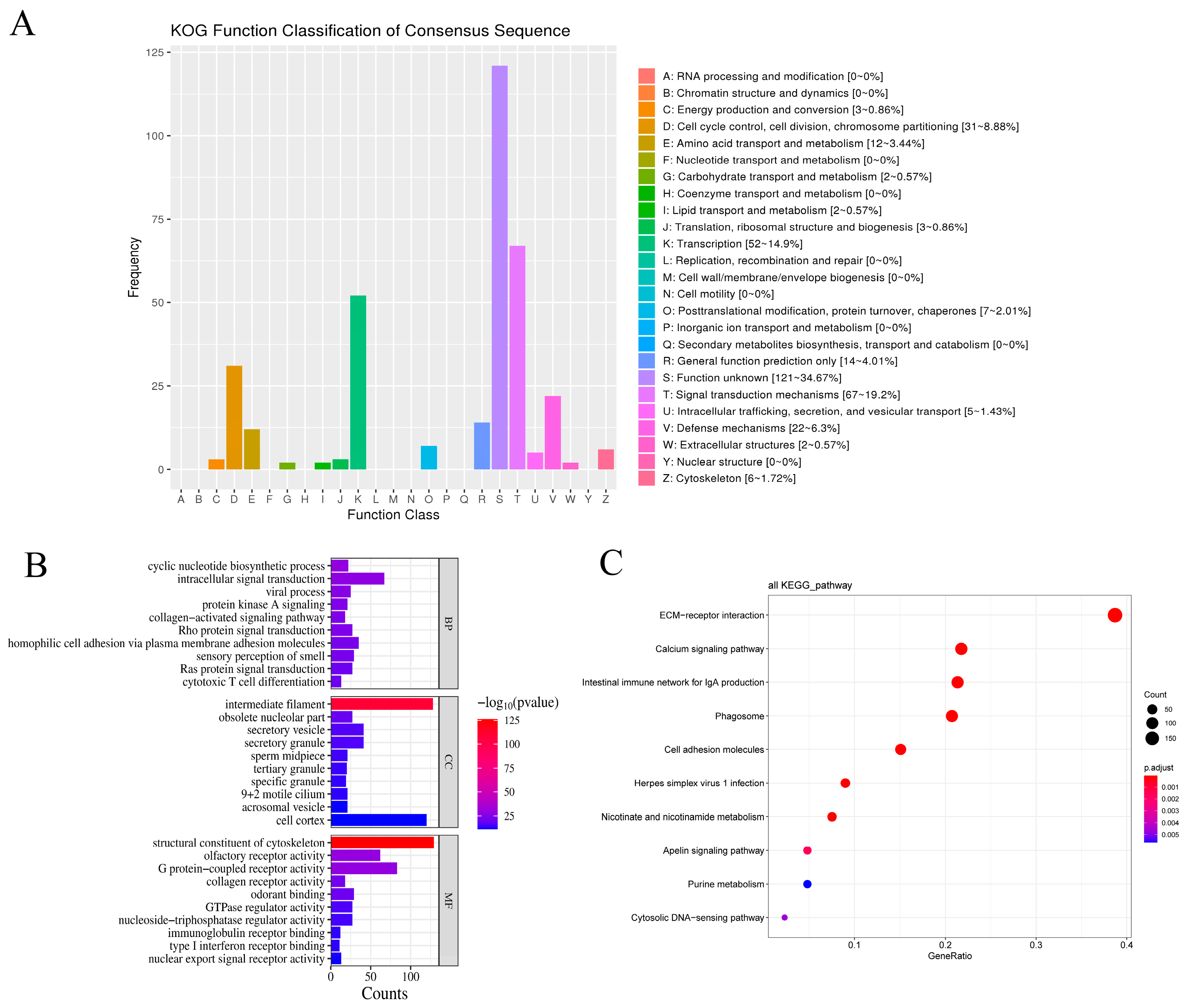
3.8.1. Functional Annotation of Candidate Genes
3.8.2. Combining Functional Annotation and a Literature Search to Explore Candidate Gene–Trait Associations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Davis, T.C.; White, R.R. Breeding animals to feed people: The many roles of animal reproduction in ensuring global food security. Theriogenology 2020, 150, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Wang, Y.; Zhu, Y.; Liu, X.; Liu, L.; Wang, J.; Tan, X.; Wang, Y.; Xing, S.; Luo, N.; et al. Genomic insights into the contribution of de novo lipogenesis to intramuscular fat deposition in chicken. J. Adv. Res. 2024, 65, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Hedman, H.D.; Zhang, L.; Butt, B.; Papias, P.; Trostle, J.A.; Eisenberg, J.N.S. “Chicken dumping”: Motivations and perceptions in shifting poultry production practices. One Health 2021, 13, 100296. [Google Scholar] [CrossRef] [PubMed]
- Chomchuen, K.; Tuntiyasawasdikul, V.; Chankitisakul, V.; Boonkum, W. Genetic Evaluation of Body Weights and Egg Production Traits Using a Multi-Trait Animal Model and Selection Index in Thai Native Synthetic Chickens (Kaimook e-san2). Animals 2022, 12, 335. [Google Scholar] [CrossRef]
- Miyumo, S.A.; Wasike, C.B.; Ilatsia, E.D.; Bennewitz, J.; Chagunda, M.G.G. Genetic and phenotypic correlations among feed efficiency, immune and production traits in indigenous chicken of Kenya. Front. Genet. 2022, 13, 1070304. [Google Scholar] [CrossRef]
- Li, H.W.; Wang, R.J.; Wang, Z.Y.; Li, X.W.; Wang, Z.Y.; Zhang, Y.; Su, R.; Liu, Z.; Li, J. The research progress of genomic selection in livestock. Yi Chuan 2017, 39, 377–387. [Google Scholar] [CrossRef]
- Liu, T.; Luo, C.; Ma, J.; Wang, Y.; Shu, D.; Su, G.; Qu, H. High-Throughput Sequencing With the Preselection of Markers Is a Good Alternative to SNP Chips for Genomic Prediction in Broilers. Front. Genet. 2020, 11, 108. [Google Scholar] [CrossRef]
- Miller, W.; Makova, K.D.; Nekrutenko, A.; Hardison, R.C. Comparative genomics. Annu. Rev. Genom. Hum. Genet. 2004, 5, 15–56. [Google Scholar] [CrossRef]
- Dillman, A.R.; Macchietto, M.; Porter, C.F.; Rogers, A.; Williams, B.; Antoshechkin, I.; Lee, M.M.; Goodwin, Z.; Lu, X.; Lewis, E.E.; et al. Comparative genomics of Steinernema reveals deeply conserved gene regulatory networks. Genome Biol. 2015, 16, 200. [Google Scholar] [CrossRef]
- Ellegren, H. Comparative genomics and the study of evolution by natural selection. Mol. Ecol. 2008, 17, 4586–4596. [Google Scholar] [CrossRef]
- Abdelmanova, A.S.; Dotsev, A.V.; Romanov, M.N.; Stanishevskaya, O.I.; Gladyr, E.A.; Rodionov, A.N.; Vetokh, A.N.; Volkova, N.A.; Fedorova, E.S.; Gusev, I.V.; et al. Unveiling Comparative Genomic Trajectories of Selection and Key Candidate Genes in Egg-Type Russian White and Meat-Type White Cornish Chickens. Biology 2021, 10, 876. [Google Scholar] [CrossRef] [PubMed]
- Assiri, A.; Vallejo-Trujillo, A.; Al-Abri, M.; Bahbahani, H.; Almathen, F.; Ahbara, A.; Al Marzooqi, W.; Tijjani, A.; Lawal, R.; Hanotte, O. Comparative genomics reveals common diversity and adaptation to harsh environments in the Arabian Peninsula indigenous chickens. Anim. Genet. 2025, 56, e70014. [Google Scholar] [CrossRef]
- He, M.; He, Y.; Zhang, K.; Lu, X.; Zhang, X.; Gao, B.; Fan, Y.; Zhao, H.; Jha, R.; Huda, M.N.; et al. Comparison of buckwheat genomes reveals the genetic basis of metabolomic divergence and ecotype differentiation. New Phytol. 2022, 235, 1927–1943. [Google Scholar] [CrossRef]
- Liu, C.; Gao, J.; Cui, X.; Li, Z.; Chen, L.; Yuan, Y.; Zhang, Y.; Mei, L.; Zhao, L.; Cai, D.; et al. A towering genome: Experimentally validated adaptations to high blood pressure and extreme stature in the giraffe. Sci. Adv. 2021, 7, eabe9459. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Ren, Y.; Wu, J.; Cui, T.; Dong, R.; Huang, C.; Feng, Z.; Zhang, T.; Yang, P.; Yuan, J.; et al. Author Correction: Evolution and expression patterns of the neo-sex chromosomes of the crested ibis. Nat. Commun. 2024, 15, 9278. [Google Scholar] [CrossRef]
- Lin, H.; Yu, M.; Wang, X.; Zhang, X.H. Comparative genomic analysis reveals the evolution and environmental adaptation strategies of vibrios. BMC Genom. 2018, 19, 135. [Google Scholar] [CrossRef]
- Bradbury, L.M.; Niehaus, T.D.; Hanson, A.D. Comparative genomics approaches to understanding and manipulating plant metabolism. Curr. Opin. Biotechnol. 2013, 24, 278–284. [Google Scholar] [CrossRef]
- Martinez-Morales, J.R. Toward understanding the evolution of vertebrate gene regulatory networks: Comparative genomics and epigenomic approaches. Brief. Funct. Genom. 2016, 15, 315–321. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Qu, K.; Liu, A.; Yin, M.; Mu, W.; Wu, S.; Hu, H.; Chen, J.; Su, X.; Dou, Q.; Ren, G. A genome assembly for Orinus kokonorica provides insights into the origin, adaptive evolution and further diversification of two closely related grass genera. Commun. Biol. 2023, 6, 1223. [Google Scholar] [CrossRef]
- Feng, X.; Wang, Y.; Zhang, N.; Gao, S.; Wu, J.; Liu, R.; Huang, Y.; Zhang, J.; Qi, Y. Comparative phylogenetic analysis of CBL reveals the gene family evolution and functional divergence in Saccharum spontaneum. BMC Plant Biol. 2021, 21, 395. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Han, M.V.; Thomas, G.W.; Lugo-Martinez, J.; Hahn, M.W. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol. Biol. Evol. 2013, 30, 1987–1997. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Zwaenepoel, A.; Van De Peer, Y. wgd-simple command line tools for the analysis of ancient whole-genome duplications. Bioinformatics 2019, 35, 2153–2155. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Tang, H.; Krishnakumar, V.; Zeng, X.; Xu, Z.; Taranto, A.; Lomas, J.S.; Zhang, Y.; Huang, Y.; Wang, Y.; Yim, W.C.; et al. JCVI: A versatile toolkit for comparative genomics analysis. iMeta 2024, 3, e211. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Ellinghaus, D.; Kurtz, S.; Willhoeft, U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinform. 2008, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wang, H. LTR_FINDER: An efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007, 35, W265–W268. [Google Scholar] [CrossRef]
- Ou, S.; Jiang, N. LTR_retriever: A Highly Accurate and Sensitive Program for Identification of Long Terminal Repeat Retrotransposons. Plant Physiol. 2018, 176, 1410–1422. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. TIG 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Jin, C.F.; Chen, Y.J.; Yang, Z.Q.; Shi, K.; Chen, C.K. A genome-wide association study of growth trait-related single nucleotide polymorphisms in Chinese Yancheng chickens. Genet. Mol. Res. GMR 2015, 14, 15783–15792. [Google Scholar] [CrossRef] [PubMed]
- Reyer, H.; Hawken, R.; Murani, E.; Ponsuksili, S.; Wimmers, K. The genetics of feed conversion efficiency traits in a commercial broiler line. Sci. Rep. 2015, 5, 16387. [Google Scholar] [CrossRef]
- Scott, J.D. Cyclic nucleotide-dependent protein kinases. Pharmacol. Ther. 1991, 50, 123–145. [Google Scholar] [CrossRef]
- Cao, Y. Identification of Key Genes for Growth Traits in Guoshi Chicken and Development of Molecular Markers. Master Thesis, Henan Agricultural University, Zhengzhou, China, 2024. [Google Scholar] [CrossRef]
- Chen, T. Etermination of Growth Traits and SNP Analysis of Related Genes in Guangxi Ma Chicken. Master’s Thesis, Guangxi University, Nanning, China, 2023. [Google Scholar] [CrossRef]
- Yang, G.; Deng, Y.; Yao, Y.; Xiao, B.; Qu, X.; Guo, S.; He, C. Correlation analysis between growth traits and productive performance of two generations of white-feather Xuefeng black-bone chicken. J. Econ. Anim. 2021, 25, 14–19. [Google Scholar] [CrossRef]
- Tan, Y.; Cao, H.; Dong, X.; Mao, H.; Lu, L.; Jiang, J.; Ma, Y.; Yi, Z. Genome-wide association studies on growth and carcass traits in Ninghai yellow chickens (Gallus gallus domesticus) and Guangxi yellow chickens based on 600 K SNP chips technology. Agric. Biotech. 2019, 27, 1434–1444. [Google Scholar]
- Baéza, E.; Arnould, C.; Jlali, M.; Chartrin, P.; Gigaud, V.; Mercerand, F.; Durand, C.; Méteau, K.; Le Bihan-Duval, E.; Berri, C. Influence of increasing slaughter age of chickens on meat quality, welfare, and technical and economic results. J. Anim. Sci. 2012, 90, 2003–2013. [Google Scholar] [CrossRef]
- Zhang, Y.; Gu, T.; Xu, S.; Wang, J.; Zhu, X. Anti-Liver Fibrosis Role of miRNA-96-5p via Targeting FN1 and Inhibiting ECM-Receptor Interaction Pathway. Appl. Biochem. Biotechnol. 2023, 195, 6840–6855. [Google Scholar] [CrossRef] [PubMed]
- Sever, S. Role of actin cytoskeleton in podocytes. Pediatr. Nephrol. 2021, 36, 2607–2614. [Google Scholar] [CrossRef]
- Ye, M.H.; Chen, J.L.; Zhao, G.P.; Zheng, M.Q.; Wen, J. Associations of A-FABP and H-FABP markers with the content of intramuscular fat in Beijing-You chicken. Anim. Biotechnol. 2010, 21, 14–24. [Google Scholar] [CrossRef]
- Wang, J.L.; Zhu, Q.; Liu, Y.P.; Du, H.R. Associations between SNP of chicken PRKAB2 gene and slaughter and meat quality traits. Yi Chuan 2008, 30, 1033–1038. [Google Scholar] [CrossRef]
- Jiang, X.; Wu, T.; Yang, F.; Wei, Y.; Chen, G.; Cai, Y.; Jiao, T.; Gu, L.; Zhao, S. Effect of quinoa on expression of meat quality-related genes in Luhua chicken. China Feed 2024, 21, 19–25. [Google Scholar] [CrossRef]
- You, X.; Liu, Y.; Jiang, X.; Du, H.; Liu, Z.; Zhu, Q. Relationships between single nucleotide polymorphisms of the H-FABP gene and slaughter and meat quality traits in chicken. Biochem. Genet. 2009, 47, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, T.; Kamon, J.; Waki, H.; Imai, Y.; Shimozawa, N.; Hioki, K.; Uchida, S.; Ito, Y.; Takakuwa, K.; Matsui, J.; et al. Globular adiponectin protected ob/ob mice from diabetes and ApoE-deficient mice from atherosclerosis. J. Biol. Chem. 2003, 278, 2461–2468. [Google Scholar] [CrossRef] [PubMed]
- Makanjuola, B.O.; Olori, V.E.; Mrode, R.A. Modeling genetic components of hatch of fertile in broiler breeders. Poult. Sci. 2021, 100, 101062. [Google Scholar] [CrossRef] [PubMed]
- Weis, W.I.; Kobilka, B.K. The molecular basis of G protein-coupled receptor activation. Annu. Rev. Biochem. 2018, 87, 897–919. [Google Scholar] [CrossRef]
- Millar, R.P.; Newton, C.L. The year in G protein-coupled receptor research. Mol. Endocrinol. 2010, 24, 261–274. [Google Scholar] [CrossRef]
- Wu, X.; Wang, J.; Yan, M.; Li, H.; Chen, K.; Tang, Q.; Zhu, W.; Yu, Y. Genetic effects of GNRHR and IGF-1 genes on the reproductive traits in wenchang chicken. Acta Vet. Zootech. Sin. 2007, 01, 31–35. [Google Scholar]
- Calvello, R.; Cianciulli, A.; Mitolo, V.; Porro, A.; Panaro, M.A. Conservation of Intronic Sequences in Vertebrate Mitochondrial Solute Carrier Genes (Zebrafish, Chicken, Mouse and Human). Non-Coding RNA 2019, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Nie, C.; Zhang, J.; Li, X.; Zhu, T.; Guan, Z.; Chen, Y.; Wang, L.; Lv, X.Z.; Yang, W.; et al. Identification of candidate genomic regions for chicken egg number traits based on genome-wide association study. BMC Genom. 2021, 22, 610. [Google Scholar] [CrossRef]
- Wang, D.; Tan, L.; Zhi, Y.; Bu, L.; Wang, Y.; Wang, Z.; Guo, Y.; Tian, W.; Xu, C.; Li, D.; et al. Genome-wide variation study and inter-tissue communication analysis unveil regulatory mechanisms of egg-laying performance in chickens. Nat. Commun. 2024, 15, 7069. [Google Scholar] [CrossRef]
- Cao, H.; Tan, Y.; Dong, X.; Mao, H.; Ma, Y.; Lu, L.; Jiang, J.; Yi, Z. Genome-wide association studies on reproductive traits in Ninghai and Guangxi yellow chickens using 600 K SNP array. Chin. J. Anim. Sci. 2019, 55, 68–73. [Google Scholar] [CrossRef]
- Idoko-Akoh, A.; Goldhill, D.H.; Sheppard, C.M.; Bialy, D.; Quantrill, J.L.; Sukhova, K.; Brown, J.C.; Richardson, S.; Campbell, C.; Taylor, L.; et al. Creating resistance to avian influenza infection through genome editing of the ANP32 gene family. Nat. Commun. 2023, 14, 6136. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.E.; Walker, M. Genetics of Insulin Resistance and the Metabolic Syndrome. Curr. Cardiol. Rep. 2016, 18, 75. [Google Scholar] [CrossRef]
- Rivas, T.; Goodrich, J.A.; Kugel, J.F. The Herpes Simplex Virus 1 Protein ICP4 Acts as both an Activator and a Repressor of Host Genome Transcription during Infection. Mol. Cell. Biol. 2021, 41, e0017121. [Google Scholar] [CrossRef]
- Reina-Campos, M.; Scharping, N.E.; Goldrath, A.W. CD8(+) T cell metabolism in infection and cancer. Nat. Rev. Immunol. 2021, 21, 718–738. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, B.; Wen, J.; Li, Q.; Zhao, G. Genome-Wide Association Study and Pathway Analysis for Heterophil/Lymphocyte (H/L) Ratio in Chicken. Genes 2020, 11, 1005. [Google Scholar] [CrossRef]
- Walugembe, M.; Amuzu-Aweh, E.N.; Botchway, P.K.; Naazie, A.; Aning, G.; Wang, Y.; Saelao, P.; Kelly, T.; Gallardo, R.A.; Zhou, H.; et al. Genetic Basis of Response of Ghanaian Local Chickens to Infection With a Lentogenic Newcastle Disease Virus. Front. Genet. 2020, 11, 739. [Google Scholar] [CrossRef] [PubMed]
- Habimana, R.; Ngeno, K.; Okeno, T.O.; Hirwa, C.A.; Keambou Tiambo, C.; Yao, N.K. Genome-Wide Association Study of Growth Performance and Immune Response to Newcastle Disease Virus of Indigenous Chicken in Rwanda. Front. Genet. 2021, 12, 723980. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Xu, C.; Wang, G.; Liang, Y.; Wan, T.; Zhang, Y.; Xu, X.; Yu, L.; Che, Z.; Han, Q.; et al. Novel TBX22 mutations in Chinese nonsyndromic cleft lip/palate families. J. Genet. 2018, 97, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Chai, D.; Sobhani, N.; Sun, N.; Neeli, P.; Zheng, J.; Tian, H. C1QBP regulates mitochondrial plasticity to impact tumor progression and antitumor immune response. Front. Physiol. 2022, 13, 1012112. [Google Scholar] [CrossRef]
- Van Marle-Köster, E.; Visser, C. Unintended consequences of selection for increased production on the health and welfare of livestock. Arch. Anim. Breed. 2021, 64, 177–185. [Google Scholar] [CrossRef]
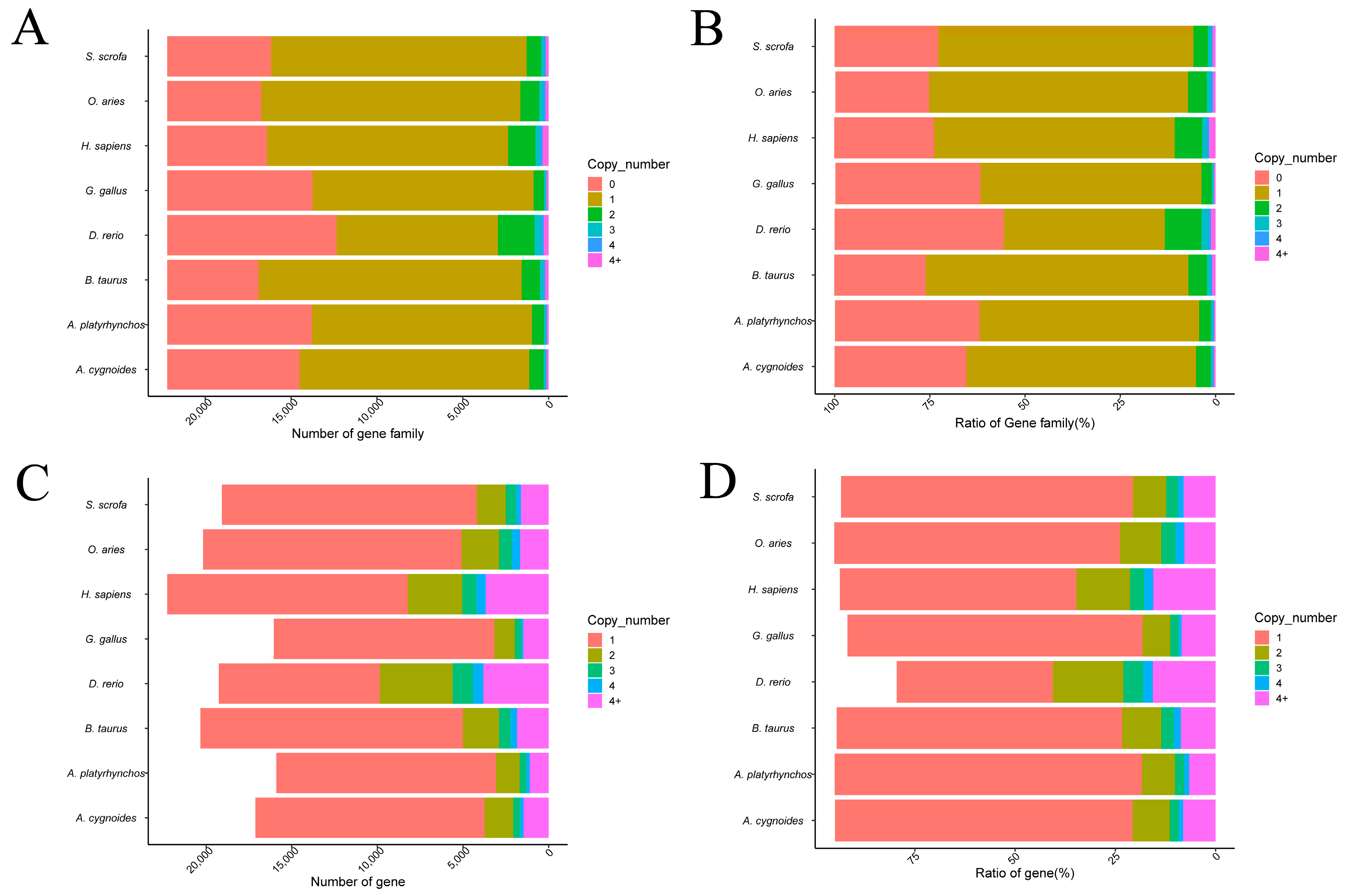


| Item | A. cygnoides | A. platyrhynchos | B. taurus | D. rerio | G. gallus | H. sapiens | O. aries | S. scrofa |
|---|---|---|---|---|---|---|---|---|
| Number of genes | 18,047 | 16,739 | 21,535 | 24,154 | 17,489 | 23,772 | 21,208 | 20,447 |
| Number of genes in orthogroups | 17,125 | 15,899 | 20,334 | 19,263 | 16,050 | 22,271 | 20,182 | 19,079 |
| Number of unassigned genes | 922 | 840 | 1201 | 4891 | 1439 | 1501 | 1026 | 1368 |
| Percentage of genes in orthogroups | 94.90 | 95.00 | 94.40 | 79.80 | 91.80 | 93.70 | 95.20 | 93.30 |
| Percentage of Unassigned genes | 5.10 | 5.00 | 5.60 | 20.20 | 8.20 | 6.30 | 4.80 | 6.70 |
| Number of orthogroups containing species | 14,520 | 13,796 | 16,888 | 12,352 | 13,742 | 16,397 | 16,747 | 16,169 |
| Percentage of othogroups containing species | 65.40 | 62.10 | 76.00 | 55.60 | 61.90 | 73.80 | 75.40 | 72.80 |
| Number of species- specific orthogroups | 148 | 71 | 68 | 751 | 127 | 275 | 35 | 82 |
| Number of genes in species-specific orthogroups | 741 | 258 | 314 | 4017 | 824 | 1563 | 179 | 432 |
| Percentage of genes in species-specific orthogroups | 4.10 | 1.50 | 1.50 | 16.60 | 4.70 | 6.60 | 0.8 | 2.10 |
| Gene | Branch p-Value | Locus Information | Gene Family |
|---|---|---|---|
| GTSF1 | 0.00065 | 8, L, 0.995 ** 46, E, 0.965 * 83, Q, 0.992 ** | OG0002511 |
| GRAP2 | 0.0034 | 20, S, 0.983 * 21, G, 0.990 ** | OG0004707 |
| RCN1L | 0.036 | 16, L, 0.952 * | OG0004041 |
| AGBL4 | 0.0454 | 64, G, 0.963 * | OG0005294 |
| ACBD6 | 0.0016 | 11, Y, 0.985 * 589, R, 0.969 * 590, K, 0.990 ** | OG0003321 |
| RP11-6L6.2 | 0.0013 | 145, Q, 0.974 * | OG0004058 |
| MYRF | 0.00017 | 16, Q, 0.961 * 31, L, 0.976 * 36, A, 0.971 * | OG0004190 |
| GCFC2 | 0.00002 | 14, P, 0.960 * 19, G, 0.957 * 69, Q, 0.983 * | OG0004841 |
| CFAP410 | 0.045 | 410, E, 0.960 * 411, L, 0.962 * | OG0005392 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, F.; Chen, H.; Chang, C.; Zhou, J.; Zhang, H. Comparative Genomic Analysis Across Multiple Species to Identify Candidate Genes Associated with Important Traits in Chickens. Genes 2025, 16, 627. https://doi.org/10.3390/genes16060627
Zhang F, Chen H, Chang C, Zhou J, Zhang H. Comparative Genomic Analysis Across Multiple Species to Identify Candidate Genes Associated with Important Traits in Chickens. Genes. 2025; 16(6):627. https://doi.org/10.3390/genes16060627
Chicago/Turabian StyleZhang, Fuyang, Hengcong Chen, Cheng Chang, Jiamei Zhou, and Hui Zhang. 2025. "Comparative Genomic Analysis Across Multiple Species to Identify Candidate Genes Associated with Important Traits in Chickens" Genes 16, no. 6: 627. https://doi.org/10.3390/genes16060627
APA StyleZhang, F., Chen, H., Chang, C., Zhou, J., & Zhang, H. (2025). Comparative Genomic Analysis Across Multiple Species to Identify Candidate Genes Associated with Important Traits in Chickens. Genes, 16(6), 627. https://doi.org/10.3390/genes16060627




