The Role of Non-Coding RNAs in ALS
Abstract
1. Introduction
2. Epidemiology
3. Etiology
4. Epigenetics and ALS
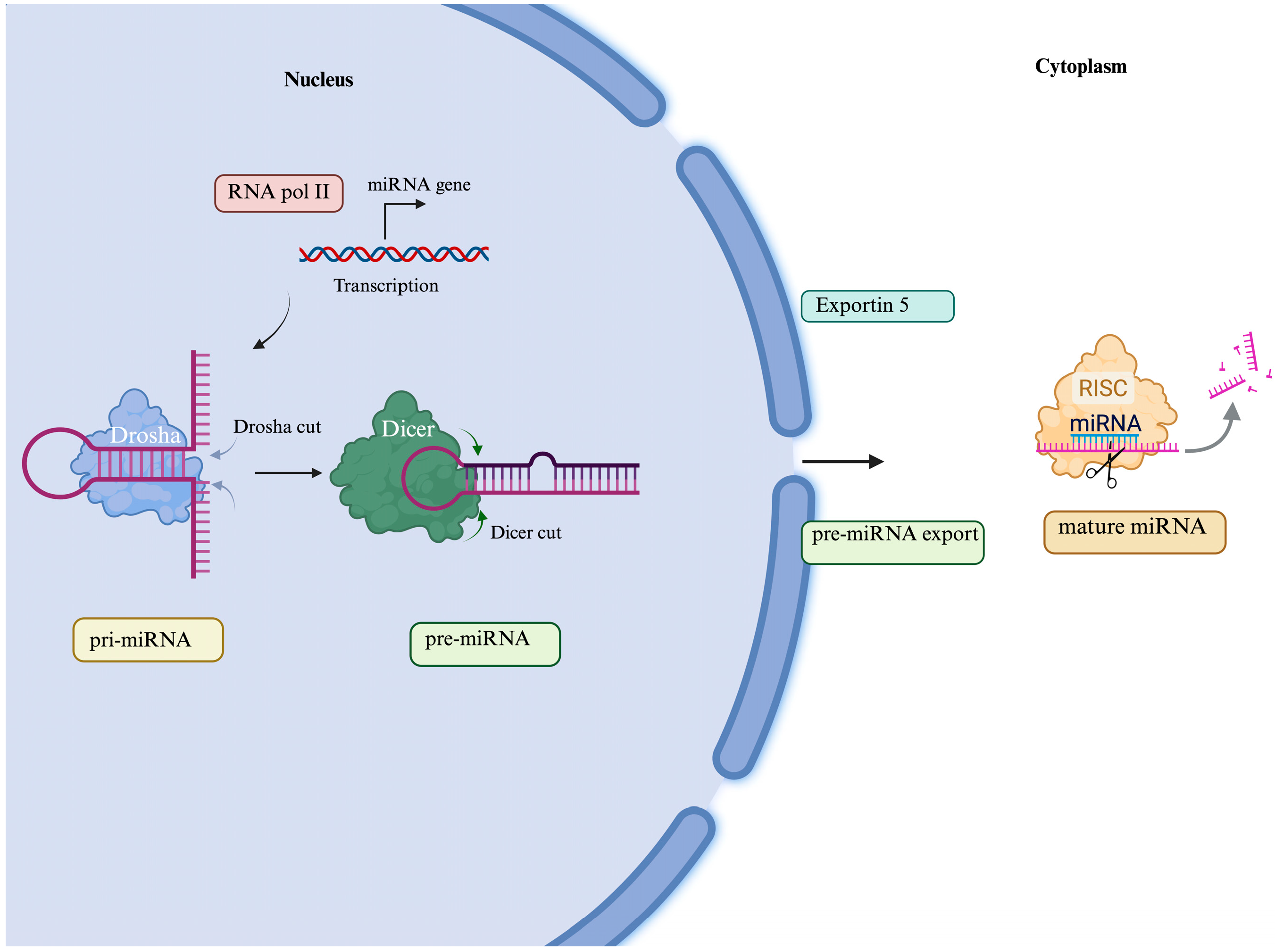
5. MicroRNA and ALS
6. miRNAs as Biomarkers and Therapeutic Targets
7. miRNAs and Cytoplasmic Inclusions
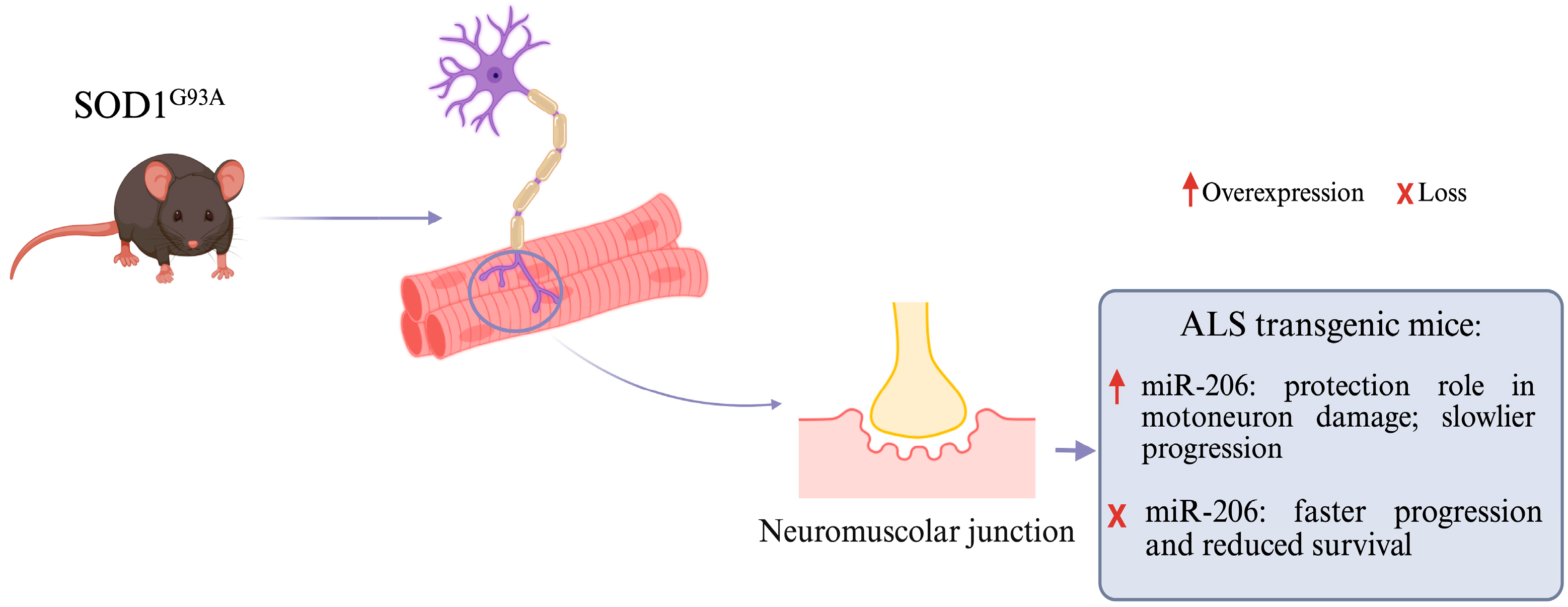
8. miRNAs and Neuromuscular Junctions
9. miRNAs and Neuroinflammation
10. miRNAs, Neuronal Survival, and Apoptosis in ALS
11. Long Non-Coding RNA and ALS
12. LncRNAs as Biomarker in ALS
13. LncRNAs as Potential Therapeutic Targets
14. Circular RNAs and ASL
15. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Goutman, S.A.; Chen, K.S.; Paez-Colasante, X.; Feldman, E.L. Emerging Understanding of the Genotype–Phenotype Relationship in Amyotrophic Lateral Sclerosis. Handb. Clin. Neurol. 2018, 148, 603–623. [Google Scholar] [PubMed]
- Feldman, E.L.; Goutman, S.A.; Petri, S.; Mazzini, L.; Savelieff, M.G.; Shaw, P.J.; Sobue, G. Amyotrophic Lateral Sclerosis. Lancet 2022, 400, 1363–1380. [Google Scholar] [CrossRef]
- De Marchi, F.; Tondo, G.; Corrado, L.; Menegon, F.; Aprile, D.; Anselmi, M.; D’Alfonso, S.; Comi, C.; Mazzini, L. Neuroinflammatory Pathways in the ALS-FTD Continuum: A Focus on Genetic Variants. Genes 2023, 14, 1658. [Google Scholar] [CrossRef]
- Loeffler, J.; Picchiarelli, G.; Dupuis, L.; Gonzalez De Aguilar, J. The Role of Skeletal Muscle in Amyotrophic Lateral Sclerosis. Brain Pathol. 2016, 26, 227–236. [Google Scholar] [CrossRef]
- Masrori, P.; Van Damme, P. Amyotrophic Lateral Sclerosis: A Clinical Review. Eur. J. Neurol. 2020, 27, 1918–1929. [Google Scholar] [CrossRef] [PubMed]
- Logroscino, G.; Traynor, B.J.; Hardiman, O.; Chio, A.; Mitchell, D.; Swingler, R.J.; Millul, A.; Benn, E.; Beghi, E. Incidence of Amyotrophic Lateral Sclerosis in Europe. J. Neurol. Neurosurg. Psychiatry 2010, 81, 385–390. [Google Scholar] [CrossRef]
- Marin, B.; Boumédiene, F.; Logroscino, G.; Couratier, P.; Babron, M.-C.; Leutenegger, A.L.; Copetti, M.; Preux, P.-M.; Beghi, E. Variation in Worldwide Incidence of Amyotrophic Lateral Sclerosis: A Meta-Analysis. Int. J. Epidemiol. 2016, 46, 57–74. [Google Scholar] [CrossRef]
- Wang, Y.; Patani, R. Novel Therapeutic Targets for Amyotrophic Lateral Sclerosis: Ribonucleoproteins and Cellular Autonomy. Expert Opin. Ther. Targets 2020, 24, 971–984. [Google Scholar] [CrossRef]
- Manjaly, Z.R.; Scott, K.M.; Abhinav, K.; Wijesekera, L.; Ganesalingam, J.; Goldstein, L.H.; Janssen, A.; Dougherty, A.; Willey, E.; Stanton, B.R.; et al. The Sex Ratio in Amyotrophic Lateral Sclerosis: A Population Based Study. Amyotroph. Lateral Scler. 2010, 11, 439–442. [Google Scholar] [CrossRef]
- Blasco, H.; Guennoc, A.-M.; Veyrat-Durebex, C.; Gordon, P.H.; Andres, C.R.; Camu, W.; Corcia, P. Amyotrophic Lateral Sclerosis: A Hormonal Condition? Amyotroph. Lateral Scler. 2012, 13, 585–588. [Google Scholar] [CrossRef]
- Dardiotis, E.; Siokas, V.; Sokratous, M.; Tsouris, Z.; Michalopoulou, A.; Andravizou, A.; Dastamani, M.; Ralli, S.; Vinceti, M.; Tsatsakis, A.; et al. Genetic Polymorphisms in Amyotrophic Lateral Sclerosis: Evidence for Implication in Detoxification Pathways of Environmental Toxicants. Environ. Int. 2018, 116, 122–135. [Google Scholar] [CrossRef]
- Dardiotis, E.; Xiromerisiou, G.; Hadjichristodoulou, C.; Tsatsakis, A.M.; Wilks, M.F.; Hadjigeorgiou, G.M. The Interplay between Environmental and Genetic Factors in Parkinson’s Disease Susceptibility: The Evidence for Pesticides. Toxicology 2013, 307, 17–23. [Google Scholar] [CrossRef]
- Pappalardo, X.G.; Jansen, G.; Amaradio, M.; Costanza, J.; Umeton, R.; Guarino, F.; De Pinto, V.; Oliver, S.G.; Messina, A.; Nicosia, G. Inferring Gene Regulatory Networks of ALS from Blood Transcriptome Profiles. Heliyon 2024, 10, e40696. [Google Scholar] [CrossRef]
- Dardiotis, E.; Siokas, V.; Sokratous, M.; Tsouris, Z.; Aloizou, A.-M.; Florou, D.; Dastamani, M.; Mentis, A.-F.A.; Brotis, A.G. Body Mass Index and Survival from Amyotrophic Lateral Sclerosis. Neurol. Clin. Pr. 2018, 8, 437–444. [Google Scholar] [CrossRef]
- Jaenisch, R.; Bird, A. Epigenetic Regulation of Gene Expression: How the Genome Integrates Intrinsic and Environmental Signals. Nat. Genet. 2003, 33, 245–254. [Google Scholar] [CrossRef]
- Sokratous, M.; Dardiotis, E.; Tsouris, Z.; Bellou, E.; Michalopoulou, A.; Siokas, V.; Arseniou, S.; Stamati, T.; Tsivgoulis, G.; Bogdanos, D.; et al. Deciphering the Role of DNA Methylation in Multiple Sclerosis: Emerging Issues. Autoimmun. Highlights 2016, 7, 12. [Google Scholar] [CrossRef][Green Version]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a Big Role in Gene Regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef]
- Ravnik-Glavač, M.; Glavač, D. Circulating RNAs as Potential Biomarkers in Amyotrophic Lateral Sclerosis. Int. J. Mol. Sci. 2020, 21, 1714. [Google Scholar] [CrossRef]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of Small RNAs in Animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; How Huang, K.; Jen Lee, M.; Galas, D.J.; Wang, K. The MicroRNA Spectrum in 12 Body Fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of MicroRNAs in Serum: A Novel Class of Biomarkers for Diagnosis of Cancer and Other Diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Freischmidt, A.; Müller, K.; Zondler, L.; Weydt, P.; Mayer, B.; von Arnim, C.A.F.; Hübers, A.; Dorst, J.; Otto, M.; Holzmann, K.; et al. Serum MicroRNAs in Sporadic Amyotrophic Lateral Sclerosis. Neurobiol. Aging 2015, 36, 2660.e15–2660.e20. [Google Scholar] [CrossRef]
- Butovsky, O.; Siddiqui, S.; Gabriely, G.; Lanser, A.J.; Dake, B.; Murugaiyan, G.; Doykan, C.E.; Wu, P.M.; Gali, R.R.; Iyer, L.K.; et al. Modulating Inflammatory Monocytes with a Unique MicroRNA Gene Signature Ameliorates Murine ALS. J. Clin. Investig. 2012, 122, 3063–3087. [Google Scholar] [CrossRef]
- Wakabayashi, K.; Mori, F.; Kakita, A.; Takahashi, H.; Utsumi, J.; Sasaki, H. Analysis of MicroRNA from Archived Formalin-Fixed Paraffin-Embedded Specimens of Amyotrophic Lateral Sclerosis. Acta Neuropathol. Commun. 2014, 2, 173. [Google Scholar] [CrossRef]
- Foggin, S.; Mesquita-Ribeiro, R.; Dajas-Bailador, F.; Layfield, R. Biological Significance of MicroRNA Biomarkers in ALS—Innocent Bystanders or Disease Culprits? Front. Neurol. 2019, 10, 578. [Google Scholar] [CrossRef]
- Kawahara, Y.; Mieda-Sato, A. TDP-43 Promotes MicroRNA Biogenesis as a Component of the Drosha and Dicer Complexes. Proc. Natl. Acad. Sci. USA 2012, 109, 3347–3352. [Google Scholar] [CrossRef]
- Williams, A.H.; Valdez, G.; Moresi, V.; Qi, X.; McAnally, J.; Elliott, J.L.; Bassel-Duby, R.; Sanes, J.R.; Olson, E.N. MicroRNA-206 Delays ALS Progression and Promotes Regeneration of Neuromuscular Synapses in Mice. Science 2009, 326, 1549–1554. [Google Scholar] [CrossRef]
- Toivonen, J.M.; Manzano, R.; Oliván, S.; Zaragoza, P.; García-Redondo, A.; Osta, R. MicroRNA-206: A Potential Circulating Biomarker Candidate for Amyotrophic Lateral Sclerosis. PLoS ONE 2014, 9, e89065. [Google Scholar] [CrossRef]
- Sempere, L.F.; Freemantle, S.; Pitha-Rowe, I.; Moss, E.; Dmitrovsky, E.; Ambros, V. Expression Profiling of Mammalian MicroRNAs Uncovers a Subset of Brain-Expressed MicroRNAs with Possible Roles in Murine and Human Neuronal Differentiation. Genome Biol. 2004, 5, R13. [Google Scholar] [CrossRef]
- Liu, J.; Zhou, F.; Guan, Y.; Meng, F.; Zhao, Z.; Su, Q.; Bao, W.; Wang, X.; Zhao, J.; Huo, Z.; et al. The Biogenesis of MiRNAs and Their Role in the Development of Amyotrophic Lateral Sclerosis. Cells 2022, 11, 572. [Google Scholar] [CrossRef]
- Butovsky, O.; Jedrychowski, M.P.; Cialic, R.; Krasemann, S.; Murugaiyan, G.; Fanek, Z.; Greco, D.J.; Wu, P.M.; Doykan, C.E.; Kiner, O.; et al. Targeting miR-155 Restores Abnormal Microglia and Attenuates Disease in SOD1 Mice. Ann. Neurol. 2015, 77, 75–99. [Google Scholar] [CrossRef] [PubMed]
- Koval, E.D.; Shaner, C.; Zhang, P.; du Maine, X.; Fischer, K.; Tay, J.; Chau, B.N.; Wu, G.F.; Miller, T.M. Method for Widespread MicroRNA-155 Inhibition Prolongs Survival in ALS-Model Mice. Hum. Mol. Genet. 2013, 22, 4127–4135. [Google Scholar] [CrossRef] [PubMed]
- Parisi, C.; Arisi, I.; D’Ambrosi, N.; Storti, A.E.; Brandi, R.; D’Onofrio, M.; Volonté, C. Dysregulated MicroRNAs in Amyotrophic Lateral Sclerosis Microglia Modulate Genes Linked to Neuroinflammation. Cell Death. Dis. 2013, 4, e959. [Google Scholar] [CrossRef]
- Cunha, C.; Santos, C.; Gomes, C.; Fernandes, A.; Correia, A.M.; Sebastião, A.M.; Vaz, A.R.; Brites, D. Downregulated Glia Interplay and Increased MiRNA-155 as Promising Markers to Track ALS at an Early Stage. Mol. Neurobiol. 2017, 55, 4207–4224. [Google Scholar] [CrossRef]
- Russell, A.P.; Wada, S.; Vergani, L.; Hock, M.B.; Lamon, S.; Léger, B.; Ushida, T.; Cartoni, R.; Wadley, G.D.; Hespel, P.; et al. Disruption of Skeletal Muscle Mitochondrial Network Genes and MiRNAs in Amyotrophic Lateral Sclerosis. Neurobiol. Dis. 2013, 49, 107–117. [Google Scholar] [CrossRef]
- Li, C.; Chen, Y.; Chen, X.; Wei, Q.; Ou, R.; Gu, X.; Cao, B.; Shang, H. MicroRNA-183-5p Is Stress-inducible and Protects Neurons against Cell Death in Amyotrophic Lateral Sclerosis. J. Cell Mol. Med. 2020, 24, 8614–8622. [Google Scholar] [CrossRef]
- Brooks, B.R.; Miller, R.G.; Swash, M.; Munsat, T.L. El Escorial Revisited: Revised Criteria for the Diagnosis of Amyotrophic Lateral Sclerosis. Amyotroph. Lateral Scler. Other Mot. Neuron Disord. 2000, 1, 293–299. [Google Scholar] [CrossRef]
- Brown, R.H.; Al-Chalabi, A. Amyotrophic Lateral Sclerosis. N. Engl. J. Med. 2017, 377, 162–172. [Google Scholar] [CrossRef]
- Zoing, M.C.; Burke, D.; Pamphlett, R.; Kiernan, M.C. Riluzole Therapy for Motor Neurone Disease: An Early Australian Experience (1996–2002). J. Clin. Neurosci. 2006, 13, 78–83. [Google Scholar] [CrossRef]
- Biomarkers Definitions Working Group. Biomarkers and Surrogate Endpoints: Preferred Definitions and Conceptual Framework. Clin. Pharmacol. Ther. 2001, 69, 89–95. [Google Scholar] [CrossRef]
- Cloutier, F.; Marrero, A.; O’Connell, C.; Morin, P.J. MicroRNAs as Potential Circulating Biomarkers for Amyotrophic Lateral Sclerosis. J. Mol. Neurosci. 2015, 56, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Freischmidt, A.; Müller, K.; Zondler, L.; Weydt, P.; Volk, A.E.; Božič, A.L.; Walter, M.; Bonin, M.; Mayer, B.; von Arnim, C.A.F.; et al. Serum MicroRNAs in Patients with Genetic Amyotrophic Lateral Sclerosis and Pre-Manifest Mutation Carriers. Brain 2014, 137, 2938–2950. [Google Scholar] [CrossRef]
- Paez-Colasante, X.; Figueroa-Romero, C.; Sakowski, S.A.; Goutman, S.A.; Feldman, E.L. Amyotrophic Lateral Sclerosis: Mechanisms and Therapeutics in the Epigenomic Era. Nat. Rev. Neurol. 2015, 11, 266–279. [Google Scholar] [CrossRef]
- Blasco, H.; Corcia, P.; Moreau, C.; Veau, S.; Fournier, C.; Vourc’h, P.; Emond, P.; Gordon, P.; Pradat, P.-F.; Praline, J.; et al. 1H-NMR-Based Metabolomic Profiling of CSF in Early Amyotrophic Lateral Sclerosis. PLoS ONE 2010, 5, e13223. [Google Scholar] [CrossRef]
- Pradat, P.-F.; Dib, M. Biomarkers in Amyotrophic Lateral Sclerosis: Facts and Future Horizons. Mol. Diagn. Ther. 2009, 13, 115–125. [Google Scholar] [CrossRef]
- Rothstein, J.D.; Kuncl, R.; Chaudhry, V.; Clawson, L.; Cornblath, D.R.; Coyle, J.T.; Drachman, D.B. Excitatory Amino Acids in Amyotrophic Lateral Sclerosis: An Update. Ann. Neurol. 1991, 30, 224–225. [Google Scholar] [CrossRef]
- Shaw, P.J.; Forrest, V.; Ince, P.G.; Richardson, J.P.; Wastell, H.J. CSF and Plasma Amino Acid Levels in Motor Neuron Disease: Elevation of CSF Glutamate in a Subset of Patients. Neurodegeneration 1995, 4, 209–216. [Google Scholar] [CrossRef]
- Spreux-Varoquaux, O.; Bensimon, G.; Lacomblez, L.; Salachas, F.; Pradat, P.F.; Le Forestier, N.; Marouan, A.; Dib, M.; Meininger, V. Glutamate Levels in Cerebrospinal Fluid in Amyotrophic Lateral Sclerosis: A Reappraisal Using a New HPLC Method with Coulometric Detection in a Large Cohort of Patients. J. Neurol. Sci. 2002, 193, 73–78. [Google Scholar] [CrossRef]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 Complexes Carry a Population of Circulating MicroRNAs Independent of Vesicles in Human Plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef]
- Gallo, A.; Tandon, M.; Alevizos, I.; Illei, G.G. The Majority of MicroRNAs Detectable in Serum and Saliva Is Concentrated in Exosomes. PLoS ONE 2012, 7, e30679. [Google Scholar] [CrossRef]
- Wagner, J.; Riwanto, M.; Besler, C.; Knau, A.; Fichtlscherer, S.; Röxe, T.; Zeiher, A.M.; Landmesser, U.; Dimmeler, S. Characterization of Levels and Cellular Transfer of Circulating Lipoprotein-Bound MicroRNAs. Arter. Thromb. Vasc. Biol. 2013, 33, 1392–1400. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Li, C.; Ge, H.; Jiang, Y.; Li, Y. Circulating MicroRNA: A Novel Potential Biomarker for Early Diagnosis of Intracranial Aneurysm Rupture a Case Control Study. J. Transl. Med. 2013, 11, 296. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, V.M.; Scaria, V.; Maiti, S. Antagomirzymes: Oligonucleotide Enzymes That Specifically Silence MicroRNA Function. Angew. Chem. Int. Ed. 2009, 48, 2557–2560. [Google Scholar] [CrossRef]
- Bader, A.G.; Brown, D.; Winkler, M. The Promise of MicroRNA Replacement Therapy. Cancer. Res. 2010, 70, 7027–7030. [Google Scholar] [CrossRef]
- Junn, E.; Mouradian, M.M. MicroRNAs in Neurodegenerative Diseases and Their Therapeutic Potential. Pharmacol. Ther. 2012, 133, 142–150. [Google Scholar] [CrossRef]
- Foust, K.D.; Nurre, E.; Montgomery, C.L.; Hernandez, A.; Chan, C.M.; Kaspar, B.K. Intravascular AAV9 Preferentially Targets Neonatal Neurons and Adult Astrocytes. Nat. Biotechnol. 2009, 27, 59–65. [Google Scholar] [CrossRef]
- Stoica, L.; Sena-Esteves, M. Adeno Associated Viral Vector Delivered RNAi for Gene Therapy of SOD1 Amyotrophic Lateral Sclerosis. Front. Mol. Neurosci. 2016, 9, 56. [Google Scholar] [CrossRef][Green Version]
- Morlando, M.; Dini Modigliani, S.; Torrelli, G.; Rosa, A.; Di Carlo, V.; Caffarelli, E.; Bozzoni, I. FUS Stimulates MicroRNA Biogenesis by Facilitating Co-Transcriptional Drosha Recruitment. EMBO J. 2012, 31, 4502–4510. [Google Scholar] [CrossRef]
- Conforti, F.L.; Sproviero, W.; Simone, I.L.; Mazzei, R.; Valentino, P.; Ungaro, C.; Magariello, A.; Patitucci, A.; La Bella, V.; Sprovieri, T.; et al. TARDBP Gene Mutations in South Italian Patients with Amyotrophic Lateral Sclerosis. J. Neurol. Neurosurg. Psychiatry 2011, 82, 587–588. [Google Scholar] [CrossRef]
- Ruffo, P.; Catalano, S.; La Bella, V.; Conforti, F.L. Deregulation of Plasma MicroRNA Expression in a TARDBP-ALS Family. Biomolecules 2023, 13, 706. [Google Scholar] [CrossRef]
- Kim, H.K.; Lee, Y.S.; Sivaprasad, U.; Malhotra, A.; Dutta, A. Muscle-Specific MicroRNA MiR-206 Promotes Muscle Differentiation. J. Cell Biol. 2006, 174, 677–687. [Google Scholar] [CrossRef]
- Goljanek-Whysall, K.; Pais, H.; Rathjen, T.; Sweetman, D.; Dalmay, T.; Münsterberg, A. Regulation of Multiple Target Genes by MiR-1 and MiR-206 Is Pivotal for C2C12 Myoblast Differentiation. J. Cell Sci. 2012, 125, 3590–3600. [Google Scholar] [CrossRef]
- O’Rourke, J.R.; Georges, S.A.; Seay, H.R.; Tapscott, S.J.; McManus, M.T.; Goldhamer, D.J.; Swanson, M.S.; Harfe, B.D. Essential Role for Dicer during Skeletal Muscle Development. Dev. Biol. 2007, 311, 359–368. [Google Scholar] [CrossRef]
- Cohen, T.J.; Waddell, D.S.; Barrientos, T.; Lu, Z.; Feng, G.; Cox, G.A.; Bodine, S.C.; Yao, T.-P. The Histone Deacetylase HDAC4 Connects Neural Activity to Muscle Transcriptional Reprogramming. J. Biol. Chem. 2007, 282, 33752–33759. [Google Scholar] [CrossRef]
- Tang, H.; Macpherson, P.; Marvin, M.; Meadows, E.; Klein, W.H.; Yang, X.-J.; Goldman, D. A Histone Deacetylase 4/Myogenin Positive Feedback Loop Coordinates Denervation-Dependent Gene Induction and Suppression. Mol. Biol. Cell 2009, 20, 1120–1131. [Google Scholar] [CrossRef]
- Taetzsch, T.; Tenga, M.J.; Valdez, G. Muscle Fibers Secrete FGFBP1 to Slow Degeneration of Neuromuscular Synapses during Aging and Progression of ALS. J. Neurosci. 2017, 37, 70–82. [Google Scholar] [CrossRef]
- Di Pietro, L.; Baranzini, M.; Berardinelli, M.G.; Lattanzi, W.; Monforte, M.; Tasca, G.; Conte, A.; Logroscino, G.; Michetti, F.; Ricci, E.; et al. Potential Therapeutic Targets for ALS: MIR206, MIR208b and MIR499 Are Modulated during Disease Progression in the Skeletal Muscle of Patients. Sci. Rep. 2017, 7, 9538. [Google Scholar] [CrossRef] [PubMed]
- Ceppi, M.; Pereira, P.M.; Dunand-Sauthier, I.; Barras, E.; Reith, W.; Santos, M.A.; Pierre, P. MicroRNA-155 Modulates the Interleukin-1 Signaling Pathway in Activated Human Monocyte-Derived Dendritic Cells. Proc. Natl. Acad. Sci. USA 2009, 106, 2735–2740. [Google Scholar] [CrossRef]
- O’Connell, R.M.; Kahn, D.; Gibson, W.S.J.; Round, J.L.; Scholz, R.L.; Chaudhuri, A.A.; Kahn, M.E.; Rao, D.S.; Baltimore, D. MicroRNA-155 Promotes Autoimmune Inflammation by Enhancing Inflammatory T Cell Development. Immunity 2010, 33, 607–619. [Google Scholar] [CrossRef]
- O’Connell, R.M.; Taganov, K.D.; Boldin, M.P.; Cheng, G.; Baltimore, D. MicroRNA-155 Is Induced during the Macrophage Inflammatory Response. Proc. Natl. Acad. Sci. USA 2007, 104, 1604–1609. [Google Scholar] [CrossRef]
- Kloske, C.M.; Gearon, M.D.; Weekman, E.M.; Rogers, C.; Patel, E.; Bachstetter, A.; Nelson, P.T.; Wilcock, D.M. Association between APOE Genotype and Microglial Cell Morphology. J. Neuropathol. Exp. Neurol. 2023, 82, 620–630. [Google Scholar] [CrossRef] [PubMed]
- Parisi, C.; Napoli, G.; Amadio, S.; Spalloni, A.; Apolloni, S.; Longone, P.; Volonté, C. MicroRNA-125b Regulates Microglia Activation and Motor Neuron Death in ALS. Cell Death. Differ. 2016, 23, 531–541. [Google Scholar] [CrossRef]
- Li, C.; Chen, Y.; Chen, X.; Wei, Q.; Cao, B.; Shang, H. Downregulation of MicroRNA-193b-3p Promotes Autophagy and Cell Survival by Targeting TSC1/MTOR Signaling in NSC-34 Cells. Front. Mol. Neurosci. 2017, 10, 160. [Google Scholar] [CrossRef]
- Vaz, A.R.; Vizinha, D.; Morais, H.; Colaço, A.R.; Loch-Neckel, G.; Barbosa, M.; Brites, D. Overexpression of MiR-124 in Motor Neurons Plays a Key Role in ALS Pathological Processes. Int. J. Mol. Sci. 2021, 22, 6128. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long Non-Coding RNAs: Definitions, Functions, Challenges and Recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Matsumoto, A.; Nakayama, K.I. Hidden Peptides Encoded by Putative Noncoding RNAs. Cell. Struct. Funct. 2018, 43, 75–83. [Google Scholar] [CrossRef]
- Li, K.; Wang, Z. Speckles and Paraspeckles Coordinate to Regulate HSV-1 Genes Transcription. Commun. Biol. 2021, 4, 1207. [Google Scholar] [CrossRef]
- Li, K.; Wang, Z. Non-Coding RNAs: Key Players in T Cell Exhaustion. Front. Immunol. 2022, 13, 959729. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, S.; Li, K. LncRNA NEAT1 Induces Autophagy through Epigenetic Regulation of Autophagy-related Gene Expression in Neuroglial Cells. J. Cell. Physiol. 2022, 237, 824–832. [Google Scholar] [CrossRef]
- Wang, Z.; Li, K.; Huang, W. Long Non-Coding RNA NEAT1-Centric Gene Regulation. Cell. Mol. Life Sci. 2020, 77, 3769–3779. [Google Scholar] [CrossRef]
- Wang, Z.; Fan, P.; Zhao, Y.; Zhang, S.; Lu, J.; Xie, W.; Jiang, Y.; Lei, F.; Xu, N.; Zhang, Y. NEAT1 Modulates Herpes Simplex Virus-1 Replication by Regulating Viral Gene Transcription. Cell. Mol. Life Sci. 2017, 74, 1117–1131. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, Z.; Mao, Y.; Li, B.; Zhu, Y.; Zhang, S.; Wang, S.; Jiang, Y.; Xu, N.; Xie, Y.; et al. NEAT1 Regulates Microtubule Stabilization via FZD3/GSK3β/P-Tau Pathway in SH-SY5Y Cells and APP/PS1 Mice. Aging 2020, 12, 23233–23250. [Google Scholar] [CrossRef]
- Yang, T.; Wang, Y.; Liao, W.; Zhang, S.; Wang, S.; Xu, N.; Xie, W.; Luo, C.; Wang, Y.; Wang, Z.; et al. Down-Regulation of EPB41L4A-AS1 Mediated the Brain Aging and Neurodegenerative Diseases via Damaging Synthesis of NAD+ and ATP. Cell. Biosci. 2021, 11, 192. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, Y.; Zhang, Y. Viral LncRNA: A Regulatory Molecule for Controlling Virus Life Cycle. Non-Coding RNA Res. 2017, 2, 38–44. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, Y.; Xu, N.; Zhang, S.; Wang, S.; Mao, Y.; Zhu, Y.; Li, B.; Jiang, Y.; Tan, Y.; et al. NEAT1 Regulates Neuroglial Cell Mediating Aβ Clearance via the Epigenetic Regulation of Endocytosis-Related Genes Expression. Cell. Mol. Life Sci. 2019, 76, 3005–3018. [Google Scholar] [CrossRef]
- Pappalardo, X.G.; Risiglione, P.; Zinghirino, F.; Ostuni, A.; Luciano, D.; Bisaccia, F.; De Pinto, V.; Guarino, F.; Messina, A. Human VDAC Pseudogenes: An Emerging Role for VDAC1P8 Pseudogene in Acute Myeloid Leukemia. Biol. Res. 2023, 56, 33. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Sasaki, Y.T.F.; Ideue, T.; Sano, M.; Mituyama, T.; Hirose, T. MENε/β Noncoding RNAs Are Essential for Structural Integrity of Nuclear Paraspeckles. Proc. Natl. Acad. Sci. USA 2009, 106, 2525–2530. [Google Scholar] [CrossRef]
- Nakagawa, S.; Yamazaki, T.; Hirose, T. Molecular Dissection of Nuclear Paraspeckles: Towards Understanding the Emerging World of the RNP Milieu. Open Biol. 2018, 8, 150. [Google Scholar] [CrossRef]
- Nishimoto, Y.; Nakagawa, S.; Okano, H. NEAT1 LncRNA and Amyotrophic Lateral Sclerosis. Neurochem. Int. 2021, 150, 105175. [Google Scholar] [CrossRef]
- An, H.; Skelt, L.; Notaro, A.; Highley, J.R.; Fox, A.H.; La Bella, V.; Buchman, V.L.; Shelkovnikova, T.A. ALS-Linked FUS Mutations Confer Loss and Gain of Function in the Nucleus by Promoting Excessive Formation of Dysfunctional Paraspeckles. Acta Neuropathol. Commun. 2019, 7, 7. [Google Scholar] [CrossRef] [PubMed]
- Colombrita, C.; Onesto, E.; Megiorni, F.; Pizzuti, A.; Baralle, F.E.; Buratti, E.; Silani, V.; Ratti, A. TDP-43 and FUS RNA-Binding Proteins Bind Distinct Sets of Cytoplasmic Messenger RNAs and Differently Regulate Their Post-Transcriptional Fate in Motoneuron-like Cells. J. Biol. Chem. 2012, 287, 15635–15647. [Google Scholar] [CrossRef] [PubMed]
- Zinszner, H.; Sok, J.; Immanuel, D.; Yin, Y.; Ron, D. TLS (FUS) Binds RNA in Vivo and Engages in Nucleo-Cytoplasmic Shuttling. J. Cell Sci. 1997, 110, 1741–1750. [Google Scholar] [CrossRef]
- Fujii, R.; Takumi, T. TLS Facilitates Transport of MRNA Encoding an Actin-Stabilizing Protein to Dendritic Spines. J. Cell Sci. 2005, 118, 5755–5765. [Google Scholar] [CrossRef]
- Vance, C.; Rogelj, B.; Hortobágyi, T.; De Vos, K.J.; Nishimura, A.L.; Sreedharan, J.; Hu, X.; Smith, B.; Ruddy, D.; Wright, P.; et al. Mutations in FUS, an RNA Processing Protein, Cause Familial Amyotrophic Lateral Sclerosis Type 6. Science 2009, 323, 1208–1211. [Google Scholar] [CrossRef]
- Gerbino, V.; Carrì, M.T.; Cozzolino, M.; Achsel, T. Mislocalised FUS Mutants Stall Spliceosomal SnRNPs in the Cytoplasm. Neurobiol. Dis. 2013, 55, 120–128. [Google Scholar] [CrossRef]
- Lagier-Tourenne, C.; Polymenidou, M.; Hutt, K.R.; Vu, A.Q.; Baughn, M.; Huelga, S.C.; Clutario, K.M.; Ling, S.-C.; Liang, T.Y.; Mazur, C.; et al. Divergent Roles of ALS-Linked Proteins FUS/TLS and TDP-43 Intersect in Processing Long Pre-MRNAs. Nat. Neurosci. 2012, 15, 1488–1497. [Google Scholar] [CrossRef]
- Tsuiji, H.; Iguchi, Y.; Furuya, A.; Kataoka, A.; Hatsuta, H.; Atsuta, N.; Tanaka, F.; Hashizume, Y.; Akatsu, H.; Murayama, S.; et al. Spliceosome Integrity Is Defective in the Motor Neuron Diseases ALS and SMA. EMBO Mol. Med. 2013, 5, 221–234. [Google Scholar] [CrossRef]
- Yamazaki, T.; Chen, S.; Yu, Y.; Yan, B.; Haertlein, T.C.; Carrasco, M.A.; Tapia, J.C.; Zhai, B.; Das, R.; Lalancette-Hebert, M.; et al. FUS-SMN Protein Interactions Link the Motor Neuron Diseases ALS and SMA. Cell Rep. 2012, 2, 799–806. [Google Scholar] [CrossRef]
- Groen, E.J.N.; Fumoto, K.; Blokhuis, A.M.; Engelen-Lee, J.; Zhou, Y.; van den Heuvel, D.M.A.; Koppers, M.; van Diggelen, F.; van Heest, J.; Demmers, J.A.A.; et al. ALS-Associated Mutations in FUS Disrupt the Axonal Distribution and Function of SMN. Hum. Mol. Genet. 2013, 22, 3690–3704. [Google Scholar] [CrossRef]
- Armstrong, G.A.B.; Drapeau, P. Loss and Gain of FUS Function Impair Neuromuscular Synaptic Transmission in a Genetic Model of ALS. Hum. Mol. Genet. 2013, 22, 4282–4292. [Google Scholar] [CrossRef] [PubMed]
- Polymenidou, M.; Lagier-Tourenne, C.; Hutt, K.R.; Bennett, C.F.; Cleveland, D.W.; Yeo, G.W. Misregulated RNA Processing in Amyotrophic Lateral Sclerosis. Brain Res. 2012, 1462, 3–15. [Google Scholar] [CrossRef]
- Renton, A.E.; Majounie, E.; Waite, A.; Simón-Sánchez, J.; Rollinson, S.; Gibbs, J.R.; Schymick, J.C.; Laaksovirta, H.; van Swieten, J.C.; Myllykangas, L.; et al. A Hexanucleotide Repeat Expansion in C9ORF72 Is the Cause of Chromosome 9p21-Linked ALS-FTD. Neuron 2011, 72, 257–268. [Google Scholar] [CrossRef]
- Majounie, E.; Renton, A.E.; Mok, K.; Dopper, E.G.; Waite, A.; Rollinson, S.; Chiò, A.; Restagno, G.; Nicolaou, N.; Simon-Sanchez, J.; et al. Frequency of the C9orf72 Hexanucleotide Repeat Expansion in Patients with Amyotrophic Lateral Sclerosis and Frontotemporal Dementia: A Cross-Sectional Study. Lancet Neurol. 2012, 11, 323–330. [Google Scholar] [CrossRef]
- Krishnan, G.; Raitcheva, D.; Bartlett, D.; Prudencio, M.; McKenna-Yasek, D.M.; Douthwright, C.; Oskarsson, B.E.; Ladha, S.; King, O.D.; Barmada, S.J.; et al. Poly(GR) and Poly(GA) in Cerebrospinal Fluid as Potential Biomarkers for C9ORF72-ALS/FTD. Nat. Commun. 2022, 13, 2799. [Google Scholar] [CrossRef]
- Suzuki, H.; Shibagaki, Y.; Hattori, S.; Matsuoka, M. C9-ALS/FTD-Linked Proline–Arginine Dipeptide Repeat Protein Associates with Paraspeckle Components and Increases Paraspeckle Formation. Cell Death. Dis. 2019, 10, 746. [Google Scholar] [CrossRef]
- Laneve, P.; Tollis, P.; Caffarelli, E. RNA Deregulation in Amyotrophic Lateral Sclerosis: The Noncoding Perspective. Int. J. Mol. Sci. 2021, 22, 10285. [Google Scholar] [CrossRef]
- Laffita-Mesa, J.M.; Paucar, M.; Svenningsson, P. Ataxin-2 Gene: A Powerful Modulator of Neurological Disorders. Curr. Opin. Neurol. 2021, 34, 578–588. [Google Scholar] [CrossRef]
- Ostrowski, L.; Hall, A.; Mekhail, K. Ataxin-2: From RNA Control to Human Health and Disease. Genes 2017, 8, 157. [Google Scholar] [CrossRef]
- Elden, A.C.; Kim, H.-J.; Hart, M.P.; Chen-Plotkin, A.S.; Johnson, B.S.; Fang, X.; Armakola, M.; Geser, F.; Greene, R.; Lu, M.M.; et al. Ataxin-2 Intermediate-Length Polyglutamine Expansions Are Associated with Increased Risk for ALS. Nature 2010, 466, 1069–1075. [Google Scholar] [CrossRef]
- Li, P.P.; Sun, X.; Xia, G.; Arbez, N.; Paul, S.; Zhu, S.; Peng, H.B.; Ross, C.A.; Koeppen, A.H.; Margolis, R.L.; et al. ATXN2-AS, a Gene Antisense to ATXN2, is Associated with Spinocerebellar Ataxia Type 2 and Amyotrophic Lateral Sclerosis. Ann. Neurol. 2016, 80, 600–615. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Yang, H.; Luo, Y.; Deng, X.; Zhou, Y.; Hu, B. Long Non-Coding RNAs in Neurodegenerative Diseases. Neurochem. Int. 2021, 148, 105096. [Google Scholar] [CrossRef]
- Gagliardi, S.; Zucca, S.; Pandini, C.; Diamanti, L.; Bordoni, M.; Sproviero, D.; Arigoni, M.; Olivero, M.; Pansarasa, O.; Ceroni, M.; et al. Long Non-Coding and Coding RNAs Characterization in Peripheral Blood Mononuclear Cells and Spinal Cord from Amyotrophic Lateral Sclerosis Patients. Sci. Rep. 2018, 8, 2378. [Google Scholar] [CrossRef]
- Rey, F.; Marcuzzo, S.; Bonanno, S.; Bordoni, M.; Giallongo, T.; Malacarne, C.; Cereda, C.; Zuccotti, G.V.; Carelli, S. LncRNAs Associated with Neuronal Development and Oncogenesis Are Deregulated in SOD1-G93A Murine Model of Amyotrophic Lateral Sclerosis. Biomedicines 2021, 9, 809. [Google Scholar] [CrossRef]
- Yang, X.; Ji, Y.; Wang, W.; Zhang, L.; Chen, Z.; Yu, M.; Shen, Y.; Ding, F.; Gu, X.; Sun, H. Amyotrophic Lateral Sclerosis: Molecular Mechanisms, Biomarkers, and Therapeutic Strategies. Antioxidants 2021, 10, 1012. [Google Scholar] [CrossRef]
- Zhou, Q.; Mareljic, N.; Michaelsen, M.; Parhizkar, S.; Heindl, S.; Nuscher, B.; Farny, D.; Czuppa, M.; Schludi, C.; Graf, A.; et al. Active Poly-GA Vaccination Prevents Microglia Activation and Motor Deficits in a C9orf72 Mouse Model. EMBO Mol. Med. 2020, 12, 10919. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Y.; Zhou, S.; Dain, L.; Mei, L.; Zhu, G. Circular RNA: An Emerging Frontier in RNA Therapeutic Targets, RNA Therapeutics, and MRNA Vaccines. J. Control. Release 2022, 348, 84–94. [Google Scholar] [CrossRef]
- Talhouarne, G.J.S.; Gall, J.G. Lariat Intronic RNAs in the Cytoplasm of Xenopus Tropicalis Oocytes. RNA 2014, 20, 1476–1487. [Google Scholar] [CrossRef]
- Gardner, E.J.; Nizami, Z.F.; Talbot, C.C.; Gall, J.G. Stable Intronic Sequence RNA (SisRNA), a New Class of Noncoding RNA from the Oocyte Nucleus of Xenopus tropicalis. Genes. Dev. 2012, 26, 2550–2559. [Google Scholar] [CrossRef]
- Qian, L.; Vu, M.N.; Carter, M.; Wilkinson, M.F. A Spliced Intron Accumulates as a Lariat in the Nucleus of T Cells. Nucleic Acids Res. 1992, 20, 5345–5350. [Google Scholar] [CrossRef]
- Chen, L.-L. The Expanding Regulatory Mechanisms and Cellular Functions of Circular RNAs. Nat. Rev. Mol. Cell Biol. 2020, 21, 475–490. [Google Scholar] [CrossRef] [PubMed]
- D’Ambra, E.; Santini, T.; Vitiello, E.; D’Uva, S.; Silenzi, V.; Morlando, M.; Bozzoni, I. Circ-Hdgfrp3 Shuttles along Neurites and Is Trapped in Aggregates Formed by ALS-Associated Mutant FUS. iScience 2021, 24, 103504. [Google Scholar] [CrossRef]
- Wu, L.-S.; Cheng, W.-C.; Chen, C.-Y.; Wu, M.-C.; Wang, Y.-C.; Tseng, Y.-H.; Chuang, T.-J.; Shen, C.-K.J. Transcriptomopathies of Pre- and Post-Symptomatic Frontotemporal Dementia-like Mice with TDP-43 Depletion in Forebrain Neurons. Acta Neuropathol. Commun. 2019, 7, 50. [Google Scholar] [CrossRef]
| Gene | miRNAs | Involved in ALS Pathogenesis | Ref. |
|---|---|---|---|
| TARDBP | miR-132-5p miR-132-3p miR-124-3p miR-133a-3p | Formation of cytoplasmatic inclusion | [26] |
| SOD1 | miR-206 miR-155 miR-125b miR-365 miR-193b-3p miR-124 | Protective role in motoneuron damage; promote inflammatory response; increase of neurodegeneration | [27,28,29,30] [31,32,33,34] |
| PGC-1α | miR-23 | Skeletal muscle mitochondrial dysfunction | [35] |
| RIPK3, PDCD4 | miR-183-5p | Dysregulation of apoptotic and necrotic pathway of neuronal cells | [36] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Falduti, A.; Giovinazzo, A.; Lo Feudo, E.; Rocca, V.; Brighina, F.; Messina, A.; Conforti, F.L.; Iuliano, R. The Role of Non-Coding RNAs in ALS. Genes 2025, 16, 623. https://doi.org/10.3390/genes16060623
Falduti A, Giovinazzo A, Lo Feudo E, Rocca V, Brighina F, Messina A, Conforti FL, Iuliano R. The Role of Non-Coding RNAs in ALS. Genes. 2025; 16(6):623. https://doi.org/10.3390/genes16060623
Chicago/Turabian StyleFalduti, Alessandra, Adele Giovinazzo, Elisa Lo Feudo, Valentina Rocca, Filippo Brighina, Angela Messina, Francesca Luisa Conforti, and Rodolfo Iuliano. 2025. "The Role of Non-Coding RNAs in ALS" Genes 16, no. 6: 623. https://doi.org/10.3390/genes16060623
APA StyleFalduti, A., Giovinazzo, A., Lo Feudo, E., Rocca, V., Brighina, F., Messina, A., Conforti, F. L., & Iuliano, R. (2025). The Role of Non-Coding RNAs in ALS. Genes, 16(6), 623. https://doi.org/10.3390/genes16060623











