Abstract
Background/Objectives: In perennial plants, developing floral buds survive winter through entering a dormant state, which is induced by low temperature and abscisic acid (ABA). ABA performs vital functions in the dormancy process. ABA-insensitive 5 (ABI5) transcription factor is a key regulator in the ABA signaling pathway. However, little is known about the regulation of ABI5 in the winter dormancy of sweet cherries. Methods: We identified the sweet cherry ABI5 gene and its expression changes using gene cloning and qRT-PCR. Additionally, we validated the interaction between PavABI5 and PavCIG1/2 using Yeast One-Hybrid and Dual-Luciferase Assays. Results: In this study, we identified a basic leucine zipper (bZIP) family gene ABI5 from the sweet cherry, which was closely related to PduABI5 from Prunus dulcis, PpABI5 from Prunus persica, PmABI5 from Prunus mume, and ParABI5 from Prunus armeniaca, through phylogenetic tree analysis. The seasonal expression pattern showed that the PavABI5 level was increased during the winter dormancy stage and induced by exogenous ABA. Specifically, we found that the expression of cherry cold-induced genes (PavCIG1/2) was positively correlated with PavABI5 expression. Furthermore, PavABI5 directly bound to the ABRE elements in the PavCIG1/2 promoters to activate their expression. We further confirmed that the dormancy-associated MADS-box (DAM) genes DAM4 and DAM5 function downstream of the ABA signaling pathway to regulate bud dormancy in sweet cherries. Conclusions: Our findings suggest a putative regulatory model of ABA-mediated bud-dormancy with PavABI5.
1. Introduction
Dormancy is an adaptive strategy in perennial trees that enables them to survive winter conditions and resume growth and bloom under favorable environmental conditions. For fruit trees, it is vital to ensure the normal development of floral buds during cold winters. Consequently, plants enter a dormant state before winter to avoid freezing injury [1]. Furthermore, floral buds can also be affected by spring frosts, which will hamper their development and reduce fruit yield [2,3,4].
The dormancy process consists of three distinct physiological stages: paradormancy, endodormancy, and ecodormancy. Among these, endodormancy is crucial for the accumulation of chilling during dormancy. Once sufficient chilling has been accumulated, endodormancy is terminated, and the plant enters ecodormancy. The primary factors inducing endodormancy of perennial plants are low temperature and abscisic acid (ABA) [5,6,7,8]. Sufficient chilling accumulation is essential for dormancy release and flowering [9,10]. In response to low temperatures, C-repeat binding factor (CBF/DREB) transcription factors regulate cold accumulation and dormancy in fruit trees [11,12]. CBFs induce plant dormancy and enhance cold hardiness via affecting the expressions of dormancy-associated MADS-box (DAM) and other cold-regulated (COR) genes, including the COR, low-temperature induced (LTI), responsive to desiccation (RD), and early dehydration-inducible (ERD) genes [13,14,15]. Additionally, CBFs are involved in retarding plant growth and flowering [16,17]. For example, CBF homologous genes PavCIGs, are involved in flower bud dormancy and flowering in sweet cherry [18]. DAMs, CBF target genes, regulate the dormancy processes and chilling requirement [19,20]. The expressions of DAM1, DAM2, and DAM4 peak at the paradormancy stage, which is closely associated with growth cessation and bud set in peaches (Prunus persica). The expressions of DAM5 and DAM6 peak at the endodormancy stage and decrease toward bud break [21].
ABA functions along with low temperature during the endodormancy phase in some plant species, including apple (Malus domestica), peach (Prunus persica), and pear (Pyrus communis). ABA suppresses bud break, affects flowering time, and enhances plant cold tolerance [22,23,24,25]. The 9-cis-epoxycarotenoid dioxygenase (NCED) genes, which encode a key enzyme in abscisic acid biosynthesis, increase ABA content during the dormancy induction and maintenance stages. In contrast, upregulated CYP707A genes promote ABA catabolism when dormancy releases [8,10,26]. In the ABA signaling pathway, an important transcription factor ABA-insensitive 5 (ABI5), belonging to the basic leucine zipper (bZIP) family, is involved in seed germination, drought, and cold tolerance [27,28,29,30]. Allelic variation in TaABI5-A4 significantly affects seed dormancy in bread wheat [31]. ABI5 regulates downstream genes by binding ABRE elements in their promoters, and most of these downstream genes are stress-related [32,33,34]. Moreover, ABA increases the expression of the CBF/DREB1 transcription factors in grape buds under low-temperature conditions [35]. Although in pears, which also belong to the Rosaceae family, ABI5-like protein ABF3 has been demonstrated to activate CBF and DAM [36], the relationship between ABI5 and the CBF homologous genes CIGs remains unclear in sweet cherries.
Sweet cherries are an important economic crop throughout the world. China has become the largest consumption market for sweet cherries. At present, the total cultivation area of sweet cherries in China is about 3 million mu (approximately 200,000 hectares), with a yield of around 700,000 tons. In some warm temperate regions, the fulfillment of chilling requirements and the breaking of dormancy have become the keys indicating whether sweet cherries can flower and fruit normally. In this study, we identified an ABA signaling pathway gene, ABI5, from sweet cherries, which was closely related to PduABI5 from P. dulcis and PpABI5 from P. persica. The seasonal expression level of PavABI5 was higher during the winter dormancy stage and was induced by exogenous ABA. Moreover, ABA enhanced the expression of PavCIG1/2 in combination with low temperature, and PavABI5 directly bound to the ABRE elements of PavCIG1/2 to increase their transcriptional activity. In addition, the expression of PavCIG1/2 downstream genes, PavDAMs, was also affected by ABA. Our results clarify the potential regulatory mechanisms of ABA-mediated dormancy and cold tolerance in tree fruit species, which will be beneficial to the expansion of sweet cherry cultivation areas in warm temperate regions.
2. Materials and Methods
2.1. Plant Materials
The 8-year-old sweet cherry (P. avium L.) cultivar Royal Lee, with a 400 h chilling requirement [20], was used for this study. The trees were cultivated at the experimental field of Shanghai Jiao Tong University (31.25° N, 121.48° E). The rootstock of Royal Lee was Gisela 6. A pool of floral buds was collected from Royal Lee on 15 July, 15 August, 15 September, 15 October, 15 November, 15 December, 30 December 2020, and 15 January, 1 February, 15 February, 25 February, 5 March, and 15 October 2021. In a previous study, we described the dormancy phases of floral buds, as follows: paradormancy stage—before 1 November; endodormancy stage—1 November to 30 December; ecodormancy stage—1 January to 5 February; budbreak stage—after 5 February [20]. Three individual trees were employed as biological replicates. These buds were frozen in liquid nitrogen and stored at −80 °C.
2.2. RNA Isolation and Gene Expression Analysis
Total RNA was isolated from collected samples of Royal Lee using the RNAprep Pure Plant Kit (Tiangen, Beijing, China). The synthesis of first-strand cDNA was carried out with the PrimeScriptTM RT reagent Kit (Takara, Shiga, Japan). Gene expression analysis used the CFX96 real-time PCR system (Bio-Rad, Hercules, CA, USA) with TB GreenTM Premix Ex TapTM II (Takara, Shiga, Japan). The 2−ΔΔCT method was employed for data analysis [37]. The primers (Supplementary Table S1) for cloning and qRT-PCR were generated via Primer 5 software, with reference to the sweet cherry genome database in NCBI. GenBank accession numbers of the PavABI5, PavCIG1, and PavCIG2 genes were XM_021954506.1, XM_021947969.1, and KC543498.1, respectively. A total of 40 PCR cycles were performed, according to the following temperature program: 95 °C for 30 s, followed by 95 °C for 5 s and 60 °C for 30 s. The phylogenetic trees were achieved using MEGA 6 software via a neighbor-joining method. Homologs of PavABI5 from different species were obtained from NCBI using the BLAST (2.6.0) program.
2.3. Yeast One-Hybrid Assays
PavABI5 coding sequence was inserted into the pB42AD vector, creating a recombinant pB42AD-PavABI5 plasmid. The promoter fragments of PavCIG1/2 were ligated to the pLacZ vector. These constructs, pB42AD-PavABI5 and pLacZ-PavCIG1, as well as pB42AD-PavABI5 and pLacZ-PavCIG2, were co-transformed into EGY48 cells. Transformed colonies were grown on SD/-Leu/-Ura medium and detected on SD/-Leu/-Ura medium supplemented with X-gal.
2.4. Dual-Luciferase Assays
The PavABI5 coding region was cloned into the pRI101 vector to serve as the effector. The promoter fragments of PavCIG1/2 were ligated to the pGreen-LUC vector to generate the reporter, and were introduced into GV3101 strains (harboring the pSoup vector). The effector-reporter system was introduced into tobacco leaves via Agrobacterium tumefaciens GV3101-mediated transformation. After 72 hours post-infiltration, protein interactions were analyzed through dual-luciferase assays (Vazyme, Nanjing, China).
2.5. Subcellular Localization of PavABI5
The PavABI5 open reading frame (stop codon removed) was amplified with specific primers (Supplementary Table S1), and cloning into the PHB-GFP vector. After transferring both the fusion construct and empty vector into A. tumefaciens GV3101, we performed Agrobacterium infiltration on tobacco plants at the 5-week growth stage. The GFP fluorescence was detected using a confocal laser scanning microscope after 48–72 h.
2.6. ABA and Low Temperature Treatments
For the bud-burst experiment, the 30 cm Royal Lee floral shoots (15 January, ecodormancy stage) were treated with 200 μM ABA, placed in 2 L beakers, for 2 days, and water instead of the ABA solution was used as the control. Two days later, we placed all shoots in 2 L beakers filled with water. Thereafter, images were captured and bud-burst rates were recorded at 0, 9, 14, and 21 days. The growth conditions consisted of a 16/8-hour light/dark cycle at 25/21°C respectively, with 75% humidity and illumination at 300 μmol⋅m−2·s−1.
For the expression experiment, 200 μM ABA was used for treating the Royal Lee shoots in 2 L beakers, and water instead of the ABA solution was used as the control. Floral buds from the shoots were collected at 24, 48, and 72 h. The growth conditions were maintained at either 25 °C or 10 °C during the photoperiod, with 75% humidity and illumination at 300 μmol⋅m−2·s−1.
2.7. Protein–Protein Interaction Prediction and Expression Pattern Analysis
STRING database analysis [38] was employed to predict PavABI5-interacting proteins, while the transcriptomic data were obtained from NCBI Gene Expression Omnibus (GSE130426) [39]. The TBtools (2.083) program was used to analyze the RNA-seq data and generate heatmaps based on TPM values [40].
3. Results
3.1. Identification of Sweet Cherry PavABI5 Gene
An important gene, PavABI5, which is involved in the ABA signaling pathway, was investigated in this study. The full-length cDNA sequence of PavABI5 was cloned from floral bud tissue, revealing a 1344-bp open reading frame encoding a protein of 447 amino acids. The amino acid sequence of the bZIP domain was extremely conserved across six plant species. Three highly conserved regions (C1, C2, and C3) were located at the N-terminus of ABI5 proteins, while a C4 region was found at their C-terminal end. As a result, PavABI5 was identified as belonging to the basic leucine zipper (bZIP) transcription factor family (Figure 1A). To investigate the phylogenetic relationships of PavABI5 with ABI5 proteins from other species, a phylogenetic tree was constructed. The analysis revealed that PavABI5 was closely related to PduABI5 from almond, PpABI5 from peach, PmABI5 from Japanese apricot, and ParABI5 from apricot. These results suggested that PavABI5 might exhibit a similar function to that of ABI5 proteins across these species (Figure 1B).
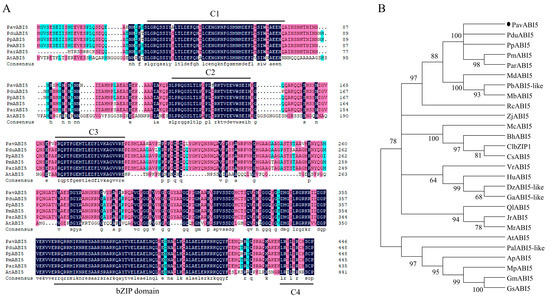
Figure 1.
Sequence and phylogenetic analysis of PavABI5. (A) Phylogenetic tree based on full-length amino acid sequence for PavABI5 proteins from sweet cherry and other plant species. The conserved domains C1–4 and bZIP are marked with black lines. Dark blue, red, and light blue backgrounds indicate 100%, 75%, and 50% identity, respectively. (B) Phylogenetic relationships among ABI5 homologues. The following ABI5 proteins are listed: PduABI5 (XP_034226254.1, Prunus dulcis), PpABI5 (XP_007203311.1, Prunus persica), PmABI5 (XP_008241247.2, Prunus mume), ParABI5 (ADL62859.1, Prunus armeniaca), MdABI5 (XP_028946642.1, Malus domestica), PbABI5-like (XP_018505200.1, Pyrus x bretschneideri), MbABI5 (TQD83332.1, Malus baccata), RcABI5 (XP_024186507.1, Rosa chinensis), ZjABI5 (XP_015898747.1, Ziziphus jujuba), McABI5 (XP_022144787.1, Momordica charantia), BhABI5 (XP_038904029.1, Benincasa hispida), ClbZIP1 (AOZ56990.1, Citrullus lanatus), CsABI5 (XP_004149224.2, Cucumis sativus), VrABI5 (XP_034693304.1, Vitis riparia), HuABI5 (XP_021290003.1, Herrania umbratica), DzABI5-like (XP_022743528.1, Durio zibethinus), GaABI5-like (KAA3465066.1, Gossypium australe), QlABI5 (XP_030925409.1, Quercus lobata), JrABI5 (XP_018813662.1, Juglans regia), MrABI5 (KAB1222563.1, Morella rubra), AtABI5 (AT2G36270, Arabidopsis thaliana), PalABI5-like (XP_028784915.1, Prosopis alba), ApABI5 (XP_027363242.1, Abrus precatorius), MpABI5 (RDY07453.1, Mucuna pruriens), GmABI5 (XP_014618516.1, Glycine max), and GsABI5 (XP_014618516.1, Glycine max).
3.2. PavCIG1 and PavCIG2 Are Downstream of PavABI5
We identified two abscisic acid response elements (ABREs) in the PavCIG1 promoter region (−2000 bp to ATG) and four ABREs in the PavCIG2 promoter region (−2000 bp to ATG) (Figure 2A). A dual-luciferase assay was conducted to investigate the relationship between PavABI5 and the PavCIG1/2 promoters. The results revealed that PavABI5 enhanced the activity of both the PavCIG1 and PavCIG2 promoters in order to activate their expressions (Figure 2B). Moreover, a Y1H assay was conducted to further confirm the interaction between PavABI5 and the PavCIG1/2 promoters. The results also indicated that PavABI5 could bind to the promoters of PavCIG1/2 (Figure 2C).
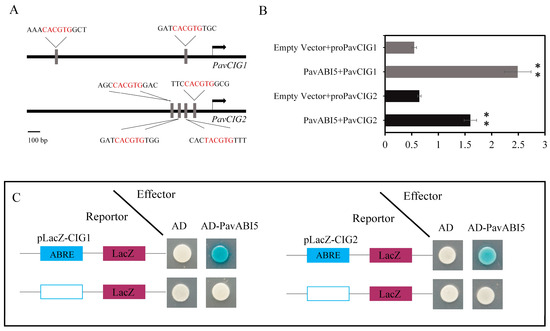
Figure 2.
Interactions between PavABI5 and PavCIG1/2. (A) Diagram of the PavCIG1/2 promoters. (B) Dual-luciferase assays for examining the interactions between PavABI5 and PavCIG1/2 promoters. Empty pGreen-LUC vector was used as a control. As measured by a Student’s t-test, an asterisk represents a significant difference of p < 0.01. (C) Yeast one-hybrid assays for examining the interactions between PavABI5 and the PavCIG1/2 promoters. Empty pLacZ vector was used as the control.
3.3. ABA Delayed Floral Budburst in Sweet Cherry
The sweet cherry flower buds collected from shoots on 15 January (side green stage) were treated with 200 μM ABA. After 14 days, the control shoots exhibited a bud-burst rate of approximately 68%. However, these flower buds treated with ABA remained in a dormant state. After 21 days, the control flower buds began to flower, with a bud-burst rate of approximately 95%, while the ABA-treated flower buds broke dormancy with a bud-burst rate of approximately 53% (Figure 3A,B).
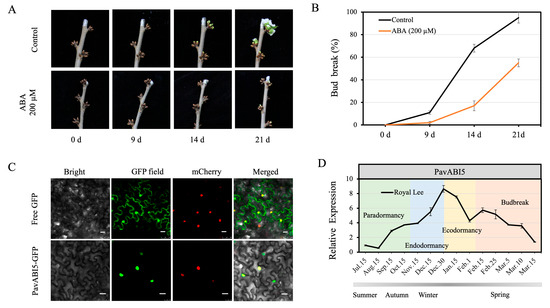
Figure 3.
ABA delayed floral bud-burst in sweet cherry. (A) Sweet cherry shoots were placed in solution (200 μM ABA or water) in 2 L beakers for 2 d; thereafter, images were recorded at 0, 9, 14, 21 d. The growth conditions were maintained at 25 °C for a 16 h light and 21 °C for an 8 h dark photoperiod, with 75% humidity and 300 μmol⋅m−2·s−1 light intensity. (B) Bud-burst rate was recorded at 0, 9, 14, 21 d after ABA treatment. (C) Subcellular localization of PavABI5. The 35S:PavABI5-GFP or 35S:GFP vectors were introduced into A. tumefaciens GV3101. The red fluorescent protein containing NLS-mCherry was co-expressed. Bars, 25 μm. (D) Seasonal expression of PavABI5 was evaluated in floral buds of Royal Lee. The error bars represent the standard deviations (SDs) of three biological replicates.
The coding regions of PavABI5 cDNA, excluding its stop codon, were inserted into the PHB-GFP vector to produce the fusion construct PavABI5-GFP. The fusion plasmids and the control vector (PHB-GFP) were introduced into A. tumefaciens GV3101. Then, they were infiltrated into the leaves of 5-week-old tobacco plants. After 48–72 h of incubation, the GFP fluorescence revealed that PavABI5 was expressed in the cell nucleus compared with the results for NLS-mCherry (Figure 3C).
To explore the expression pattern of PavABI5 across different seasons in sweet cherry, real-time quantitative PCR was performed on floral buds. As shown in Figure 3D, PavABI5 transcription was barely detectable in summer. PavABI5 expression began to accumulate from autumn, and reached a peak on 30 December during winter. In addition, our previous research suggested that PavCIG1 and PavCIG2 exhibited similar expression patterns to those of PavABI5 [18].
3.4. Gene Expression Analysis Through ABA and Low Temperature Treatments
To assess the effect of ABA and low temperature on the downstream genes, ABI5 and CIGs, we examined their expression patterns in the flower buds. As shown in Figure 4A, after a 48 h ABA treatment, the expression of PavABI5 was increased significantly at 25 °C. Moreover, the expression of PavABI5 was increased rapidly under the low temperature of 10 °C, and reached a higher level than that of the control (10 °C) during the first 24 h (Figure 4B). PavCIG1 and PavCIG2 were expressed at higher levels after ABA treatment (10 °C) than in the control (10 °C) at 24 and 48 h (Figure 4B).
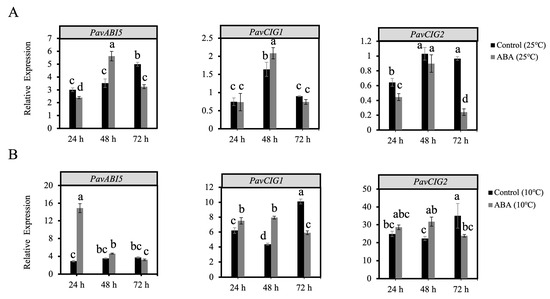
Figure 4.
Expression patterns of PavABI5 and PavCIG1/2 after ABA and low temperature treatments. (A) After ABA treatment, floral buds were collected at 24, 48, and 72 h under a temperature of 25 °C. The expressions of PavABI5 and PavCIGs were detected by qRT-PCR assay. (B) After ABA treatment, floral buds collected at 24, 48, and 72 h under a temperature of 10 °C were used to detect the expressions of PavABI5 and PavCIGs. Error bars represent the standard deviation of three biological replicates. Different letters indicate significant differences (p < 0.01).
3.5. Expression Analysis of Downstream Gene DAMs After ABA and Cold Treatments
DAMs are selectively affected by CBFs in the regulation of the bud dormancy process [15,19]. In this study, we found that the expression levels of the major genes PavDAM4 and PavDAM5 were significantly increased after ABA treatment (25 °C) at 24 h and 48 h, respectively (Figure 5A). Moreover, the expressions of both genes were significantly higher than those of the control after a 48 h ABA treatment under low temperature conditions (10 °C) (Figure 5B). However, no direct interactions were observed between PavCIG1/2 and PavDAM4/5 (Supplementary Figure S1). These results suggested that PavDAM4 and PavDAM5 may act downstream of ABA signaling to modulate dormancy.
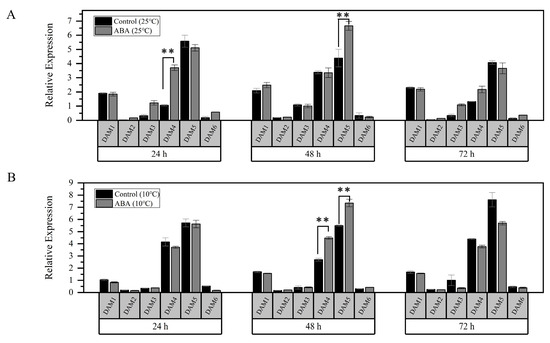
Figure 5.
Expression patterns of PavCIG1/2 downstream of gene PavDAM1-6 after ABA and low temperature treatments. (A) After ABA treatment, floral buds were collected at 24, 48, and 72 h under a temperature of 25 °C. The expressions of PavDAM1-6 were detected by qRT-PCR assay. (B) After ABA treatment, floral buds collected at 24, 48, and 72 h under a temperature of 10 °C were used to detect the expressions of PavDAM1-6. Error bars represent the standard deviation of three biological replicates. (** p ≤ 0.01, Student’s t-test).
3.6. Prediction of Protein–Protein Interactions Between PavABI5 and Other Proteins
To further explore the function of PavABI5, we constructed a protein interaction network using the STRING database (Figure 6A) [38]. As shown in Table 1, nearly 30 predicted proteins (Score > 0.7) were likely to interact with PavABI5. These proteins included ABI3/4, E3 ubiquitin-protein ligase KEG, serine/threonine-protein kinase SRK2I, E3 SUMO-protein ligase SIZ1, flowering time control protein FCA, bZIP transcription factor TGA10, and others. Then, we analyzed the expression patterns of these 30 genes using transcriptomic data from sweet cherry floral buds collected between July 2015 and February 2016 (Figure 6B) [39]. The results revealed that some genes, such as XP_021829418.1 (ABI3), XP_021808288.1 (FCA), and XP_021822943.1 (FUS3), were expressed at low levels during this period, while others, such as XP_021804893.1 (4A-2-like), XP_021813562.1 (SAPK3), and XP_021834231.1 (Cul4), were consistently expressed at high levels. Notably, similar expression patterns to that of PavABI5 were observed, including the TGA family genes TGA9 (XP_021829675.1), TGA10 (XP_021802713.1), and SLE1-like (XP_021827856.1). Additionally, the expression of E3 SUMO-protein ligase SIZ1 (XP_021831342.1) significantly increased during the winter period.
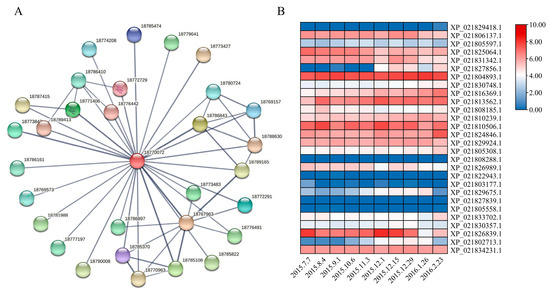
Figure 6.
The interaction prediction between PavABI5 and other proteins. (A) Prediction model diagram of the interaction between PavABI5 and other proteins through the STRING database. PavABI5 homologous protein PpABI5 (18770072, XP_020423436, Prunus persica) was used to predict the interaction proteins. (B) Seasonal expression patterns of predicted genes from 7 June 2015 to 23 February 2016 in the sweet cherry floral buds. Expression values for these genes were transformed by log2 (TPM+1). Red and white cells indicate relative higher or lower expression.

Table 1.
Predicted proteins interacting with PavABI5.
4. Discussion
ABA is a stress hormone and a central regulator of dormancy. Genes involved in ABA synthesis and catabolism play important roles in dormancy. In Arabidopsis, three NCED-related genes involved in ABA biosynthesis, NCED5, NCED6, and NCED9, have been proven to regulate seed development and dormancy [41,42]. For ABA catabolism, the expression of CYP707A genes reduces the concentration of ABA, promoting seed dormancy release during the maturation stage [43]. Low temperature can enhance dormancy by increasing ABA content and decreasing CYP707A expression in Arabidopsis seeds [44]. Similarly, ABA plays a key role in regulating bud dormancy [45,46]. It induces shoot growth cessation and initiates dormancy in autumn, with peak ABA concentrations observed in mid-winter, followed by a decline in December in apple buds [47]. In grape buds, ABA content begins to increase during para- and endodormancy, then decreases towards dormancy release [48]. The breaking of pear flower buds is inhibited after ABA applications, which affects the endodormancy induction [49]. In our research, the bud-break percentage of sweet cherries was decreased after ABA treatment compared to that of the control (Figure 3A). Recently, ABA content has been proposed as a determining factor for assessing dormancy status in sweet cherries [25,50], and genes involved in ABA-related pathways were central in the transcriptomic analysis of flower bud dormancy [39].
ABI5, a core component of the ABA signaling pathway, controls the dormancy process. The ABA receptors PYR/PYL/RCAR positively regulate seed dormancy by inhibiting the PP2C protein [51], while PP2Cs negatively regulate ABA signaling by interacting with SnRK2s and dephosphorylating them [52]. Downstream of SnRK2s, ABI5, participates in ABA-mediated seed dormancy [53,54]. Similar to other species, PavABI5 belongs to the bZIP family and contains conserved domains C1-4 (Figure 1) [55,56,57]. In gladiolus, GhABI5 is expressed in dormant organs (corm, cormel, stolon, stamen) and induced by drought and ABA [58]. In our results, the expression of PavABI5 reached a peak in mid-winter, and was strongly induced by exogenous ABA (Figure 3D and Figure 4A). The expression of PavABI5 is also affected by the sumoylation of SUMO E3 ligase SIZ1, an interaction protein (Figure 6) [59]. It is predicted that PavABI5 interacts with PavABI3 in the sweet cherries to regulate the bud-break. Meanwhile, ABI5 acts as a downstream gene of ABI3 in Arabidopsis [60]. The interactions of PavABI5 with PavFCA, PavTGA9, and PavTGA10 may be responsible for the delay in flowering time and flower development in sweet cherries [61,62].
Buds of perennial fruit trees typically enter a dormancy state during winter in order to survive the low temperatures. For perennial fruit trees such as pear and apple, dormancy induction is established by low temperature rather than by the photoperiod [5]. In the presence of ABA and low temperature, PavABI5 was expressed more rapidly and reached higher levels, whereas low temperature alone had minimal effects on its expression, suggesting that low temperature enhanced the expression of PavABI5 in an ABA-dependent manner (Figure 4). Banana MaABI5 interacts with MaC3HC4-1 to participate in the regulation of cold tolerance [29]. Moreover, ABI5 induces the expressions of ABA-responsive genes by binding to the ABA responsive elements (ABRE; ACGTGG/TC) in their promoter regions [32,59]. We found that the expressions of PavCIG1 and PavCIG2 were increased in response to exogenous ABA treatment compared with the results for the control under low temperature conditions (Figure 4B). In grapes, ABA treatment upregulates CBF1/2/3 transcripts, enhancing the freezing tolerance [63]. Similarly, the expression of cassava MeCBF1 is highly responsive to cold and ABA treatment [64]. Moreover, there were two ABRE elements in the promoter of the cold-induced gene PavCIG1 and four ABRE elements in the promoter of PavCIG2. Additional experiments proved the interactions between PavABI5 and PavCIG1/2 (Figure 2). In pears, the ABI5 homologous protein PpyABF3 activates the PpyCBF4 promoter by binding to its ABREs to regulate bud dormancy [36]. Previous research indicates that overexpressing the CBF gene results in dwarf plants, delayed bud-break, and increased cold tolerance [13,65,66,67]. Similar to the results for these species, the ectopic expression of sweet cherry PavCIG1 and PavCIG2 in Arabidopsis also delayed flowering [18]. DAMs, CBF downstream genes, are also regulated by the AREB transcription factor and function to inhibit bud growth and enhance bud dormancy [14,36,68,69]. In our results, we found that the expressions of PavDAM4 and PavDAM5 were significantly induced in response to exogenous ABA treatment under the temperatures of 25 °C and 10 °C (Figure 5). However, no direct interactions were observed between PavABI5 and PavDAM4/5 (Supplementary Figure S2). As a result, PavABI5 might be involved in regulating bud dormancy and cold tolerance via the CBF-mediated pathway in sweet cherries (Figure 7).
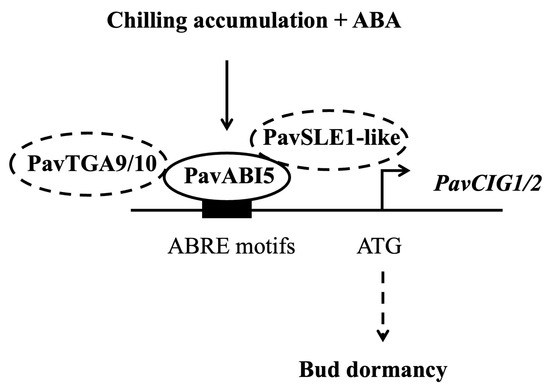
Figure 7.
A regulation model of PavABI5 in sweet cherry flower bud dormancy.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes16050596/s1, Figure S1: Yeast one-hybrid assay was performed to confirm the interactions between PavCIG1/2 and the PavDAM4/5 promoters; Figure S2: Interactions between PavABI5 and PavDAM4/5; Table S1: List of primer sequences used in this study.
Author Contributions
Conceptualization, C.Z.; methodology, J.W.; software, M.U.; validation, J.W. and L.W.; investigation, J.Z.; resources, S.J.; data curation, R.L.; writing—original draft, J.W. and L.W.; writing—review and editing, R.L.; funding acquisition, C.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (32102347), scientific research project of colleges and universities in Anhui Province (2022AH050408), the Shanghai Agriculture Applied Technology Development Program, China (G2022-02-08-00-12-F01111), and the China Agriculture Research System (CARS-30-2-08).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Arora, R.; Rowland, L.J.; Tanino, K. Induction and release of bud dormancy in woody perennials: A science comes of age. HortScience 2003, 38, 911–921. [Google Scholar] [CrossRef]
- Rodrigo, J. Spring frosts in deciduous fruit trees-morphological damage and flower hardiness. Sci. Hortic. 2000, 85, 155–173. [Google Scholar] [CrossRef]
- Salazar-Gutiérrez, M.R.; Chaves, B.; Hoogenboom, G. Freezing tolerance of apple flower buds. Sci. Hortic. 2016, 198, 344–351. [Google Scholar] [CrossRef]
- Kaya, O.; Kose, C.; Gecim, T. An exothermic process involved in the late spring frost injury to flower buds of some apricot cultivars (Prunus armenica L.). Sci. Hortic. 2018, 241, 322–328. [Google Scholar] [CrossRef]
- Heide, O.M.; Prestrud, A.K. Low temperature, but not photoperiod, controls growth cessation and dormancy induction and release in apple and pear. Tree Physiol. 2005, 25, 109–114. [Google Scholar] [CrossRef]
- Kovaleski, A.P. The potential for an increasing threat of unseasonal temperature cycles to dormant plants. New Phytol. 2024, 244, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Zeng, L.; Kong, D.; Mao, Y.; Xu, Y.; Wang, M.; Zhao, Y.; Jiang, C.; Zhang, Y.; Sun, D. Abscisic acid–induced transcription factor PsMYB306 negatively regulates tree peony bud dormancy release. Plant Physiol. 2024, 194, 2449–2471. [Google Scholar] [CrossRef]
- Zheng, C.; Halaly, T.; Acheampong, A.K.; Takebayashi, Y.; Jikumaru, Y.; Kamiya, Y.; Or, E. Abscisic acid (ABA) regulates grape bud dormancy, and dormancy release stimuli may act through modification of ABA metabolism. J. Exp. Bot. 2015, 66, 1527–1542. [Google Scholar] [CrossRef]
- Alburquerque, N.; García-Montiel, F.; Carrillo, A.; Burgos, L. Chilling and heat requirements of sweet cherry cultivars and the relationship between altitude and the probability of satisfying the chill requirements. Environ. Exp. Bot. 2008, 64, 162–170. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, L.; Ma, C.; Xu, W.; Liu, Z.; Zhang, C.; Matthew, D.W.; Wang, S. Impact of chilling accumulation and hydrogen cyanamide on floral organ development of sweet cherry in a warm region. J. Integr. Agr. 2016, 15, 2529–2538. [Google Scholar] [CrossRef]
- Artlip, T.; McDermaid, A.; Ma, Q.; Wisniewski, M. Differential gene expression in non-transgenic and transgenic “M. 26” apple overexpressing a peach CBF gene during the transition from eco-dormancy to bud break. Hortic. Res. 2019, 6, 86. [Google Scholar] [CrossRef] [PubMed]
- Barros, P.M. Insights into the Role of Almond CBF Transcription Factors in the Environmental Control of Cold Acclimation and Dormancy Break. Doctoral Dissertation, Universidade NOVA de Lisboa, Lisbon, Portugal, 2012. [Google Scholar]
- Artlip, T.S.; Wisniewski, M.E.; Norelli, J.L. Field evaluation of apple overexpressing a peach CBF gene confirms its effect on cold hardiness, dormancy, and growth. Environ. Exp. Bot. 2014, 106, 79–86. [Google Scholar] [CrossRef]
- Niu, Q.; Li, J.; Cai, D.; Qian, M.; Jia, H.; Bai, S.; Hussain, S.; Liu, G.; Teng, Y.; Zheng, X. Dormancy-associated MADS-box genes and microRNAs jointly control dormancy transition in pear (Pyrus pyrifolia white pear group) flower bud. J. Exp. Bot. 2016, 67, 239–257. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yan, X.; Yang, Q.; Ma, Y.; Yang, B.; Tian, J.; Teng, Y.; Bai, S. PpCBFs selectively regulate PpDAMs and contribute to the pear bud endodormancy process. Plant Mol. Biol. 2019, 99, 575–586. [Google Scholar] [CrossRef]
- Park, S.; Lee, C.M.; Doherty, C.J.; Gilmour, S.J.; Kim, Y.; Thomashow, M.F. Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J. 2015, 82, 193–207. [Google Scholar] [CrossRef]
- Artlip, T.S.; Wisniewski, M.E.; Arora, R.; Norelli, J.L. An apple rootstock overexpressing a peach CBF gene alters growth and flowering in the scion but does not impact cold hardiness or dormancy. Hortic. Res. 2016, 3, 16006. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X.; Sun, W.; Xu, Y.; Sabir, I.A.; Abdullah, M.; Wang, S.; Jiu, S.; Zhang, C. Cold induced genes (CIGs) regulate flower development and dormancy in Prunus avium L. Plant Sci. 2021, 313, 111061. [Google Scholar] [CrossRef]
- Zhao, K.; Zhou, Y.; Ahmad, S.; Yong, X.; Xie, X.; Han, Y.; Li, Y.; Zhang, Q. PmCBFs synthetically affect PmDAM6 by alternative promoter binding and protein complexes towards the dormancy of bud for Prunus mume. Sci. Rep. 2018, 8, 4527. [Google Scholar] [CrossRef]
- Wang, J.; Gao, Z.; Li, H.; Jiu, S.; Qu, Y.; Wang, L.; Ma, C.; Xu, W.; Wang, S.; Zhang, C. Dormancy-associated MADS-Box (DAM) genes influence chilling requirement of sweet cherries and co-regulate flower development with SOC1 gene. Int. J. Mol. Sci. 2020, 21, 921. [Google Scholar] [CrossRef]
- Li, Z.; Reighard, G.L.; Abbott, A.G.; Bielenberg, D.G. Dormancy-associated MADS genes from the EVG locus of peach [Prunus persica (L.) Batsch] have distinct seasonal and photoperiodic expression patterns. J. Exp. Bot. 2009, 60, 3521–3530. [Google Scholar] [CrossRef]
- Hu, C.; Wang, M.; Zhu, C.; Wu, S.; Li, J.; Yu, J.; Hu, Z. A transcriptional regulation of ERF15 contributes to ABA-mediated cold tolerance in tomato. Plant Cell Environ. 2024, 47, 1334–1347. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Zareen, S.; Park, J.; Khan, H.A.; Lim, C.J.; Bader, Z.E.; Hussain, S.; Chung, W.S.; Gechev, T.; Pardo, J.M.; et al. ABA INSENSITIVE 2 promotes flowering by inhibiting OST1/ABI5-dependent FLOWERING LOCUS C transcription in Arabidopsis. J. Exp. Bot. 2024, 75, 2481–2493. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, L.; Sun, W.; Wang, L.; Liu, X.; Jiu, S.; Liu, R.; Zhang, C. N6-methyladenosine RNA methylation is important for dormancy release in sweet cherry. Sci. Hortic. 2024, 338, 113725. [Google Scholar] [CrossRef]
- Vimont, N.; Schwarzenberg, A.; Domijan, M.; Donkpegan, A.S.; Beauvieux, R.; Le, D.L.; Arkoun, M.; Jamois, F.; Yvin, J.; Wigge, A.P.; et al. Fine tuning of hormonal signaling is linked to dormancy status in sweet cherry flower buds. Tree Physiol. 2021, 41, 544–561. [Google Scholar] [CrossRef]
- Zheng, C.; Acheampong, A.K.; Shi, Z.; Mugzech, A.; Halaly-Basha, T.; Shaya, F.; Sun, Y.; Colova, V.; Mosquna, A.; Ophir, R.; et al. Abscisic acid catabolism enhances dormancy release of grapevine buds. Plant Cell Environ. 2018, 41, 2490–2503. [Google Scholar] [CrossRef]
- Finkelstein, R.R.; Lynch, T.J. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 2000, 12, 599–609. [Google Scholar] [CrossRef] [PubMed]
- Mittal, A.; Gampala, S.S.; Ritchie, G.L.; Payton, P.; Burke, J.J.; Rock, C.D. Related to ABA-Insensitive3 (ABI3)/Viviparous1 and AtABI5 transcription factor coexpression in cotton enhances drought stress adaptation. Plant Biotechnol J. 2014, 12, 578–589. [Google Scholar]
- Chen, J.; Li, Y.; Li, F.; Wu, Q.; Jiang, Y.; Yuan, D. Banana MaABI5 is involved in ABA-induced cold tolerance through interaction with a RING E3 ubiquitin ligase; MaC3HC4-1. Sci. Hortic. 2018, 237, 239–246. [Google Scholar] [CrossRef]
- Zhao, H.; Nie, K.; Zhou, H.; Yan, X.; Zhan, Q.; Zheng, Y.; Song, C.P. ABI5 modulates seed germination via feedback regulation of the expression of the PYR/PYL/RCAR ABA receptor genes. New Phytol. 2020, 228, 596–608. [Google Scholar] [CrossRef]
- Han, Y.; Wang, Z.; Han, B.; Zhang, Y.; Liu, J.; Yang, Y. Allelic variation of TaABI5-A4 significantly affects seed dormancy in bread wheat. Theor. Appl. Genet. 2024, 137, 240. [Google Scholar] [CrossRef]
- Carles, C.; Bies-Etheve, N.; Aspart, L.; Léon-Kloosterziel, K.M.; Koornneef, M.; Echeverria, M.; Delseny, M. Regulation of Arabidopsis thaliana Em genes: Role of ABI5. Plant J. 2002, 30, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yang, Y.N.; Xue, L.J.; Zou, M.J.; Liu, J.Y.; Chen, F.; Xue, H.W. Rice ABI5-Like1 regulates abscisic acid and auxin responses by affecting the expression of ABRE-containing genes. Plant Physiol. 2011, 156, 1397–1409. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Deng, W.; Wang, X.; Yang, C.; Li, Z. Maize (Zea mays L.) homologue of ABA-insensitive (ABI) 5 gene plays a negative regulatory role in abiotic stresses response. Plant Growth Regul. 2012, 68, 383–393. [Google Scholar] [CrossRef]
- Rubio, S.; Noriega, X.; Pérez, F.J. Abscisic acid (ABA) and low temperatures synergistically increase the expression of CBF/DREB1 transcription factors and cold-hardiness in grapevine dormant buds. Ann. Bot. 2019, 123, 681–689. [Google Scholar] [CrossRef]
- Yang, Q.; Yang, B.; Li, J.; Wang, Y.; Tao, R.; Yang, F.; Wu, X.; Yan, X.; Ahmad, M.; Shen, J.; et al. ABA-responsive ABRE-BINDING FACTOR3 activates DAM3 expression to promote bud dormancy in Asian pear. Plant Cell Environ. 2020, 43, 1360–1375. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein–protein interaction networks.; integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Vimont, N.; Fouché, M.; Campoy, J.A.; Tong, M.; Arkoun, M.; Yvin, J.C.; Wigge, P.A.; Dirlewanger, E.; Cortijo, S.; Wenden, B. From bud formation to flowering: Transcriptomic state defines the cherry developmental phases of sweet cherry bud dormancy. BMC Genom. 2019, 20, 974. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lefebvre, V.; North, H.; Frey, A.; Sotta, B.; Seo, M.; Okamoto, M.; Nambara, E.; Marion-Poll, A. Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 2006, 45, 309–319. [Google Scholar] [CrossRef]
- Frey, A.; Effroy, D.; Lefebvre, V.; Seo, M.; Perreau, F.; Berger, A.; Sechet, J.; To, A.; North, H.M.; Marion-Poll, A. Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J. 2012, 70, 501–512. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, M.; Kuwahara, A.; Seo, M.; Kushiro, T.; Asami, T.; Hirai, N.; Kamiya, Y.; Koshiba, T.; Nambara, E. CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases.; are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 2006, 141, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Kendall, S.L.; Hellwege, A.; Marriot, P.; Whalley, C.; Graham, I.A.; Penfield, S. Induction of dormancy in Arabidopsis summer annuals requires parallel regulation of DOG1 and hormone metabolism by low temperature and CBF transcription factors. Plant Cell 2011, 23, 2568–2580. [Google Scholar] [CrossRef]
- Cooke, J.E.; Eriksson, M.E.; Junttila, O. The dynamic nature of bud dormancy in trees: Environmental control and molecular mechanisms. Plant Cell Environ. 2012, 35, 1707–1728. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Gao, J.; Im, K.J.; Chen, K.; Bressan, R.A.; Zhu, J.K. Control of plant water use by ABA induction of senescence and dormancy: An overlooked lesson from evolution. Plant Cell Physiol. 2017, 58, 1319–1327. [Google Scholar] [CrossRef]
- Guak, S.; Fuchigami, L.H. Effects of applied ABA on growth cessation, bud dormancy, cold acclimation, leaf senescence and N mobilization in apple nursery plants. J. Hortic. Sci. Biotech 2001, 76, 459–464. [Google Scholar] [CrossRef]
- Or, E.; Belausov, E.; Popilevsky, I.; Bental, Y. Changes in endogenous ABA level in relation to the dormancy cycle in grapevines grown in a hot climate. J. Hortic. Sci. Biotech 2000, 75, 190–194. [Google Scholar] [CrossRef]
- Li, J.; Xu, Y.; Niu, Q.; He, L.; Teng, Y.; Bai, S. Abscisic acid (ABA) promotes the induction and maintenance of pear (Pyrus pyrifolia white pear group) flower bud endodormancy. Int. J. Mol. Sci. 2018, 19, 310. [Google Scholar] [CrossRef]
- Chmielewski, F.M.; Gotz, K.; Homann, T.; Huschek, G.; Rawel, H. Identification of endodormancy release for cherries (Prunus avium L.) by abscisic acid and sugars. J. Hortic. 2017, 4, 1000210. [Google Scholar]
- Park, S.Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.-F.F.; et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef]
- Vlad, F.; Rubio, S.; Rodrigues, A.; Sirichandra, C.; Belin, C.; Robert, N.; Leung, J.; Rodriguez, P.L.; Laurière, C.; Merlot, S. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 2009, 21, 3170–3184. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Chen, S.; Yang, Y.; An, C. ABA-insensitive (ABI) 4 and ABI5 synergistically regulate DGAT1 expression in Arabidopsis seedlings under stress. FEBS Lett. 2013, 587, 3076–3082. [Google Scholar] [CrossRef] [PubMed]
- Utsugi, S.; Ashikawa, I.; Nakamura, S.; Shibasaka, M. TaABI5, a wheat homolog of Arabidopsis thaliana ABA insensitive 5, controls seed germination. J. Plant Res. 2020, 133, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Bensmihen, S.; Rippa, S.; Lambert, G.; Jublot, D.; Pautot, V.; Granier, F.; Giraudat, J.; Parcy, F. The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 2002, 14, 1391–1403. [Google Scholar] [CrossRef]
- Skubacz, A.; Daszkowska-Golec, A.; Szarejko, I. The role and regulation of ABI5 (ABA-Insensitive 5) in plant development.; abiotic stress responses and phytohormone crosstalk. Front. Plant Sci. 2016, 7, 1884. [Google Scholar] [CrossRef]
- Wang, Y.H.; Que, F.; Li, T.; Zhang, R.R.; Khadr, A.; Xu, Z.S.; Tian, Y.S.; Xiong, A.S. DcABF3, an ABF transcription factor from carrot.; alters stomatal density and reduces ABA sensitivity in transgenic Arabidopsis. Plant Sci. 2021, 302, 110699. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Seng, S.; Sui, J.; Vonaartis, E.; Luo, X.; Gong, B.; Liu, C.; Wu, C.; Liu, C.; Zhang, F.; et al. Gladiolus hybridus ABSCISIC ACID INSENSITIVE 5 (GhABI5) is an important transcription factor in ABA signaling that can enhance Gladiolus corm dormancy and Arabidopsis seed dormancy. Front. Plant Sci. 2015, 6, 960. [Google Scholar] [CrossRef]
- Miura, K.; Lee, J.; Jin, J.B.; Yoo, C.Y.; Miura, T.; Hasegawa, P.M. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 5418–5423. [Google Scholar] [CrossRef]
- Lopez-Molina, L.; Mongrand, S.; McLachlin, D.T.; Chait, B.T.; Chua, N.H. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 2002, 32, 317–328. [Google Scholar] [CrossRef]
- Wang, Y.; Tao, Z.; Wang, W.; Filiault, D.; Qiu, C.; Wang, C.; Wang, H.; Rehman, S.; Shi, J.; Zhang, Y.; et al. Molecular variation in a functionally divergent homolog of FCA regulates flowering time in Arabidopsis thaliana. Nat. Commun. 2020, 11, 5830. [Google Scholar] [CrossRef]
- Murmu, J.; Bush, M.J.; DeLong, C.; Li, S.; Xu, M.; Khan, M.; Malcolmson, C.; Fobert, P.R.; Zachgo, S.; Hepworth, S.R. Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for anther development. Plant Physiol. 2010, 154, 1492–1504. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Siddiqua, M.; Braybrook, S.; Nassuth, A. Three grape CBF/DREB1 genes respond to low temperature, drought and abscisic acid. Plant Cell Environ. 2006, 29, 1410–1421. [Google Scholar] [CrossRef]
- An, D.; Ma, Q.; Wang, H.; Yang, J.; Zhou, W.; Zhang, P. Cassava C-repeat binding factor 1 gene responds to low temperature and enhances cold tolerance when overexpressed in Arabidopsis and cassava. Plant Mol. Biol. 2017, 94, 109–124. [Google Scholar] [CrossRef] [PubMed]
- Siddiqua, M.; Nassuth, A. Vitis CBF1 and Vitis CBF4 differ in their effect on Arabidopsis abiotic stress tolerance.; development and gene expression. Plant Cell Environ. 2011, 34, 1345–1359. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, J.; Guo, H.; He, X.; Wu, W.; Du, J.; Zhang, Z.; An, X. Characterization of two highly similar CBF/DREB1-like genes, PhCBF4a and PhCBF4b, in Populus hopeiensis. Plant Physiol. Bioch. 2014, 83, 107–116. [Google Scholar] [CrossRef]
- Wisniewski, M.; Norelli, J.; Artlip, T. Overexpression of a peach CBF gene in apple: A model for understanding the integration of growth, dormancy, and cold hardiness in woody plants. Front. Plant Sci. 2015, 6, 85. [Google Scholar] [CrossRef]
- Tuan, P.A.; Bai, S.; Saito, T.; Ito, A.; Moriguchi, T. Dormancy-Associated MADS-Box (DAM) and the abscisic acid pathway regulate pear endodormancy through a feedback mechanism. Plant Cell Physiol. 2017, 58, 1378–1390. [Google Scholar] [CrossRef]
- Yamane, H.; Wada, M.; Honda, C.; Matsuura, T.; Ikeda, Y.; Hirayama, T.; Osako, Y.; Gao-Takai, M.; Kojima, M.; Sakakibara, H.; et al. Overexpression of Prunus DAM6 inhibits growth.; represses bud break competency of dormant buds and delays bud outgrowth in apple plants. PLoS ONE 2019, 14, e0214788. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).