Reduced Genetic Diversity of Key Fertility and Vector Competency Related Genes in Anopheles gambiae s.l. Across Sub-Saharan Africa
Abstract
1. Introduction
2. Results
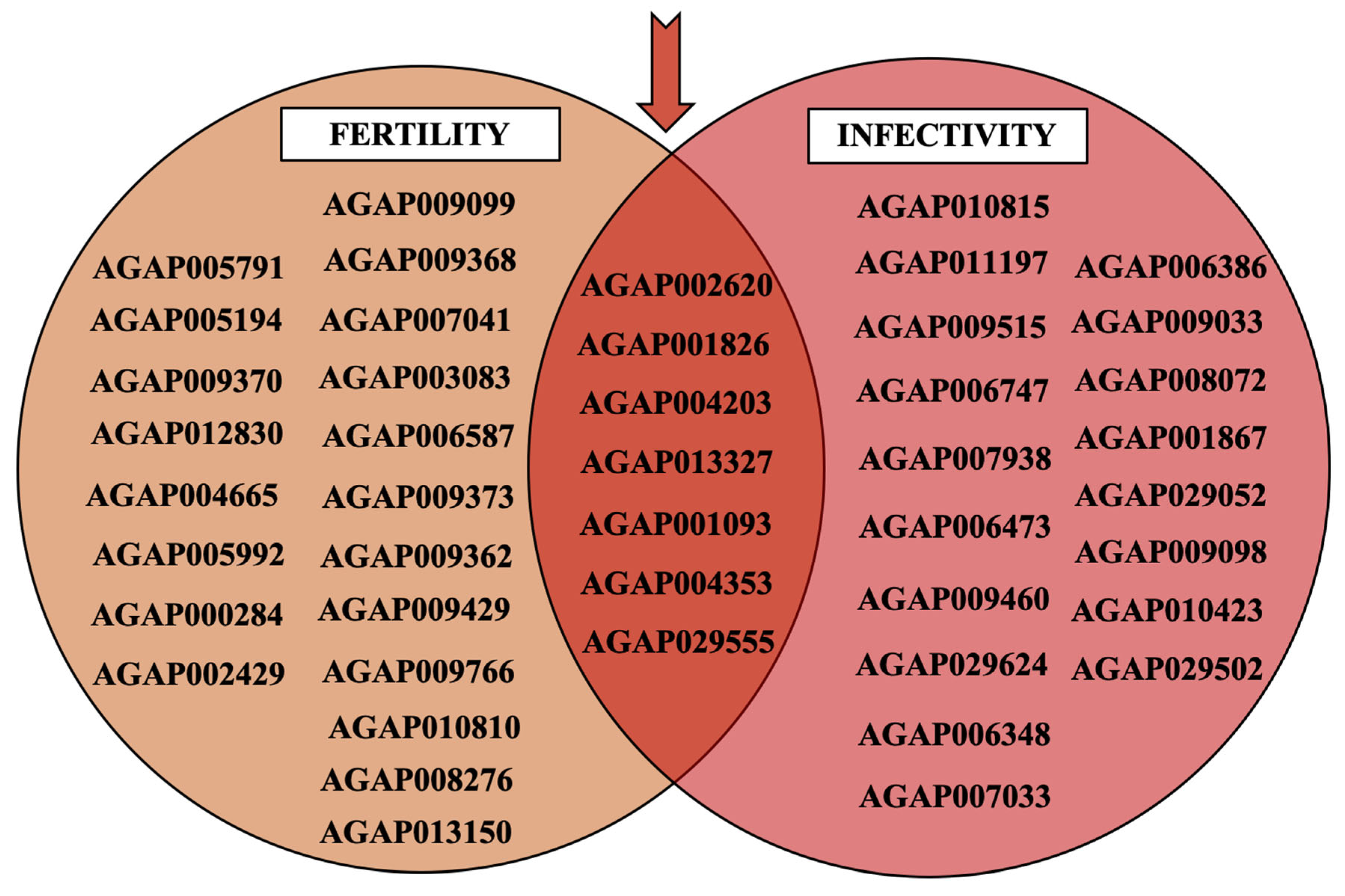
2.1. Potential Targetable Genes Involved in Anopheles gambiae s.l. Fertility and Plasmodium Infectivity
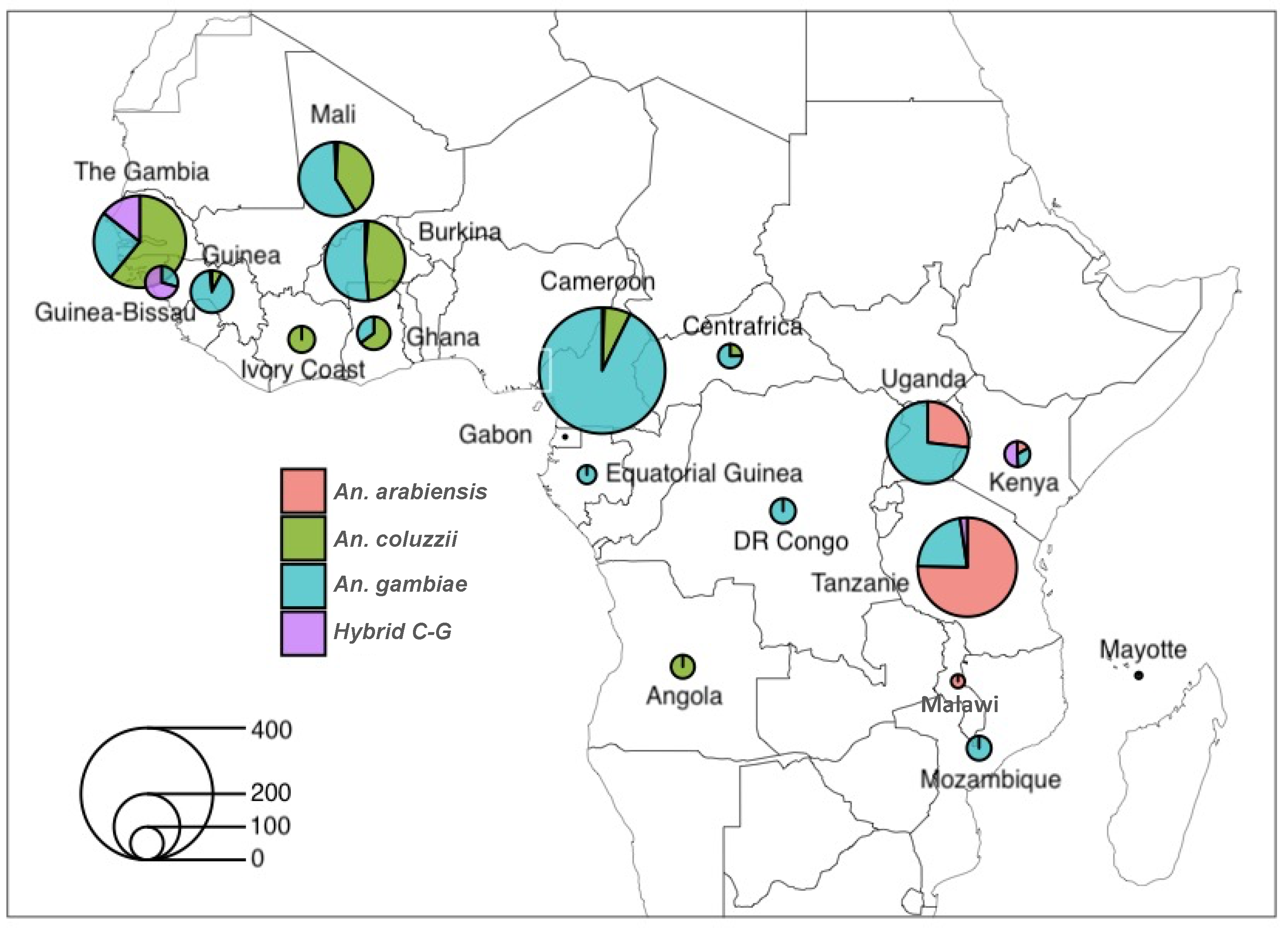
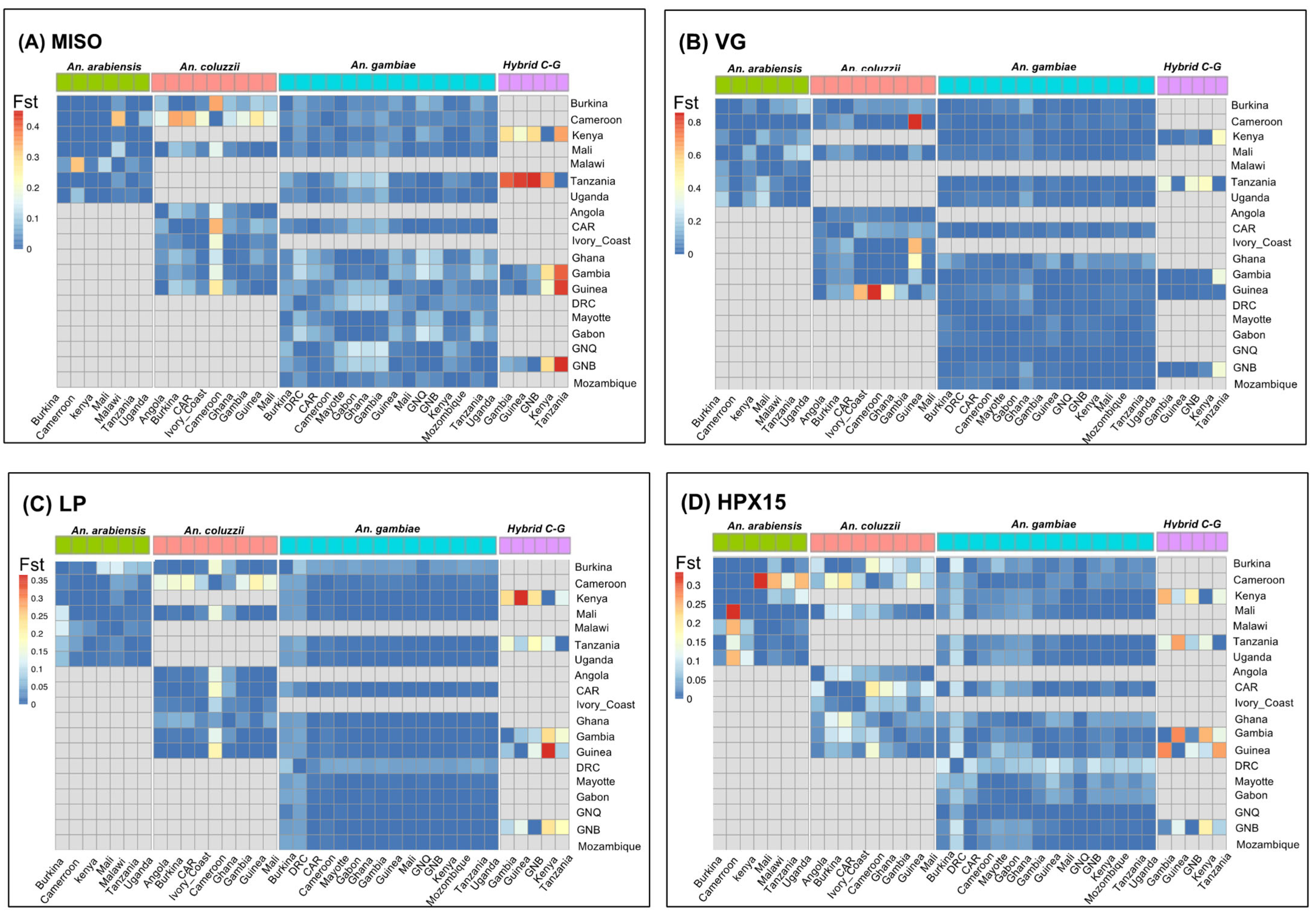
2.2. Low Population Differentiation in the Four Targetable Genes Involved in Anopheles gambiae s.l. Fertility and Plasmodium Infectivity
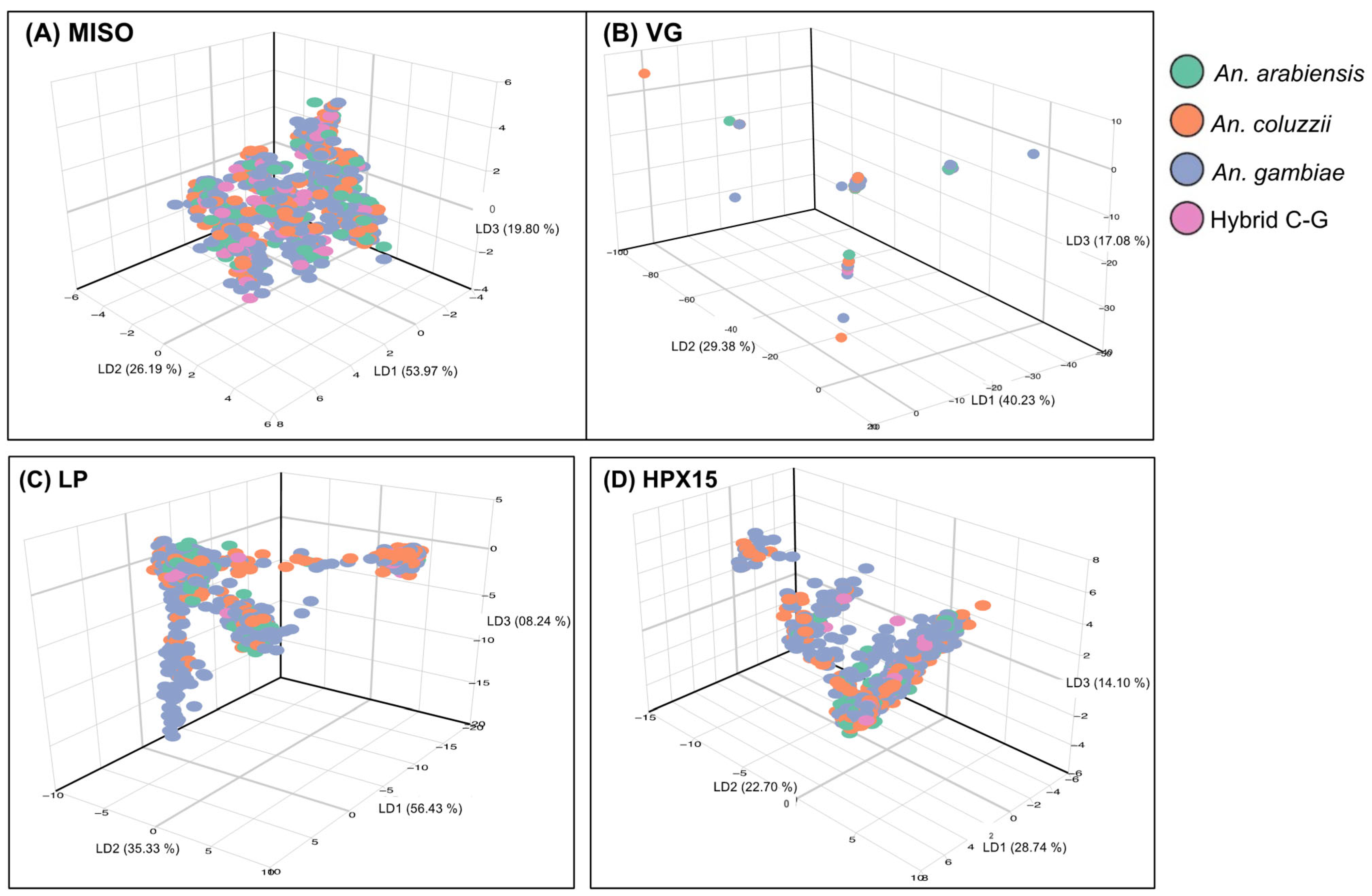
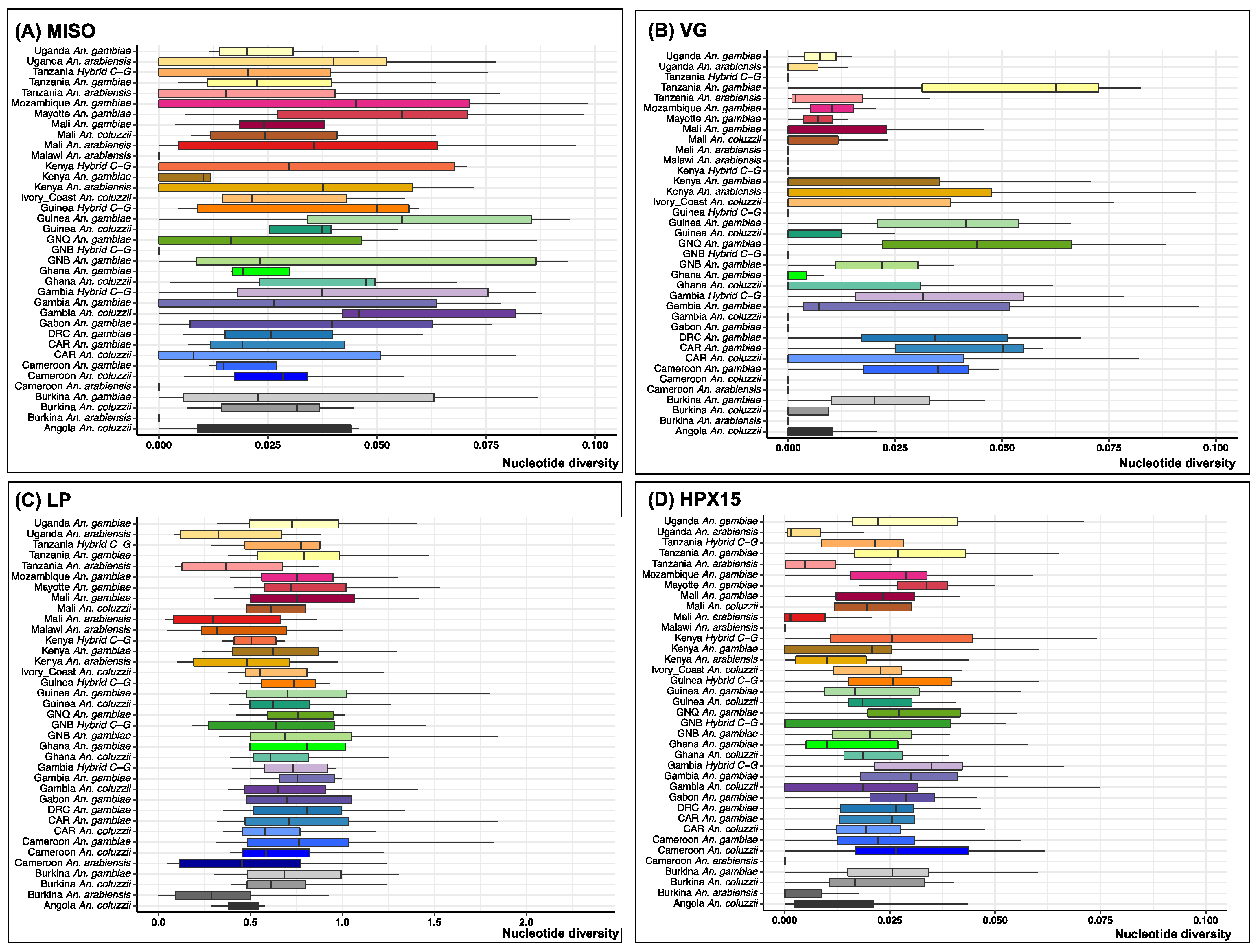
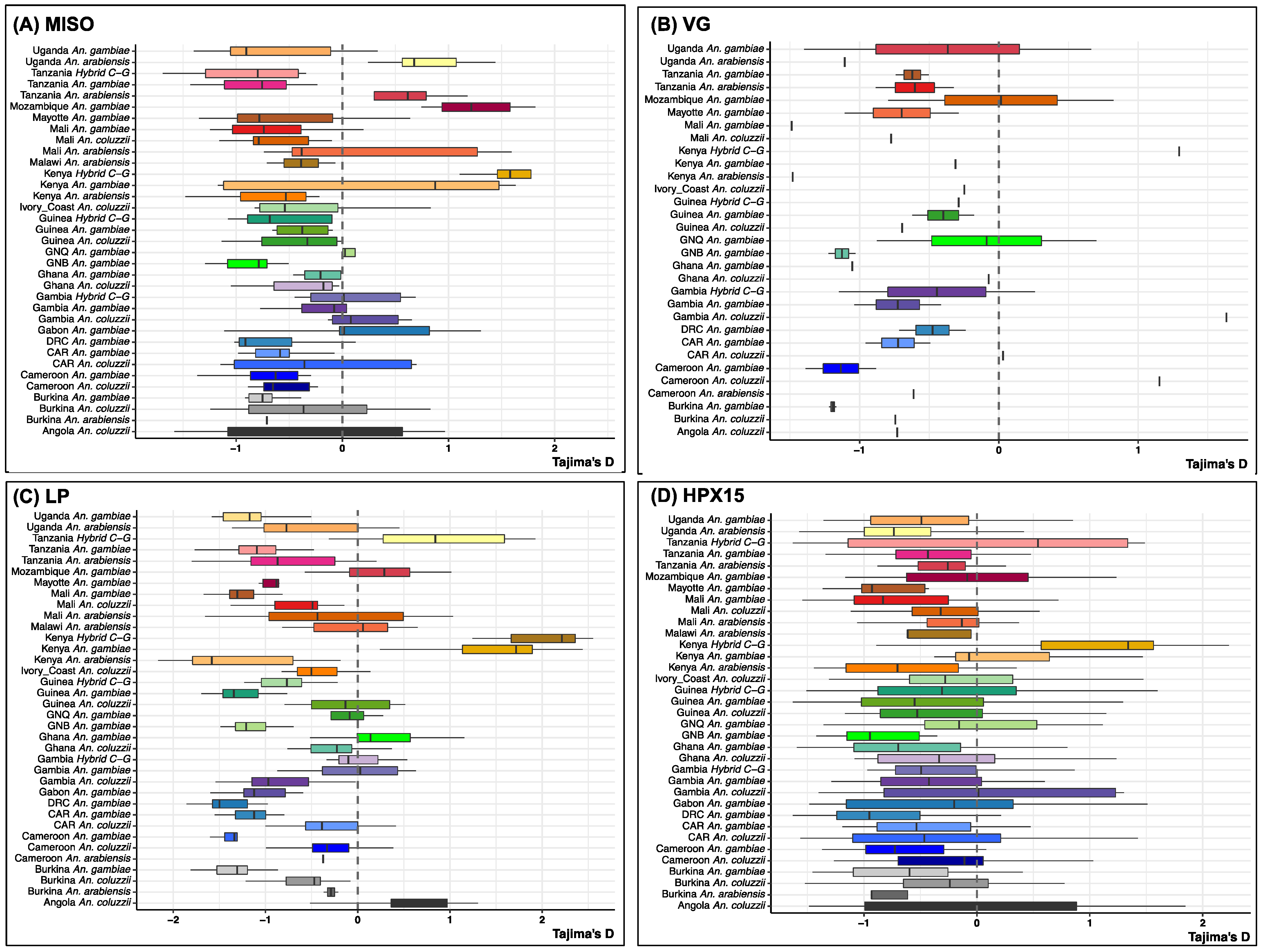
2.3. Low Genetic Diversity of the Selected Targetable Genes
3. Discussion
4. Conclusions
5. Methods
5.1. Selection of Potential Targetable Genes Involved in Anopheles gambiae s.s PEST Fertility and Plasmodium Infectivity
5.2. SNP Data Used
5.3. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. World Malaria Report 2024—Addressing Inequity in the Global Malaria Response. Available online: https://www.who.int/publications/i/item/9789240104440 (accessed on 3 February 2025).
- Bhatt, S.; Weiss, D.J.; Cameron, E.; Bisanzio, D.; Mappin, B.; Dalrymple, U.; Battle, K.; Moyes, C.L.; Henry, A.; Eckhoff, P.A.; et al. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature 2015, 526, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Ranson, H.; N’guessan, R.; Lines, J.; Moiroux, N.; Nkuni, Z.; Corbel, V. Pyrethroid resistance in African anopheline mosquitoes: What are the implications for malaria control? Trends Parasitol. 2011, 27, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Ranson, H.; Lissenden, N. Insecticide Resistance in African Anopheles Mosquitoes: A Worsening Situation that Needs Urgent Action to Maintain Malaria Control. Trends Parasitol. 2016, 32, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Hemingway, J.; Ranson, H.; Magill, A.; Kolaczinski, J.; Fornadel, C.; Gimnig, J.; Coetzee, M.; Simard, F.; Roch, D.K.; Hinzoumbe, C.K.; et al. Averting a malaria disaster: Will insecticide resistance derail malaria control? Lancet 2016, 387, 1785–1788. [Google Scholar] [CrossRef]
- Riveron, J.M.; Tchouakui, M.; Mugenzi, L.; Menze, B.D.; Chiang, M.-C.; Wondji, C.S. Insecticide Resistance in Malaria Vectors: An Update at a Global Scale. In Towards Malaria Elimination—A Leap Forward; IntechOpen: London, UK, 2018. [Google Scholar] [CrossRef]
- Hancock, P.A.; Hendriks, C.J.M.; Tangena, J.-A.; Gibson, H.; Hemingway, J.; Coleman, M.; Gething, P.W.; Cameron, E.; Bhatt, S.; Moyes, C.L. Mapping trends in insecticide resistance phenotypes in African malaria vectors. PLoS Biol. 2020, 18, e3000633. [Google Scholar] [CrossRef]
- Sougoufara, S.; Doucouré, S.; Backé Sembéne, P.M.; Harry, M.; Sokhna, C. Challenges for malaria vector control in sub-Saharan Africa: Resistance and behavioral adaptations in Anopheles populations. J. Vector Borne Dis. 2017, 54, 4–15. [Google Scholar] [CrossRef]
- Sougoufara, S.; Ottih, E.C.; Tripet, F. The need for new vector control approaches targeting outdoor biting Anopheline malaria vector communities. Parasit. Vectors 2020, 13, 295. [Google Scholar] [CrossRef]
- Sanou, A.; Nelli, L.; Guelbéogo, W.M.; Cissé, F.; Tapsoba, M.; Ouédraogo, P.; Sagnon, N.; Ranson, H.; Matthiopoulos, J.; Ferguson, H.M. Insecticide resistance and behavioural adaptation as a response to long-lasting insecticidal net deployment in malaria vectors in the Cascades region of Burkina Faso. Sci. Rep. 2021, 11, 17569. [Google Scholar] [CrossRef]
- Mashatola, T.; Ndo, C.; Koekemoer, L.L.; Dandalo, L.C.; Wood, O.R.; Malakoane, L.; Poumachu, Y.; Lobb, L.N.; Kaiser, M.; Bourtzis, K.; et al. A review on the progress of sex-separation techniques for sterile insect technique applications against Anopheles arabiensis. Parasit. Vectors 2018, 11, 646. [Google Scholar] [CrossRef]
- Yao, F.A.; Millogo, A.-A.; Epopa, P.S.; North, A.; Noulin, F.; Dao, K.; Drabo, M.; Guissou, C.; Kekele, S.; Namountougou, M.; et al. Mark-release-recapture experiment in Burkina Faso demonstrates reduced fitness and dispersal of genetically-modified sterile malaria mosquitoes. Nat. Commun. 2022, 13, 796. [Google Scholar] [CrossRef]
- Atella, G.C.; Silva-Neto, M.A.C.; Golodne, D.M.; Arefin, S.; Shahabuddin, M. Anopheles gambiae lipophorin: Characterization and role in lipid transport to developing oocyte. Insect Biochem. Mol. Biol. 2006, 36, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Rogers, D.W.; Whitten, M.M.A.; Thailayil, J.; Soichot, J.; Levashina, E.A.; Catteruccia, F. Molecular and cellular components of the mating machinery in Anopheles gambiae females. Proc. Natl. Acad. Sci. USA 2008, 105, 19390–19395. [Google Scholar] [CrossRef] [PubMed]
- Rono, M.K.; Whitten, M.M.A.; Oulad-Abdelghani, M.; Levashina, E.A.; Marois, E. The major yolk protein vitellogenin interferes with the anti-plasmodium response in the malaria mosquito Anopheles gambiae. PLoS Biol. 2010, 8, e1000434. [Google Scholar] [CrossRef] [PubMed]
- Baldini, F.; Gabrieli, P.; Rogers, D.W.; Catteruccia, F. Function and composition of male accessory gland secretions in Anopheles gambiae: A comparison with other insect vectors of infectious diseases. Pathog. Glob. Health 2012, 106, 82–93. [Google Scholar] [CrossRef]
- Baldini, F.; Gabrieli, P.; South, A.; Valim, C.; Mancini, F.; Catteruccia, F. The interaction between a sexually transferred steroid hormone and a female protein regulates oogenesis in the malaria mosquito Anopheles gambiae. PLoS Biol. 2013, 11, e1001695. [Google Scholar] [CrossRef]
- Le, B.V.; Nguyen, J.B.; Logarajah, S.; Wang, B.; Marcus, J.; Williams, H.P.; Catteruccia, F.; Baxter, R.H.G. Characterization of Anopheles gambiae transglutaminase 3 (AgTG3) and its native substrate Plugin. J. Biol. Chem. 2013, 288, 4844–4853. [Google Scholar] [CrossRef]
- Gabrieli, P.; Kakani, E.G.; Mitchell, S.N.; Mameli, E.; Want, E.J.; Mariezcurrena Anton, A.; Serrao, A.; Baldini, F.; Catteruccia, F. Sexual transfer of the steroid hormone 20E induces the postmating switch in Anopheles gambiae. Proc. Natl. Acad. Sci. USA 2014, 111, 16353–16358. [Google Scholar] [CrossRef]
- Shaw, W.R.; Teodori, E.; Mitchell, S.N.; Baldini, F.; Gabrieli, P.; Rogers, D.W.; Catteruccia, F. Mating activates the heme peroxidase HPX15 in the sperm storage organ to ensure fertility in Anopheles gambiae. Proc. Natl. Acad. Sci. USA 2014, 111, 5854–5859. [Google Scholar] [CrossRef]
- Kajla, M.; Choudhury, T.P.; Kakani, P.; Gupta, K.; Dhawan, R.; Gupta, L.; Kumar, S. Silencing of Anopheles stephensi Heme Peroxidase HPX15 Activates Diverse Immune Pathways to Regulate the Growth of Midgut Bacteria. Front. Microbiol. 2016, 7, 1351. [Google Scholar] [CrossRef]
- Kajla, M.; Kakani, P.; Choudhury, T.P.; Kumar, V.; Gupta, K.; Dhawan, R.; Gupta, L.; Kumar, S. Anopheles stephensi Heme Peroxidase HPX15 Suppresses Midgut Immunity to Support Plasmodium Development. Front. Immunol. 2017, 8, 249. [Google Scholar] [CrossRef]
- Cui, Y.; Niu, G.; Li, V.L.; Wang, X.; Li, J. Analysis of blood-induced Anopheles gambiae midgut proteins and sexual stage Plasmodium falciparum interaction reveals mosquito genes important for malaria transmission. Sci. Rep. 2020, 10, 14316. [Google Scholar] [CrossRef] [PubMed]
- Marcenac, P.; Shaw, W.R.; Kakani, E.G.; Mitchell, S.N.; South, A.; Werling, K.; Marrogi, E.; Abernathy, D.G.; Yerbanga, R.S.; Dabiré, R.K.; et al. A mating-induced reproductive gene promotes Anopheles tolerance to Plasmodium falciparum infection. PLoS Pathog. 2020, 16, e1008908. [Google Scholar] [CrossRef] [PubMed]
- Le, B.V.; Klöck, C.; Schatz, A.; Nguyen, J.B.; Kakani, E.G.; Catteruccia, F.; Khosla, C.; Baxter, R.H.G. Dihydroisoxazole inhibitors of Anopheles gambiae seminal transglutaminase AgTG3. Malar. J. 2014, 13, 210. [Google Scholar] [CrossRef]
- Bhattacharya, R.; Rose, P.; Burley, S.; Prlić, A. Impact of genetic variation on three dimensional structure and function of proteins. PLoS ONE 2017, 12, e0171355. [Google Scholar] [CrossRef]
- Anopheles Gambiae 1000 Genomes Consortium; Data Analysis Group; Partner Working Group. Genetic diversity of the African malaria vector Anopheles gambiae. Nature 2017, 552, 96–100. [Google Scholar] [CrossRef]
- Amambua-Ngwa, A.; Amenga-Etego, L.; Kamau, E.; Amato, R.; Ghansah, A.; Golassa, L.; Randrianarivelojosia, M.; Ishengoma, D.; Apinjoh, T.; Maïga-Ascofaré, O.; et al. Major subpopulations of Plasmodium falciparum in sub-Saharan Africa. Science 2019, 365, 813–816. [Google Scholar] [CrossRef]
- Anopheles gambiae 1000 Genomes Consortium. Genome variation and population structure among 1142 mosquitoes of the African malaria vector species Anopheles gambiae and Anopheles coluzzii. Genome Res. 2020, 30, 1533–1546. [Google Scholar] [CrossRef]
- Diallo, M.; Hamid-Adiamoh, M.; Sy, O.; Sarr, P.C.; Manneh, J.; Ndiath, M.O.; Gaye, O.; Faye, O.; Konaté, L.; Sesay, A.K.; et al. Evolution of the Pyrethroids Target-Site Resistance Mechanisms in Senegal: Early Stage of the Vgsc-1014F and Vgsc-1014S Allelic Frequencies Shift. Genes 2021, 12, 1948. [Google Scholar] [CrossRef]
- Diallo, M.; Kolley, E.S.; Dia, A.K.; Oboh, M.A.; Seck, F.; Manneh, J.; Sesay, A.K.; Diédhiou, S.M.; Sarr, P.C.; Sy, O.; et al. Evolution of the Ace-1 and Gste2 Mutations and Their Potential Impact on the Use of Carbamate and Organophosphates in IRS for Controlling Anopheles gambiae s.l., the Major Malaria Mosquito in Senegal. Pathogens 2022, 11, 1021. [Google Scholar] [CrossRef]
- Ishengoma, D.S.; Saidi, Q.; Sibley, C.H.; Roper, C.; Alifrangis, M. Deployment and utilization of next-generation sequencing of Plasmodium falciparum to guide anti-malarial drug policy decisions in sub-Saharan Africa: Opportunities and challenges. Malar. J. 2019, 18, 267. [Google Scholar] [CrossRef]
- Höglund, J. Genetic diversity in changing environments. In Evolutionary Conservation Genetics; Höglund, J., Ed.; Oxford University Press: Oxford, UK, 2009; pp. 60–80. [Google Scholar] [CrossRef]
- Cardoso, J.G.R.; Andersen, M.R.; Herrgård, M.J.; Sonnenschein, N. Analysis of genetic variation and potential applications in genome-scale metabolic modeling. Front. Bioeng. Biotechnol. 2015, 3, 13. [Google Scholar] [CrossRef] [PubMed]
- Birader, K. Genetic Diversity and the Adaptation of Species to Changing Environments. J. Biodivers. Endanger. Species 2023, 11, 3. [Google Scholar] [CrossRef]
- Costantini, C.; Sagnon, N.; della Torre, A.; Coluzzi, M. Mosquito behavioural aspects of vector-human interactions in the Anopheles gambiae complex. Parassitologia 1999, 41, 209–217. [Google Scholar]
- Main, B.J.; Lee, Y.; Ferguson, H.M.; Kreppel, K.S.; Kihonda, A.; Govella, N.J.; Collier, T.C.; Cornel, A.J.; Eskin, E.; Kang, E.Y.; et al. The Genetic Basis of Host Preference and Resting Behavior in the Major African Malaria Vector, Anopheles arabiensis. PLoS Genet. 2016, 12, e1006303. [Google Scholar] [CrossRef]
- Ashine, T.; Getachew, D.; Demisse, M.; Lobo, N.F.; Tadesse, F.G. Anopheles arabiensis. Trends Parasitol. 2023, 40, 91–92. [Google Scholar] [CrossRef]
- Coetzee, M.; Craig, M.; le Sueur, D.; Coetzee, M.; Craig, M.; le Sueur, D. Distribution of African Malaria Mosquitoes Belonging to the Anopheles gambiae Complex. Parasitol. Today 2000, 16, 74–77. [Google Scholar] [CrossRef]
- Koenraadt, C.J.M.; Githeko, A.K.; Takken, W. The effects of rainfall and evapotranspiration on the temporal dynamics of Anopheles gambiae s.s. and Anopheles arabiensis in a Kenyan village. Acta Trop. 2004, 90, 141–153. [Google Scholar] [CrossRef]
- Gray, E.M.; Bradley, T.J. Physiology of desiccation resistance in Anopheles gambiae and Anopheles arabiensis. Am. J. Trop. Med. Hyg. 2005, 73, 553–559. [Google Scholar] [CrossRef]
- Sinka, M.E.; Bangs, M.J.; Manguin, S.; Coetzee, M.; Mbogo, C.M.; Hemingway, J.; Patil, A.P.; Temperley, W.H.; Gething, P.W.; Kabaria, C.W.; et al. The dominant Anopheles vectors of human malaria in Africa, Europe and the Middle East: Occurrence data, distribution maps and bionomic précis. Parasit. Vectors 2010, 3, 117. [Google Scholar] [CrossRef]
- Ng’habi, K.R.; Knols, B.G.; Lee, Y.; Ferguson, H.M.; Lanzaro, G.C. Population genetic structure of Anopheles arabiensis and Anopheles gambiae in a malaria endemic region of southern Tanzania. Malar. J. 2011, 10, 289. [Google Scholar] [CrossRef]
- Derua, Y.A.; Alifrangis, M.; Hosea, K.M.; Meyrowitsch, D.W.; Magesa, S.M.; Pedersen, E.M.; Simonsen, P.E. Change in composition of the Anopheles gambiae complex and its possible implications for the transmission of malaria and lymphatic filariasis in north-eastern Tanzania. Malar. J. 2012, 11, 188. [Google Scholar] [CrossRef] [PubMed]
- Coetzee, M.; Hunt, R.H.; Wilkerson, R.; Della Torre, A.; Coulibaly, M.B.; Besansky, N.J. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa 2013, 3619, 246–274. [Google Scholar] [CrossRef] [PubMed]
- Verkhivker, G.M. Leveraging Structural Diversity and Allosteric Regulatory Mechanisms of Protein Kinases in the Discovery of Small Molecule Inhibitors. Curr. Med. Chem. 2017, 24, 4838–4872. [Google Scholar] [CrossRef] [PubMed]
- Oboh, M.A.; Asmorom, N.; Falade, C.; Ojurongbe, O.; Thomas, B.N. High genetic and haplotype diversity in vaccine candidate Pfceltos but not Pfrh5 among malaria-infected children in Ibadan, Nigeria. PeerJ 2023, 11, e16519. [Google Scholar] [CrossRef]
- Koama, B.; Namountougou, M.; Sanou, R.; Ndo, S.; Ouattara, A.; Dabiré, R.K.; Malone, D.; Diabaté, A. The sterilizing effect of pyriproxyfen on the malaria vector Anopheles gambiae: Physiological impact on ovaries development. Malar. J. 2015, 14, 101. [Google Scholar] [CrossRef]
- Paton, D.G.; Childs, L.M.; Itoe, M.A.; Holmdahl, I.E.; Buckee, C.O.; Catteruccia, F. Exposing Anopheles mosquitoes to antimalarials blocks Plasmodium parasite transmission. Nature 2019, 567, 239–243. [Google Scholar] [CrossRef]
- Paton, D.G.; Probst, A.S.; Ma, E.; Adams, K.L.; Shaw, W.R.; Singh, N.; Bopp, S.; Volkman, S.K.; Hien, D.F.S.; Paré, P.S.L.; et al. Using an antimalarial in mosquitoes overcomes Anopheles and Plasmodium resistance to malaria control strategies. PLoS Pathog. 2022, 18, e1010609. [Google Scholar] [CrossRef]
- Pondeville, E.; Maria, A.; Jacques, J.-C.; Bourgouin, C.; Dauphin-Villemant, C. Anopheles gambiae males produce and transfer the vitellogenic steroid hormone 20-hydroxyecdysone to females during mating. Proc. Natl. Acad. Sci. USA 2008, 105, 19631–19636. [Google Scholar] [CrossRef]
- Rogers, D.W.; Baldini, F.; Battaglia, F.; Panico, M.; Dell, A.; Morris, H.R.; Catteruccia, F. Transglutaminase-mediated semen coagulation controls sperm storage in the malaria mosquito. PLOS Biol. 2009, 7, e1000272. [Google Scholar] [CrossRef]
- Dottorini, T.; Nicolaides, L.; Ranson, H.; Rogers, D.W.; Crisanti, A.; Catteruccia, F. A genome-wide analysis in Anopheles gambiae mosquitoes reveals 46 male accessory gland genes, possible modulators of female behavior. Proc. Natl. Acad. Sci. USA 2007, 104, 16215–16220. [Google Scholar] [CrossRef]
- Liang, J.; Hodge, J.M.; Sharakhov, I.V. Asymmetric phenotypes of sterile hybrid males from reciprocal crosses between species of the Anopheles gambiae Complex. Front. Ecol. Evol. 2021, 9, 660207. [Google Scholar] [CrossRef]
- Thailayil, J.; Gabrieli, P.; Caputo, B.; Bascuñán, P.; South, A.; Diabate, A.; Dabire, R.; Della Torre, A.; Catteruccia, F. Analysis of natural female post-mating responses of Anopheles gambiae and Anopheles coluzzii unravels similarities and differences in their reproductive ecology. Sci. Rep. 2018, 8, 6594. [Google Scholar] [CrossRef]
- Kumar, S.; Molina-Cruz, A.; Gupta, L.; Rodrigues, J.; Barillas-Mury, C. A peroxidase/dual oxidase system modulates midgut epithelial immunity in Anopheles gambiae. Science 2010, 327, 1644–1648. [Google Scholar] [CrossRef] [PubMed]
- Bascuñán, P.; Gabrieli, P.; Mameli, E.; Catteruccia, F. Mating-regulated atrial proteases control reinsemination rates in Anopheles gambiae Females. Sci. Rep. 2020, 10, 21974. [Google Scholar] [CrossRef]
- Peirce, M.J.; Mitchell, S.N.; Kakani, E.G.; Scarpelli, P.; South, A.; Shaw, W.R.; Werling, K.L.; Gabrieli, P.; Marcenac, P.; Bordoni, M.; et al. JNK signaling regulates oviposition in the malaria vector Anopheles gambiae. Sci. Rep. 2020, 10, 14344. [Google Scholar] [CrossRef]
- Mendes, A.M.; Schlegelmilch, T.; Cohuet, A.; Awono-Ambene, P.; De Iorio, M.; Fontenille, D.; Morlais, I.; Christophides, G.K.; Kafatos, F.C.; Vlachou, D. Conserved mosquito/parasite interactions affect development of Plasmodium falciparum in Africa. PLoS Pathog. 2008, 4, e1000069. [Google Scholar] [CrossRef]
- Vlachou, D.; Schlegelmilch, T.; Christophides, G.K.; Kafatos, F.C. Functional genomic analysis of midgut epithelial responses in Anopheles during Plasmodium Invasion. Curr. Biol. 2005, 15, 1185–1195. [Google Scholar] [CrossRef]
- Upton, L.M.; Povelones, M.; Christophides, G.K. Anopheles gambiae blood feeding initiates an anticipatory defense response to Plasmodium berghei. J. Innate Immun. 2015, 7, 74–86. [Google Scholar] [CrossRef]
- Green, E.; Jaouen, E.; Klug, D.; Olmo, R.P.; Gautier, A.; Blandin, S.; Marois, E. A population modification gene drive targeting both saglin and lipophorin impairs Plasmodium transmission in Anopheles mosquitoes. Elife 2023, 12, e93142. [Google Scholar] [CrossRef]
- Christophides, G.K.; Zdobnov, E.; Barillas-Mury, C.; Birney, E.; Blandin, S.; Blass, C.; Brey, P.T.; Collins, F.H.; Danielli, A.; Dimopoulos, G.; et al. Immunity-related genes and gene families in Anopheles gambiae. Science 2002, 298, 159–165. [Google Scholar] [CrossRef]
- Wormald, S.; Hilton, D.J. Inhibitors of cytokine signal transduction. J Biol Chem 2004, 279, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Frolet, C.; Thoma, M.; Blandin, S.; Hoffmann, J.A.; Levashina, E.A. Boosting NF-kappaB-dependent basal immunity of Anopheles gambiae aborts development of Plasmodium berghei. Immunity 2006, 25, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Garver, L.S.; Dong, Y.; Dimopoulos, G. Caspar controls resistance to Plasmodium falciparum in diverse anopheline species. PLOS Pathog. 2009, 5, e1000335. [Google Scholar] [CrossRef]
- Gupta, L.; Molina-Cruz, A.; Kumar, S.; Rodrigues, J.; Dixit, R.; Zamora, R.E.; Barillas-Mury, C. The STAT pathway mediates late-phase immunity against Plasmodium in the mosquito Anopheles gambiae. Cell Host Microbe 2009, 5, 498–507. [Google Scholar] [CrossRef]
- Garver, L.S.; de Almeida Oliveira, G.; Barillas-Mury, C. The JNK pathway is a key mediator of Anopheles gambiae antiplasmodial immunity. PLOS Pathog. 2013, 9, e1003622. [Google Scholar] [CrossRef]
- Dong, Y.; Das, S.; Cirimotich, C.; Souza-Neto, J.A.; McLean, K.J.; Dimopoulos, G. Engineered Anopheles immunity to Plasmodium infection. PLOS Pathog. 2011, 7, e1002458. [Google Scholar] [CrossRef]
- Fraiture, M.; Baxter, R.H.G.; Steinert, S.; Chelliah, Y.; Frolet, C.; Quispe-Tintaya, W.; Hoffmann, J.A.; Blandin, S.A.; Levashina, E.A. Two mosquito LRR proteins function as complement control factors in the TEP1-mediated killing of Plasmodium. Cell Host Microbe 2009, 5, 273–284. [Google Scholar] [CrossRef]
- Povelones, M.; Upton, L.M.; Sala, K.A.; Christophides, G.K. Structure-function analysis of the Anopheles gambiae LRIM1/APL1C complex and its interaction with complement C3-Like protein TEP1. PLOS Pathog. 2011, 7, e1002023. [Google Scholar] [CrossRef]
- de Almeida Oliveira, G.; Lieberman, J.; Barillas-Mury, C. Epithelial nitration by a peroxidase/NOX5 system mediates mosquito antiplasmodial immunity. Science 2012, 335, 856–859. [Google Scholar] [CrossRef]
- Horton, A.A.; Wang, B.; Camp, L.; Price, M.S.; Arshi, A.; Nagy, M.; Nadler, S.A.; Faeder, J.R.; Luckhart, S. The mitogen-activated protein kinome from Anopheles gambiae: Identification, phylogeny and functional characterization of the ERK, JNK and P38 MAP Kinases. BMC Genom. 2011, 12, 574. [Google Scholar] [CrossRef]
- Blandin, S.; Shiao, S.-H.; Moita, L.F.; Janse, C.J.; Waters, A.P.; Kafatos, F.C.; Levashina, E.A. Complement-like protein TEP1 Is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell 2004, 116, 661–670. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Dimopoulos, G. Anopheles fibrinogen-related proteins provide expanded pattern recognition capacity against bacteria and malaria parasites. J. Biol. Chem. 2009, 284, 9835–9844. [Google Scholar] [CrossRef] [PubMed]
- Nsango, S.E.; Pompon, J.; Xie, T.; Rademacher, A.; Fraiture, M.; Thoma, M.; Awono-Ambene, P.H.; Moyou, R.S.; Morlais, I.; Levashina, E.A. AP-1/Fos-TGase2 axis mediates wounding-induced Plasmodium falciparum Killing in Anopheles gambiae. J. Biol. Chem. 2013, 288, 16145–16154. [Google Scholar] [CrossRef]
- Adedeji, E.O.; Ogunlana, O.O.; Fatumo, S.; Beder, T.; Ajamma, Y.; Koenig, R.; Adebiyi, E. Anopheles metabolic proteins in malaria transmission, prevention and control: A review. Parasites Vectors 2020, 13, 465. [Google Scholar] [CrossRef]
- Giraldo-Calderón, G.I.; Emrich, S.J.; MacCallum, R.M.; Maslen, G.; Dialynas, E.; Topalis, P.; Ho, N.; Gesing, S.; the VectorBase Consortium; Madey, G.; et al. VectorBase: An updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2015, 43, D707–D713. [Google Scholar] [CrossRef]
- Ag3.0 (Ag1000G Phase 3) SNP Data Release | MalariaGEN. Available online: https://www.malariagen.net/data_package/ag1000g-phase3-snp/ (accessed on 6 August 2021).
- Home | MalariaGEN. Available online: https://www.malariagen.net/ (accessed on 6 August 2021).
- Sharakhova, M.V.; Hammond, M.P.; Lobo, N.F.; Krzywinski, J.; Unger, M.F.; Hillenmeyer, M.E.; Bruggner, R.V.; Birney, E.; Collins, F.H. Update of the Anopheles gambiae PEST genome assembly. Genome Biol. 2007, 8, R5. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project data processing subgroup the sequence alignment/mapformat and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call ormat and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Wright, S. The genetical structure of populations. Ann. Eugen. 1951, 15, 323–354. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F-Statistics for the analysis of population structire. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T. Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef] [PubMed]
- Jolliffe, I. Principal Component Analysis (2ed., Springer, 2002) (518s)—MVsa—PDF | PDF. Available online: https://fr.scribd.com/document/484920903/Jolliffe-I-Principal-Component-Analysis-2ed-Springer-2002-518s-MVsa-pdf (accessed on 3 February 2025).
- Goudet, J. Hierfstat, a package for R to compute and test hierarchical F-Statistics. Mol. Ecol. Notes 2005, 5, 184–186. [Google Scholar] [CrossRef]
- Pfeifer, B.; Wittelsbürger, U.; Ramos-Onsins, S.E.; Lercher, M.J. PopGenome: An efficient swiss army knife for population genomic analyses in R. Mol. Biol. Evol. 2014, 31, 1929–1936. [Google Scholar] [CrossRef]
- Nei, M.; Li, W.H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 1979, 76, 5269–5273. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Kimura, M. The Neutral Theory of Molecular Evolution; Cambridge University Press: Cambridge, UK, 1983; ISBN 978-0-521-31793-1. [Google Scholar]
- Slatkin, M. Linkage Disequilibrium—Understanding the evolutionary past and mapping the medical Future. Nat. Rev. Genet. 2008, 9, 477–485. [Google Scholar] [CrossRef]
- Shin, J.-H.; Blay, S.; McNeney, B.; Graham, J. LDheatmap: An R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J. Stat. Softw. 2006, 16, 1–9. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seck, F.; Diop, M.F.; Mané, K.; Diallo, A.; Dieng, I.; Namountougou, M.; Diabate, A.; Amambua-Ngwa, A.; Dia, I.; Assogba, B.S. Reduced Genetic Diversity of Key Fertility and Vector Competency Related Genes in Anopheles gambiae s.l. Across Sub-Saharan Africa. Genes 2025, 16, 543. https://doi.org/10.3390/genes16050543
Seck F, Diop MF, Mané K, Diallo A, Dieng I, Namountougou M, Diabate A, Amambua-Ngwa A, Dia I, Assogba BS. Reduced Genetic Diversity of Key Fertility and Vector Competency Related Genes in Anopheles gambiae s.l. Across Sub-Saharan Africa. Genes. 2025; 16(5):543. https://doi.org/10.3390/genes16050543
Chicago/Turabian StyleSeck, Fatoumata, Mouhamadou Fadel Diop, Karim Mané, Amadou Diallo, Idrissa Dieng, Moussa Namountougou, Abdoulaye Diabate, Alfred Amambua-Ngwa, Ibrahima Dia, and Benoit Sessinou Assogba. 2025. "Reduced Genetic Diversity of Key Fertility and Vector Competency Related Genes in Anopheles gambiae s.l. Across Sub-Saharan Africa" Genes 16, no. 5: 543. https://doi.org/10.3390/genes16050543
APA StyleSeck, F., Diop, M. F., Mané, K., Diallo, A., Dieng, I., Namountougou, M., Diabate, A., Amambua-Ngwa, A., Dia, I., & Assogba, B. S. (2025). Reduced Genetic Diversity of Key Fertility and Vector Competency Related Genes in Anopheles gambiae s.l. Across Sub-Saharan Africa. Genes, 16(5), 543. https://doi.org/10.3390/genes16050543





