Integrative Genomic and in Silico Analysis Reveals Mitochondrially Encoded Cytochrome C Oxidase III (MT—CO3) Overexpression and Potential Neem-Derived Inhibitors in Breast Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection and Preprocessing: TCGA-BRCA Data Retrieval
2.2. Normalization and Data Processing of RNA Sequencing
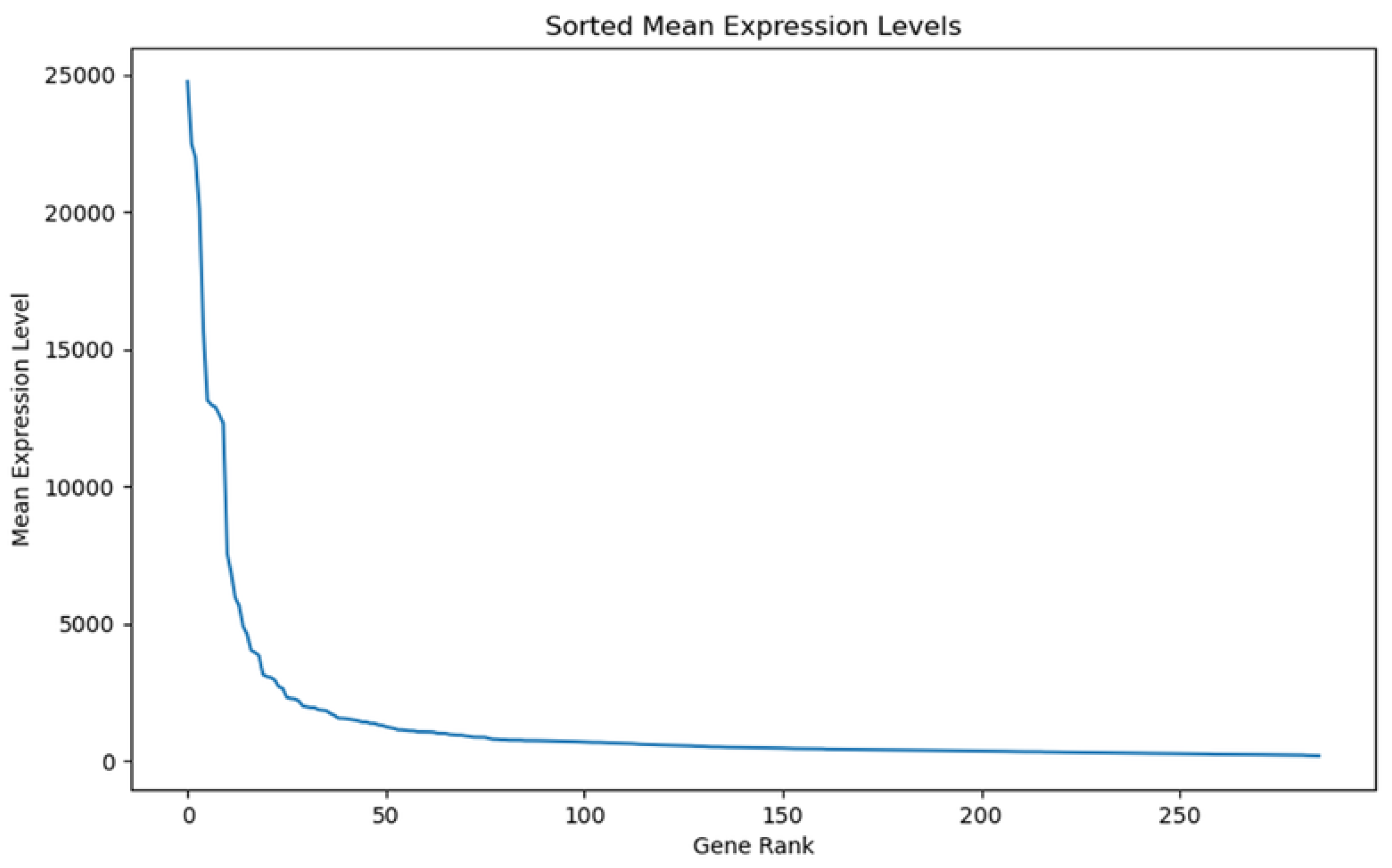
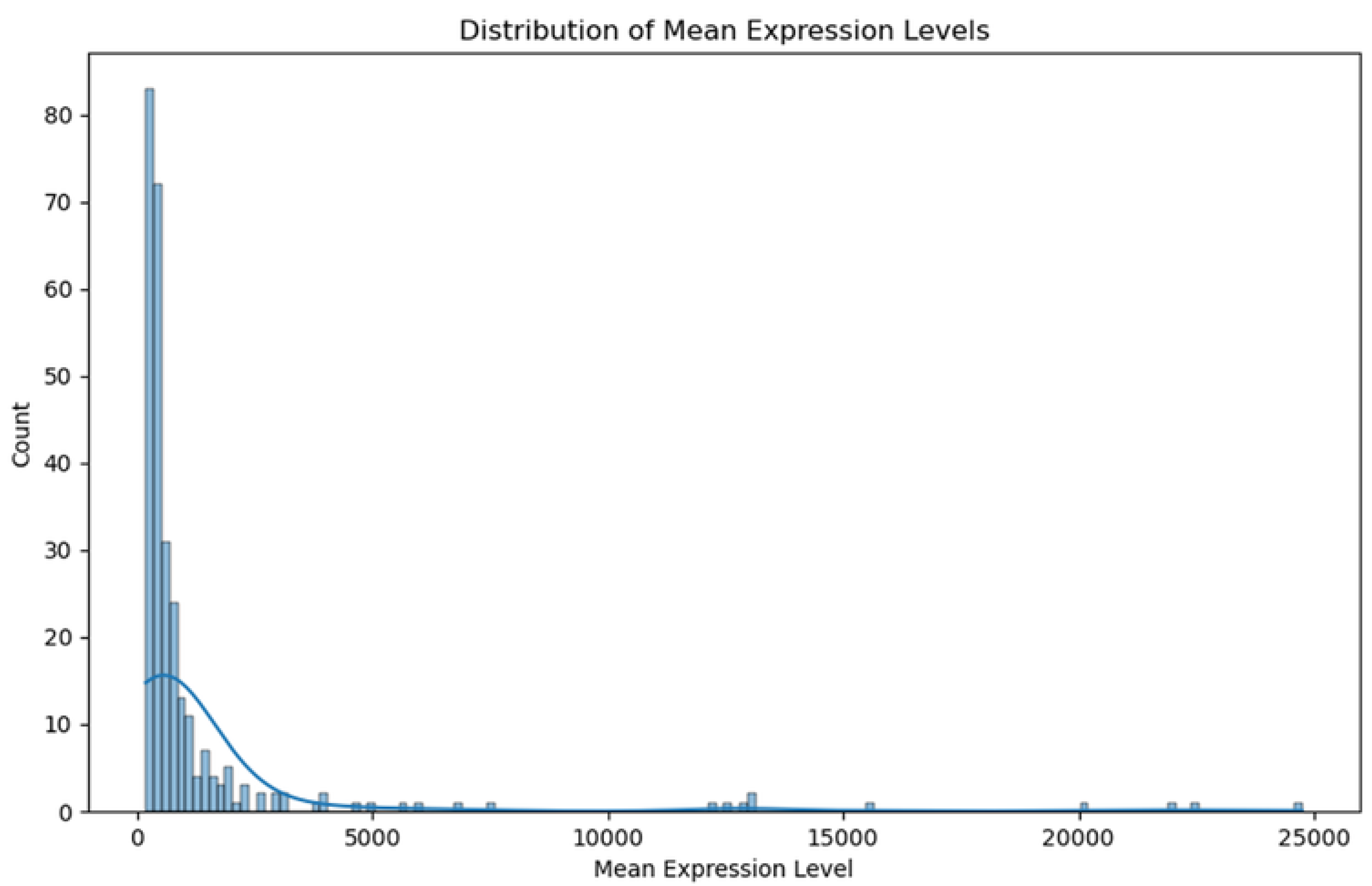
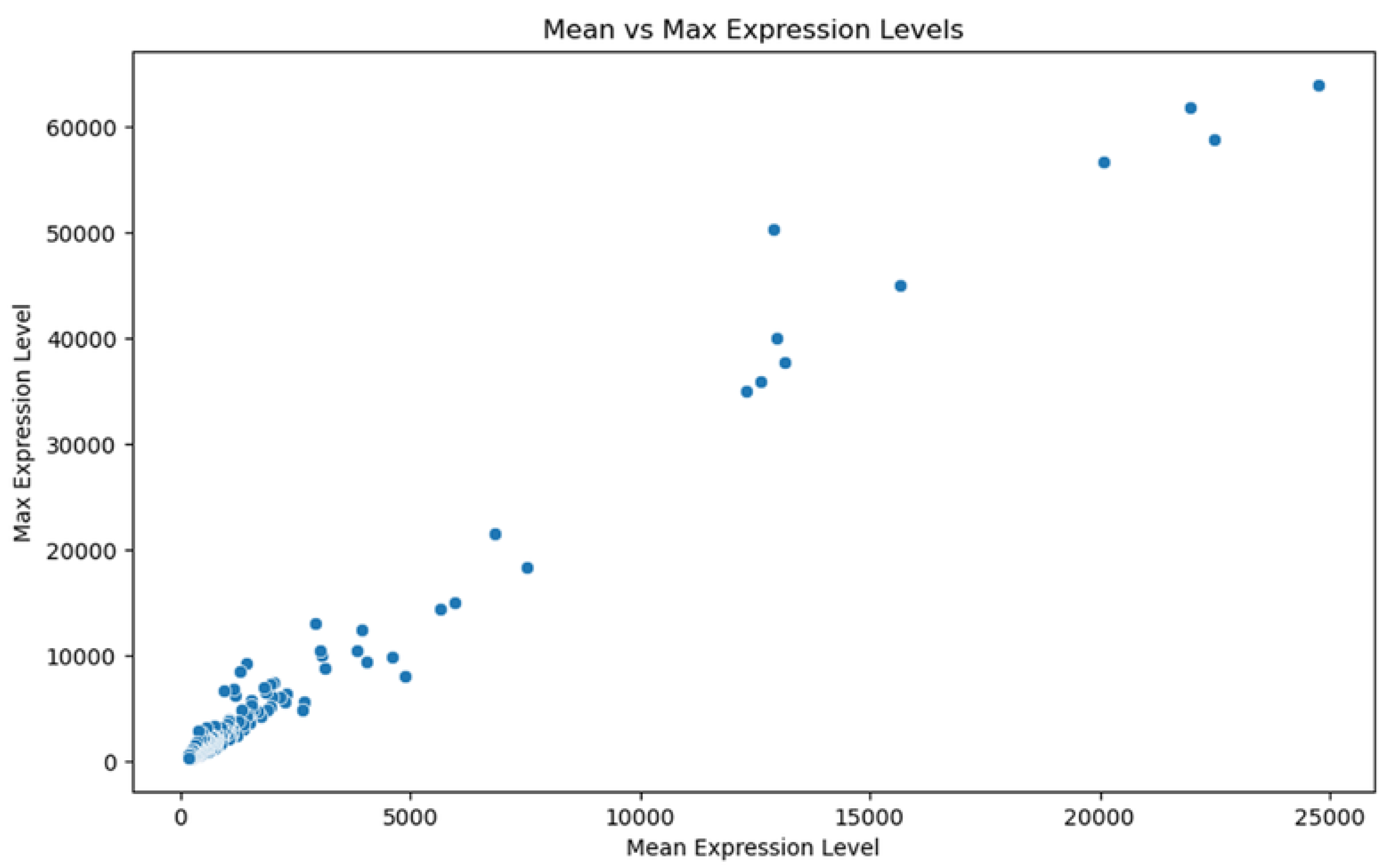
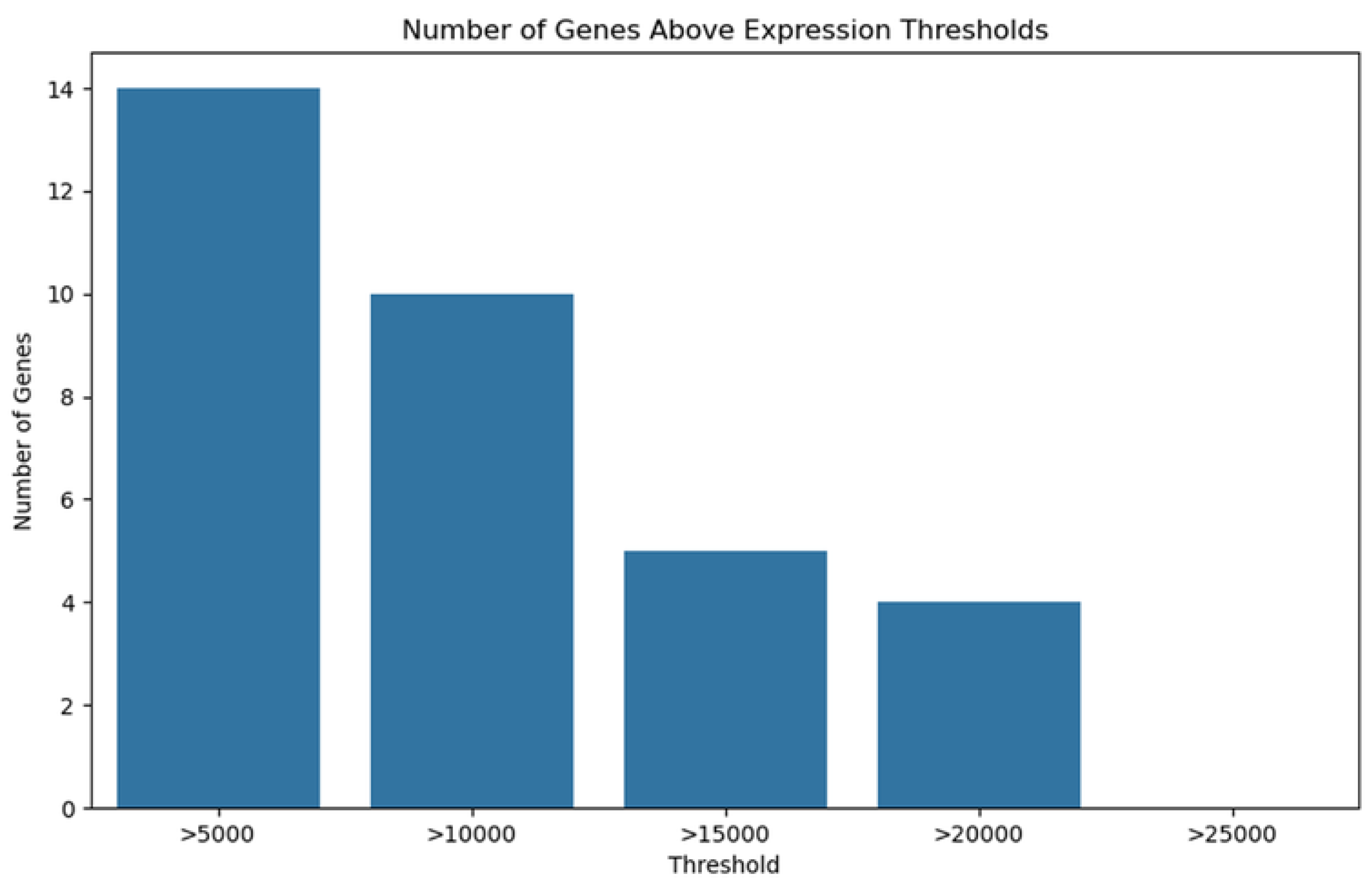
2.3. Gene Expression Analysis and Differential Expression
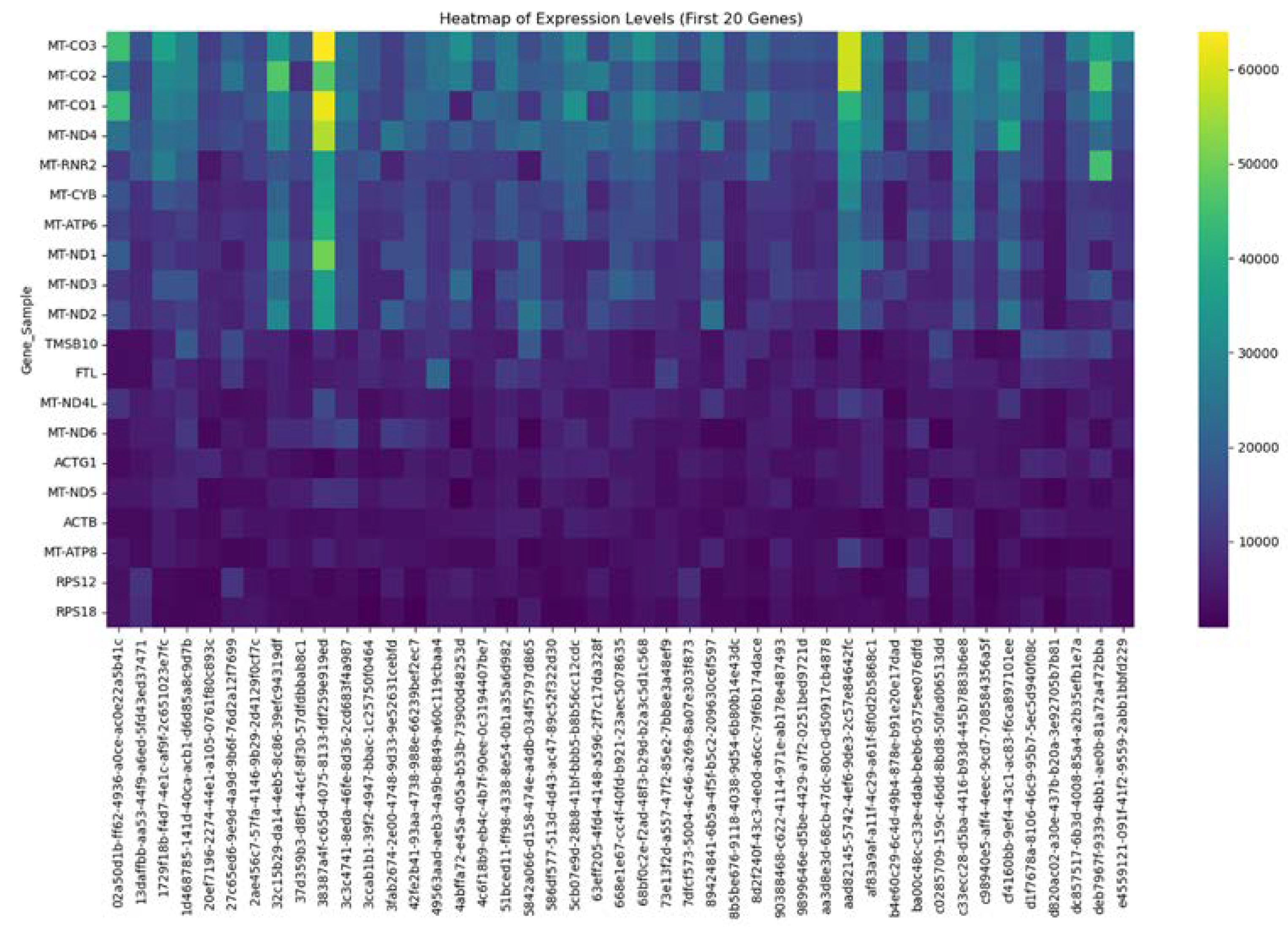
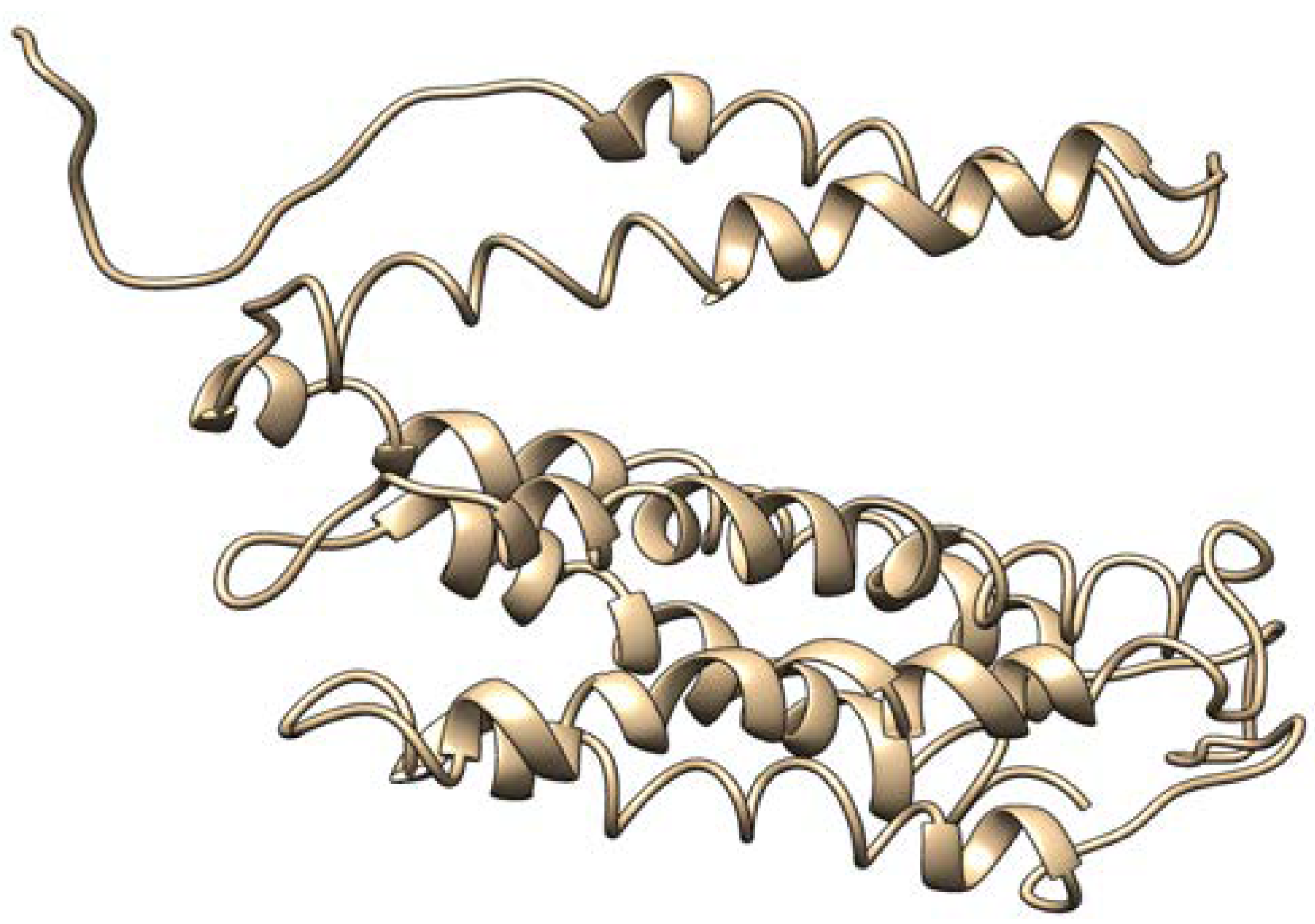
2.4. Identification of MT—CO3 as a Dominant Overexpressed Gene and Modeling of the MT—CO3 Protein
2.5. Retrieval and Translation of Sequences
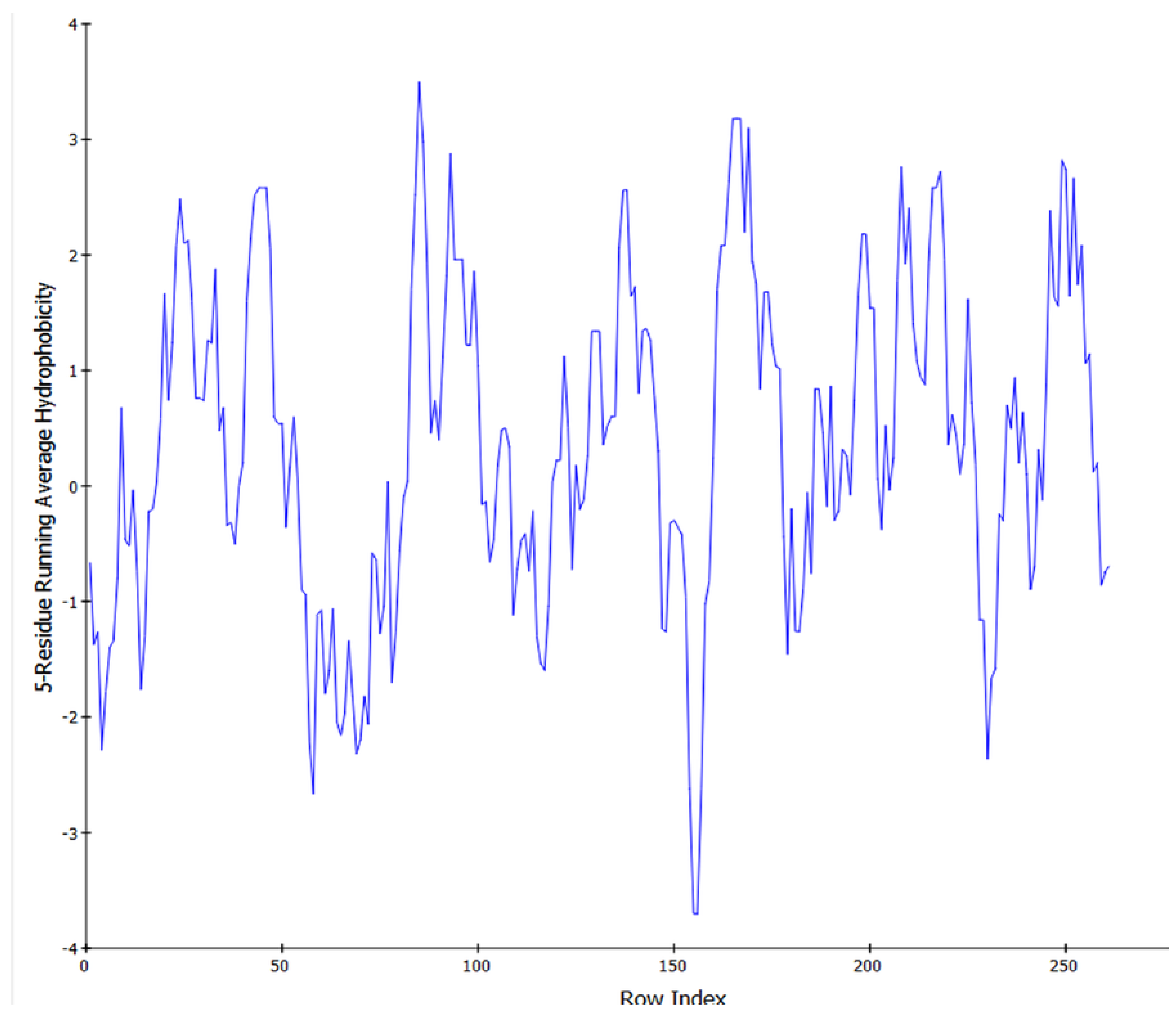
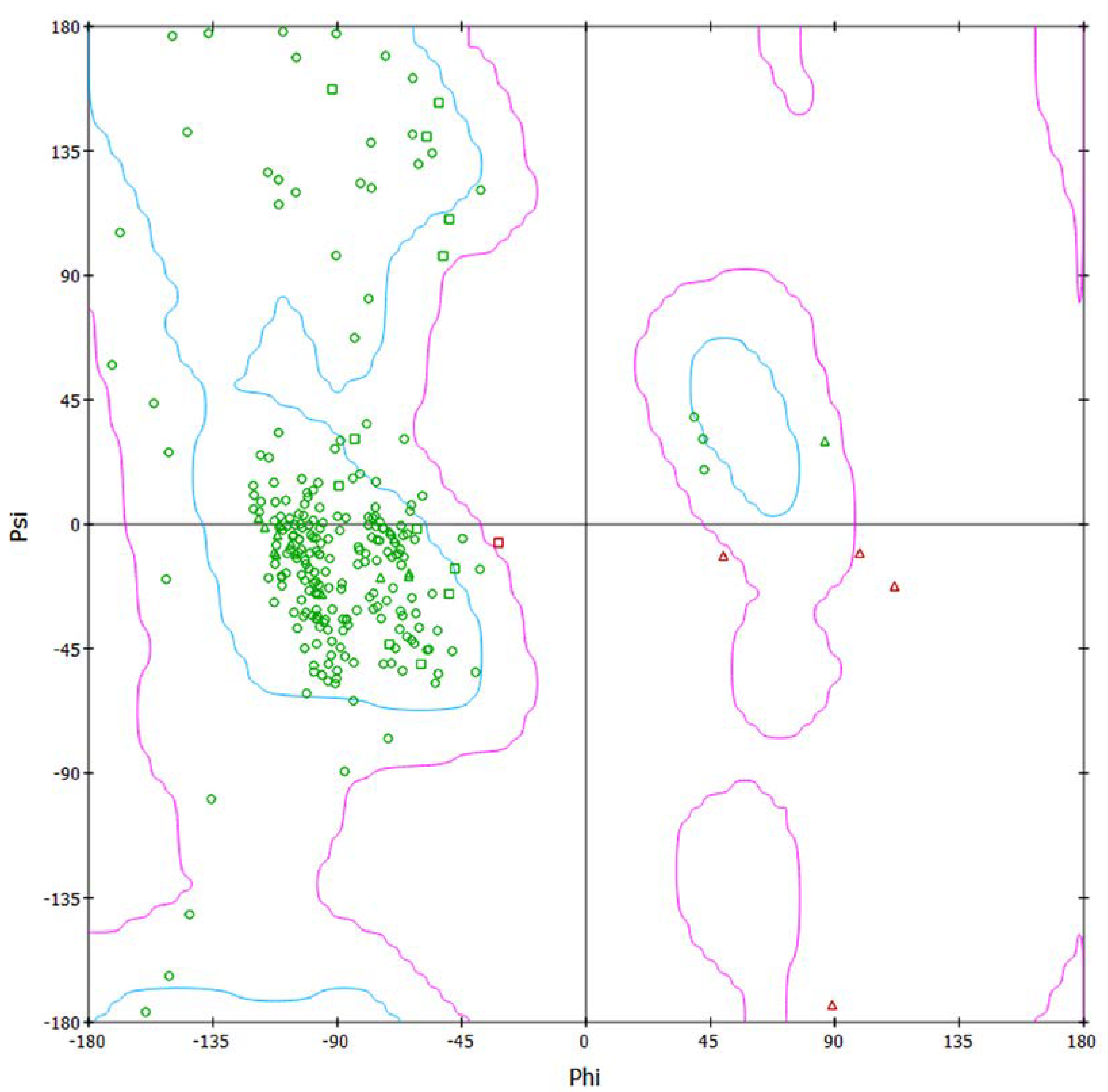
2.6. Model Validation and Structure Prediction
2.7. Analysis and Forecasting via Computation
2.8. The Identification and Selection Procedure of Binding Sites
2.9. Preparation of Ligands and Molecular Docking of Neem Compounds
2.10. Molecular Docking and Receptor Preparation
2.11. Docking, Analysis, Visualization, and Interaction
2.12. Statistical Analysis
3. Results
Neem-Derived Compounds as Potential Inhibitors of MT—CO3
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TCGA-BRCA | The Cancer Genome Atlas—Breast Cancer |
| MT—CO3 | mitochondrially encoded cytochrome C oxidase III |
| RNA | ribonucleic acid |
| GDC | genomic data commons |
| PDB | protein data bank |
| 2D | two-dimensional |
| 3D | three-dimensional |
| MMFF94 | Merck molecular force field 1994 |
References
- Aravindan, A.; Gupta, A.; Moorkoth, S.; Dhas, N. Implications of Nanotherapeutic advancements to leverage Multi-Drug Resistant Breast Cancer: The State-of-the-Art Review. J. Drug Deliv. Sci. Technol. 2024, 100, 106007. [Google Scholar] [CrossRef]
- Kalinowski, L.; Saunus, J.M.; McCart Reed, A.E.; Lakhani, S.R. Breast cancer heterogeneity in primary and metastatic disease. In Breast Cancer Metastasis and Drug Resistance: Challenges and Progress; Springer: Berlin/Heidelberg, Germany, 2019; pp. 75–104. [Google Scholar]
- Jin, P.; Jiang, J.; Zhou, L.; Huang, Z.; Nice, E.C.; Huang, C.; Fu, L. Mitochondrial adaptation in cancer drug resistance: Prevalence, mechanisms, and management. J. Hematol. Oncol. 2022, 15, 97. [Google Scholar] [CrossRef] [PubMed]
- Nakhle, J.; Rodriguez, A.M.; Vignais, M.L. Multifaceted roles of mitochondrial components and metabolites in metabolic diseases and cancer. Int. J. Mol. Sci. 2020, 21, 4405. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.; Kosnacova, H.; Chovanec, M.; Jurkovicova, D. Mitochondrial genetic and epigenetic regulations in cancer: Therapeutic potential. Int. J. Mol. Sci. 2022, 23, 7897. [Google Scholar] [CrossRef]
- Kalachaveedu, M.; Senthil, R.; Azhagiyamanavalan, S.; Ravi, R.; Meenakshisundaram, H.; Dharmarajan, A. Traditional medicine herbs as natural product matrices in cancer chemoprevention: A trans pharmacological perspective (scoping review). Phytother. Res. 2023, 37, 1539–1573. [Google Scholar] [CrossRef]
- Bose, A.; Baral, R. Neem leaf glycoprotein in cancer immunomodulation and immunotherapy. In New Look to Phytomedicine; Academic Press: Cambridge, MA, USA, 2019; pp. 391–408. [Google Scholar]
- Kapur, A.; Mehta, P.; Simmons, A.D.; Ericksen, S.S.; Mehta, G.; Palecek, S.P.; Felder, M.; Stenerson, Z.; Nayak, A.; Dominguez, J.M.A.; et al. Atovaquone: An Inhibitor of Oxidative Phosphorylation as Studied in Gynecologic Cancers. Cancers 2022, 14, 2297. [Google Scholar] [CrossRef]
- di Magno, L.; Di Pastena, F.; Bordone, R.; Coni, S.; Canettieri, G. The Mechanism of Action of Biguanides: New Answers to a Complex Question. Cancers 2022, 14, 3220. [Google Scholar] [CrossRef]
- Nagini, S.; Palrasu, M.; Bishayee, A. Limonoids from neem (Azadirachta indica A. Juss.) are potential anticancer drug candidates. Med. Res. Rev. 2024, 44, 457–496. [Google Scholar] [CrossRef]
- Cappelli, E.; Cumbo, F.; Bernasconi, A.; Canakoglu, A.; Ceri, S.; Masseroli, M.; Weitschek, E. OpenGDC: Unifying, modeling, integrating cancer genomic data and clinical metadata. Appl. Sci. 2020, 10, 6367. [Google Scholar] [CrossRef]
- Fukushima, K.; Pollock, D.D. Amalgamated cross-species transcriptomes reveal organ-specific propensity in gene expression evolution. Nat. Commun. 2020, 11, 4459. [Google Scholar] [CrossRef]
- Wang, J.; Yang, Z.; Zhang, C.; Ouyang, J.; Zhang, G.; Wu, C. A four-gene signature in the tumor microenvironment that significantly associates with the prognosis of patients with breast cancer. Gene 2020, 761, 145049. [Google Scholar] [CrossRef]
- Schneider, M.; Tognolli, M.; Bairoch, A. The Swiss-Prot protein knowledgebase and ExPASy: Providing the plant community with high quality proteomic data and tools. Plant Physiol. Biochem. 2004, 42, 1013–1021. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.; Nagarajan, K.; Gupta, A. SwissADME: An in-silico ADME examination tool. In Emerging Trends in IoT and Computing Technologies; Routledge: Oxfordshire, UK, 2022; pp. 219–225. [Google Scholar]
- Tasleem, M.; Alrehaily, A.; Almeleebia, T.M.; Alshahrani, M.Y.; Ahmad, I.; Asiri, M.; Alabdallah, N.M.; Saeed, M. Investigation of antidepressant properties of yohimbine by employing structure-based computational assessments. Curr. Issues Mol. Biol. 2021, 43, 1805–1827. [Google Scholar] [CrossRef]
- Xing, M.; Zhao, T.T.; Ren, Y.J.; Peng, N.N.; Yang, X.H.; Li, X.; Zhang, H.; Liu, G.Q.; Zhang, L.R.; Zhu, H.L. Synthesis, biological evaluation, and molecular docking studies of pyrazolyl-acylhydrazone derivatives as novel anticancer agents. Med. Chem. Res. 2014, 23, 3274–3286. [Google Scholar] [CrossRef]
- Akomolafe, S.F.; Agboola, O.E.; Ajayi, O.O.; Oyetayo, F.L.; Ajayi, O.B. Elucidating the antidiabetic potential of Chrysophyllum albidum: An integrative approach combining in vitro and in silico studies on dipeptidyl peptidase IV and adenosine deaminase inhibition. Next Res. 2024, 1, 100010. [Google Scholar] [CrossRef]
- Agboola, O.E.; Ayinla, Z.A.; Agboola, S.S.; Omolayo, E.Y.; Fadugba, A.E.; Odeghe, O.B.; Olaiya, O.E.; Oyinloye, B.E. In Silico Profiling of Neem Phytochemicals and Gut Microbiome Metabolites for Alzheimer’s Therapeutics: Targeted Inhibition of BACE1 and Elucidation of Intricate Molecular Crosstalk with Tau Oligomers, and Bacterial Gingipains. Discov. Appl. Sci. 2025, 7, 312. [Google Scholar] [CrossRef]
- Bickerton, G.R.; Paolini, G.V.; Besnard, J.; Muresan, S.; Hopkins, A.L. Quantifying the chemical beauty of drugs. Nat. Chem. 2012, 4, 90–98. [Google Scholar] [CrossRef]
- Chen, Z.; Chen, L.; Lyu, T.D.; Weng, S.; Xie, Y.; Jin, Y.; Wu, O.; Jones, M.; Kwan, K.; Makvnadi, P.; et al. Targeted mitochondrial nanomaterials in biomedicine: Advances in therapeutic strategies and imaging modalities. Acta Biomater. 2024, 186, 1–29. [Google Scholar] [CrossRef]
- Jaworska, M.; Szczudło, J.; Pietrzyk, A.; Shah, J.; Trojan, S.E.; Ostrowska, B.; Kocemba-Pilarczyk, K.A. The Warburg effect: A score for many instruments in the concert of cancer and cancer niche cells. Pharmacol. Rep. 2023, 75, 876–890. [Google Scholar] [CrossRef]
- Bedi, M.; Das, S.; Das, J.; Mukherjee, S.; Basu, A.; Saha, S.; Ghosh, A. Mitochondrial proteome analysis reveals that an augmented cytochrome c oxidase assembly and activity potentiates respiratory capacity in sarcoma. Biochem. Biophys. Res. Commun. 2024, 736, 150501. [Google Scholar] [CrossRef]
- Fiorucci, L.; Erba, F.; Santucci, R.; Sinibaldi, F. Cytochrome c interaction with cardiolipin plays a key role in cell apoptosis: Implications for human diseases. Symmetry 2022, 14, 767. [Google Scholar] [CrossRef]
- Marinko, J.T.; Huang, H.; Penn, W.D.; Capra, J.A.; Schlebach, J.P.; Sanders, C.R. Folding and misfolding of human membrane proteins in health and disease: From single molecules to cellular proteostasis. Chem. Rev. 2019, 119, 5537–5606. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; He, P.Y.; Zhang, Y.; Li, N. Natural products targeting the mitochondria in cancers. Molecules 2020, 26, 92. [Google Scholar] [CrossRef] [PubMed]
- Brylinski, M. Aromatic interactions at the ligand–protein interface: Implications for the development of docking scoring functions. Chem. Biol. Drug Des. 2018, 91, 380–390. [Google Scholar] [CrossRef]
- Patil, R.; Das, S.; Stanley, A.; Yadav, L.; Sudhakar, A.; Varma, A.K. Optimized hydrophobic interactions and hydrogen bonding at the target-ligand interface leads the pathways of drug-designing. PLoS ONE 2010, 5, e12029. [Google Scholar] [CrossRef]
- Buwalda, R.T. Molecular Aggregation in Water: The Interplay of Hydrophobic and Electrostatic Interactions. Ph.D. Thesis, University of Groningen, Groningen, The Netherlands, 2001. [Google Scholar]
- Maity, A.; Majumdar, S.; Priya, P.; De, P.; Saha, S.; Ghosh Dastidar, S. Adaptability in protein structures: Structural dynamics and implications in ligand design. J. Biomol. Struct. Dyn. 2015, 33, 298–321. [Google Scholar] [CrossRef]
- Shan, S.O.; Herschlag, D. [11] Hydrogen bonding in enzymatic catalysis: Analysis of energetic contributions. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1999; Volume 308, pp. 246–276. [Google Scholar]
- Hart, K.M.; Moeder, K.E.; Ho, C.M.; Zimmerman, M.I.; Frederick, T.E.; Bowman, G.R. Designing small molecules to target cryptic pockets yields both positive and negative allosteric modulators. PLoS ONE 2017, 12, e0178678. [Google Scholar] [CrossRef]
- Qi, C.; Di Minin, G.; Vercellino, I.; Wutz, A.; Korkhov, V.M. Structural basis of sterol recognition by human hedgehog receptor PTCH1. Sci. Adv. 2019, 5, eaaw6490. [Google Scholar] [CrossRef]
- Saleem, S.; Khan, A.; Ali, S.S.; Sayaf, A.M.; Khan, M. Molecular modeling of the multiple-drug resistant protein (MRP7) and pharmacophore modelling based virtual screening to identify novel drugs against cancer. J. Comput. Chem. Mol. Model. 2020, 4, 328–342. [Google Scholar]
- Martins, J.N.; Lima, J.C.; Basílio, N. Selective recognition of amino acids and peptides by small supramolecular receptors. Molecules 2020, 26, 106. [Google Scholar] [CrossRef]
- Li, M.; Rehman, A.U.; Liu, Y.; Chen, K.; Lu, S. Dual roles of ATP-binding site in protein kinases: Orthosteric inhibition and allosteric regulation. Adv. Protein Chem. Struct. Biol. 2021, 124, 87–119. [Google Scholar] [PubMed]
- Malmström, B.G. Cytochrome oxidase: Pathways for electron tunneling and proton transfer. JBIC J. Biol. Inorg. Chem. 1998, 3, 339–343. [Google Scholar] [CrossRef]
- Abdullah, A.; Biswas, P.; Sahabuddin, M.; Mubasharah, A.; Khan, D.A.; Hossain, A.; Roy, T.; Rafi, N.M.; Dey, D.; Hasan, M.N.; et al. Research Article Molecular Dynamics Simulation and Pharmacoinformatic Integrated Analysis of Bioactive Phytochemicals from Azadirachta indica (Neem) to Treat Diabetes Mellitus. J. Chem. 2023, 2023, 4170703. [Google Scholar] [CrossRef]
- Ribas, V.; García-Ruiz, C.; Fernández-Checa, J.C. Mitochondria, cholesterol and cancer cell metabolism. Clin. Transl. Med. 2016, 5, 22. [Google Scholar] [CrossRef]
- Roth, K.G.; Mambetsariev, I.; Kulkarni, P.; Salgia, R. The mitochondrion as an emerging therapeutic target in cancer. Trends Mol. Med. 2020, 26, 119–134. [Google Scholar] [CrossRef]
- Block, K.I.; Gyllenhaal, C.; Lowe, L.; Amedei, A.; Amin, A.R.; Amin, A.; Aquilano, K.; Arbiser, J.; Arreola, A.; Arzumanyan, A.; et al. Designing a broad-spectrum integrative approach for cancer prevention and treatment. In Seminars in Cancer Biology; Academic Press: Cambridge, MA, USA, 2015; Volume 35, pp. S276–S304. [Google Scholar]
- Agboola, O.E.; Oluwatobi, F.B.; Tugbobo, O.S.; Ayinla, Z.A.; Agboola, S.S.; Solomon, I.K.; Akinseye, J.F.; Ilesanmi, O.S.; Adeboye, T.O.; Akindojutimi, D.B. Purified Azadirachta indica leaf arginase exhibits properties with potential tumor therapeutics: An Invitro Study. J. Pharm. Sci. Res. 2023, 15, 1168–1171. [Google Scholar]
- Agboola, O.E.; Ayinla, Z.A.; Agboola, S.S.; Adegbuyi, T.A.; Akinseye, J.F.; Sijuade, A.; Egbebi, A.H.; Ilesanmi, O.S.; Agboola, A.A.; Ibrahim, O.K. Molecular mechanisms underlying the erectogenic effects of nutraceutical lunamarine, a novel PDE5 inhibitor derived from watermelon (Citrullus lanatus). Discov. Food 2024, 4, 153. [Google Scholar] [CrossRef]
- Oyinloye, B.E.; Agbebi, E.A.; Agboola, O.E.; Ubah, C.S.; Owolabi, O.V.; Aruleba, R.T.; Onikanni, S.A.; Ejeje, J.N.; Ajiboye, B.O.; Omotuyi, O.I. Skin Anti-Aging Potentials of Phytochemicals from Peperomia pellucida against Selected Metalloproteinase Targets: An In Silico Approach. Cosmetics 2023, 10, 151. [Google Scholar] [CrossRef]
- Agboola, O.E.; Agboola, S.S.; Fadugba, A.E.; Adegbuyi, A.T.; Odeghe, O.B. Computational Analysis of Curcuma longa L Compounds: Unraveling Molecular Interactions and Drug-like Properties for Novel Therapeutic Applications. Next Res. 2025, 2, 100283. [Google Scholar] [CrossRef]



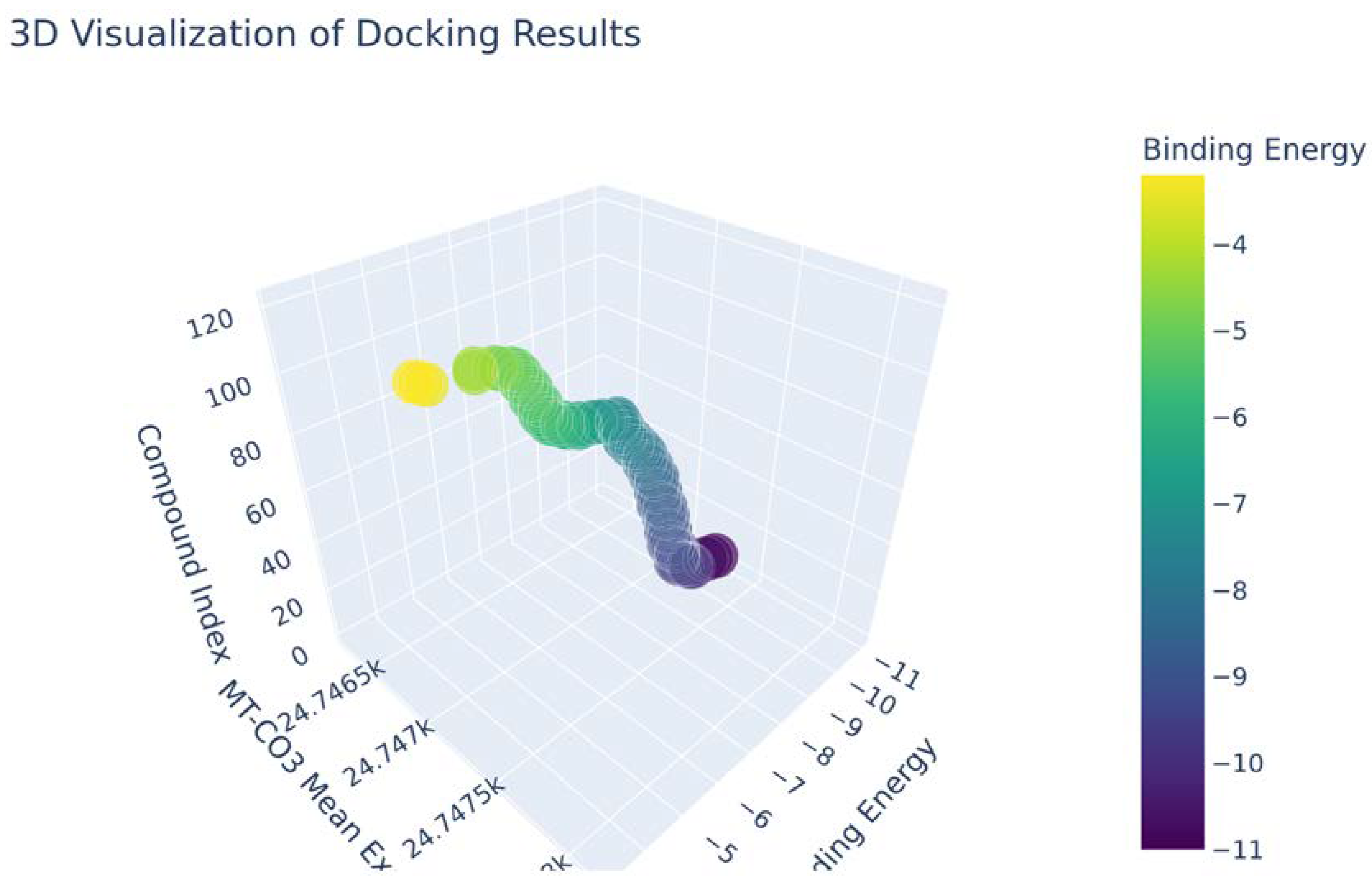
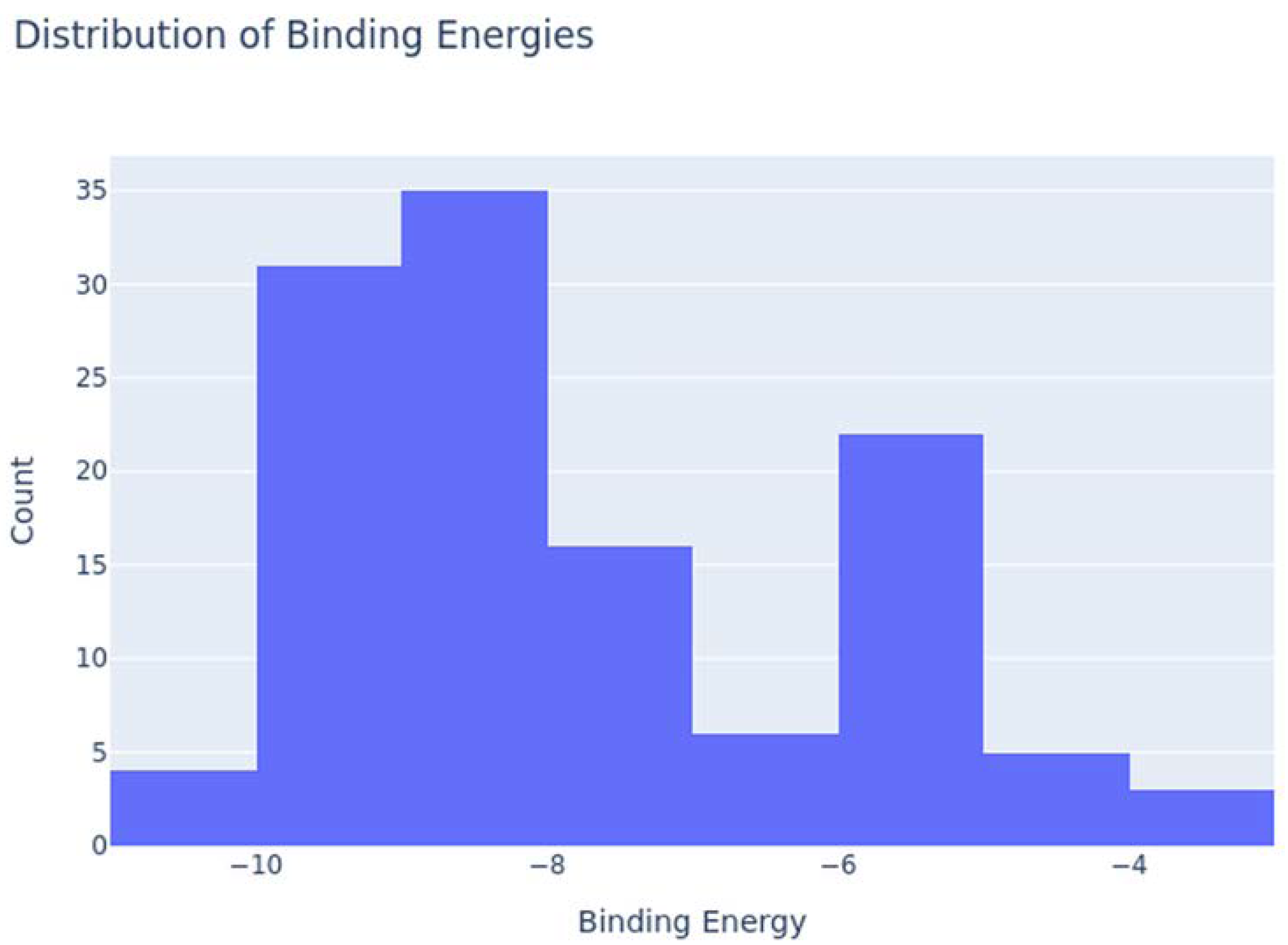

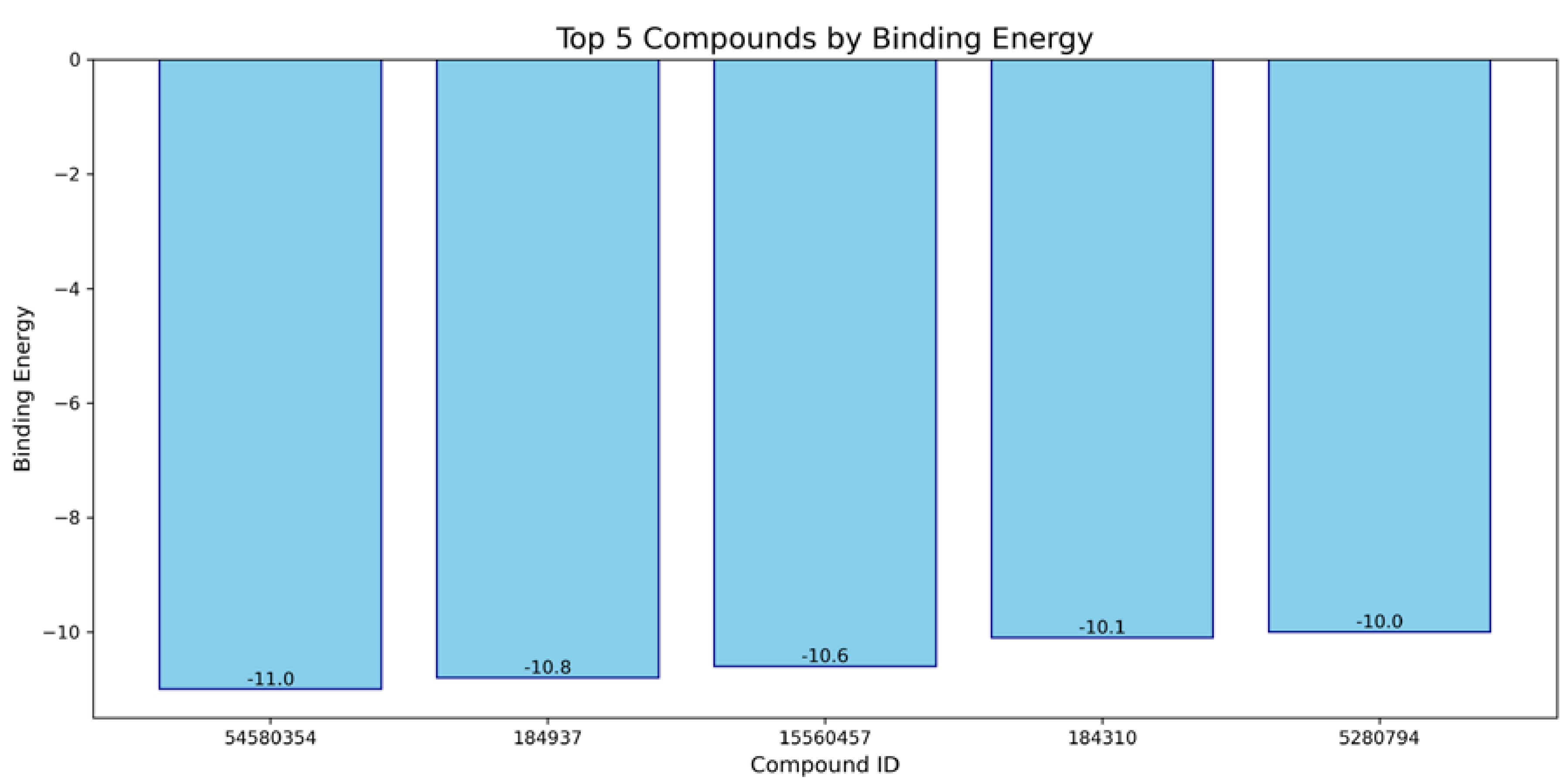
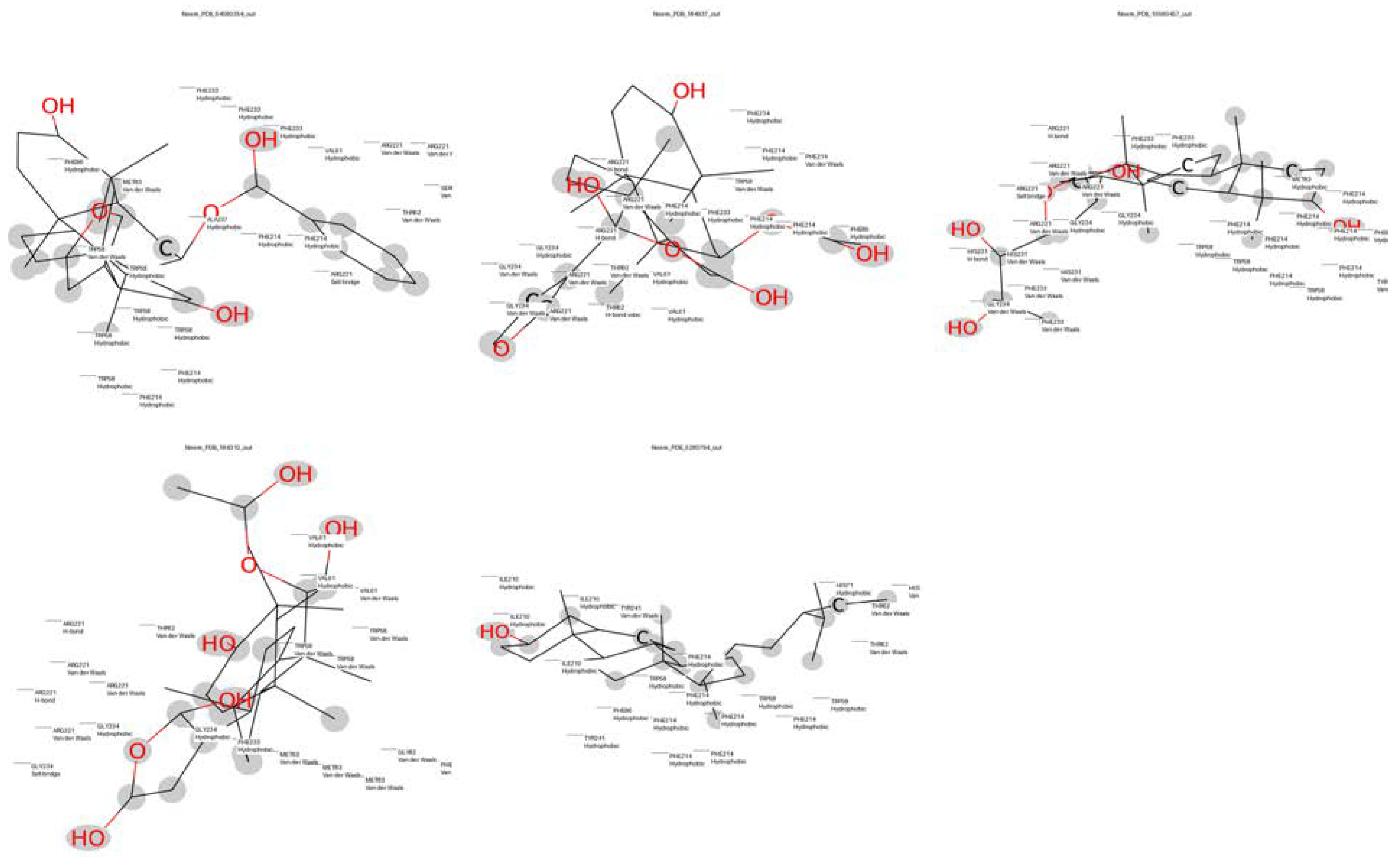
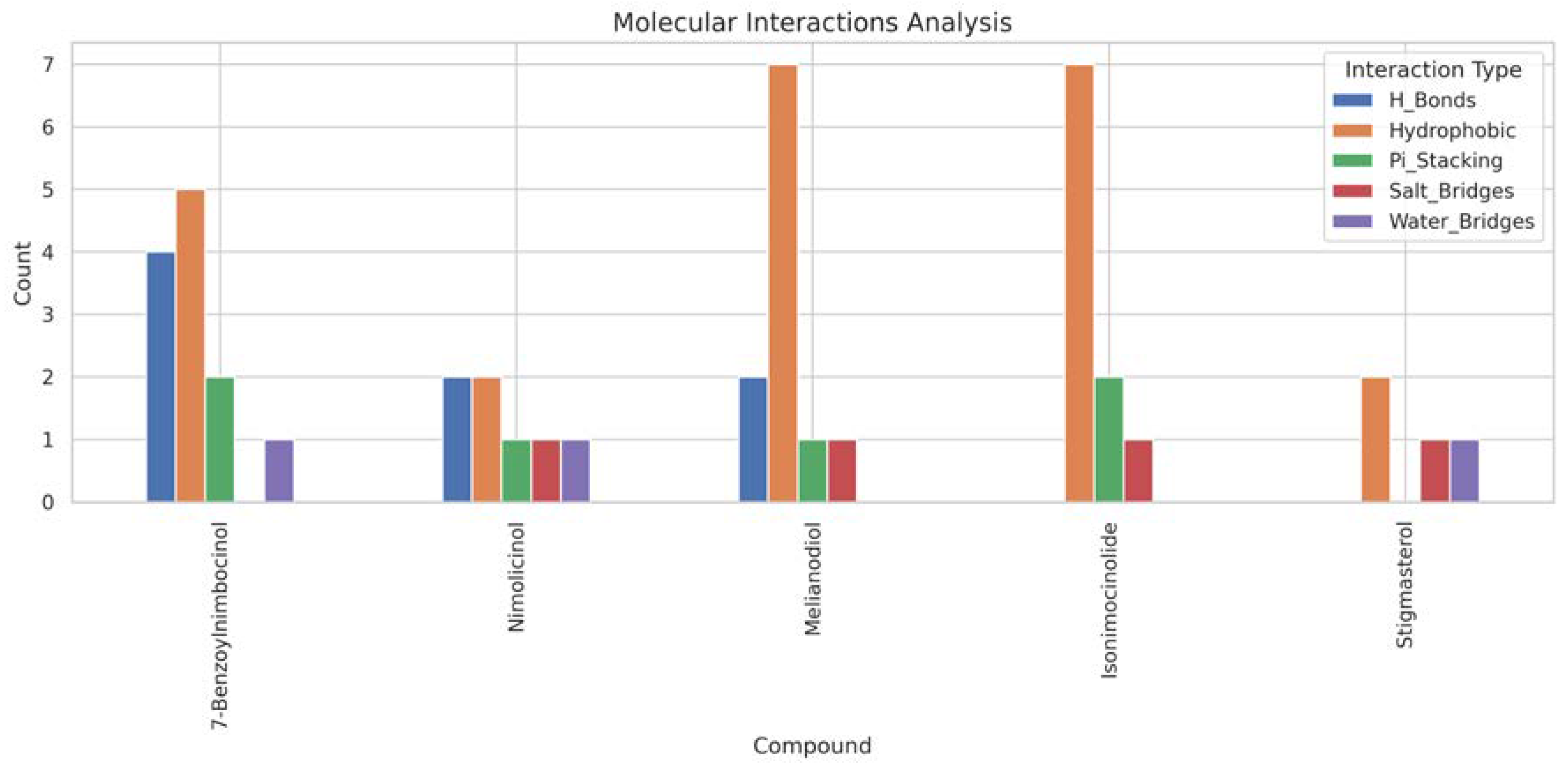


| Compound | Molecular Weight | Scaffold Weight | Scaffold Proportion | Rotatable Bonds | Binding Energy (kcal/mol) | Energy per Heavy Atom | Common Scaffold Atoms | Fragment Efficiency |
|---|---|---|---|---|---|---|---|---|
| 7-Benzoylnimbocinol | 512.26 | 442.18 | 0.86 | 3 | −11.00 | −0.29 | 14 | −0.03 |
| Nimolicinol | 482.23 | 338.15 | 0.70 | 2 | −10.80 | −0.31 | 14 | −0.03 |
| Melianodiol | 488.35 | 314.22 | 0.64 | 3 | −10.60 | −0.30 | 14 | −0.03 |
| Isonimocinolide | 484.25 | 324.17 | 0.67 | 2 | −10.10 | −0.29 | 14 | −0.03 |
| Stigmasterol | 412.37 | 230.20 | 0.56 | 5 | −10.00 | −0.33 | 14 | −0.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agboola, O.E.; Agboola, S.S.; Oyinloye, O.M.; Fadugba, A.E.; Omolayo, E.Y.; Ayinla, Z.A.; Osunsanmi, F.O.; Olaiya, O.E.; Olojo, F.O.; Ajiboye, B.O.; et al. Integrative Genomic and in Silico Analysis Reveals Mitochondrially Encoded Cytochrome C Oxidase III (MT—CO3) Overexpression and Potential Neem-Derived Inhibitors in Breast Cancer. Genes 2025, 16, 546. https://doi.org/10.3390/genes16050546
Agboola OE, Agboola SS, Oyinloye OM, Fadugba AE, Omolayo EY, Ayinla ZA, Osunsanmi FO, Olaiya OE, Olojo FO, Ajiboye BO, et al. Integrative Genomic and in Silico Analysis Reveals Mitochondrially Encoded Cytochrome C Oxidase III (MT—CO3) Overexpression and Potential Neem-Derived Inhibitors in Breast Cancer. Genes. 2025; 16(5):546. https://doi.org/10.3390/genes16050546
Chicago/Turabian StyleAgboola, Oluwaseun E., Samuel S. Agboola, Oluwatoyin M. Oyinloye, Abimbola E. Fadugba, Esther Y. Omolayo, Zainab A. Ayinla, Foluso O. Osunsanmi, Oluranti E. Olaiya, Folake O. Olojo, Basiru O. Ajiboye, and et al. 2025. "Integrative Genomic and in Silico Analysis Reveals Mitochondrially Encoded Cytochrome C Oxidase III (MT—CO3) Overexpression and Potential Neem-Derived Inhibitors in Breast Cancer" Genes 16, no. 5: 546. https://doi.org/10.3390/genes16050546
APA StyleAgboola, O. E., Agboola, S. S., Oyinloye, O. M., Fadugba, A. E., Omolayo, E. Y., Ayinla, Z. A., Osunsanmi, F. O., Olaiya, O. E., Olojo, F. O., Ajiboye, B. O., & Oyinloye, B. E. (2025). Integrative Genomic and in Silico Analysis Reveals Mitochondrially Encoded Cytochrome C Oxidase III (MT—CO3) Overexpression and Potential Neem-Derived Inhibitors in Breast Cancer. Genes, 16(5), 546. https://doi.org/10.3390/genes16050546







