Comparative Mitogenomics of Wonder Geckos (Sphaerodactylidae: Teratoscincus Strauch, 1863): Uncovering Evolutionary Insights into Protein-Coding Genes
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and DNA Extraction
2.2. Primer Design, PCR Amplification, and Sequencing
2.3. Assembly and Annotation
2.4. Bioinformatics Analyses
2.5. Phylogenetic Analysis
3. Results
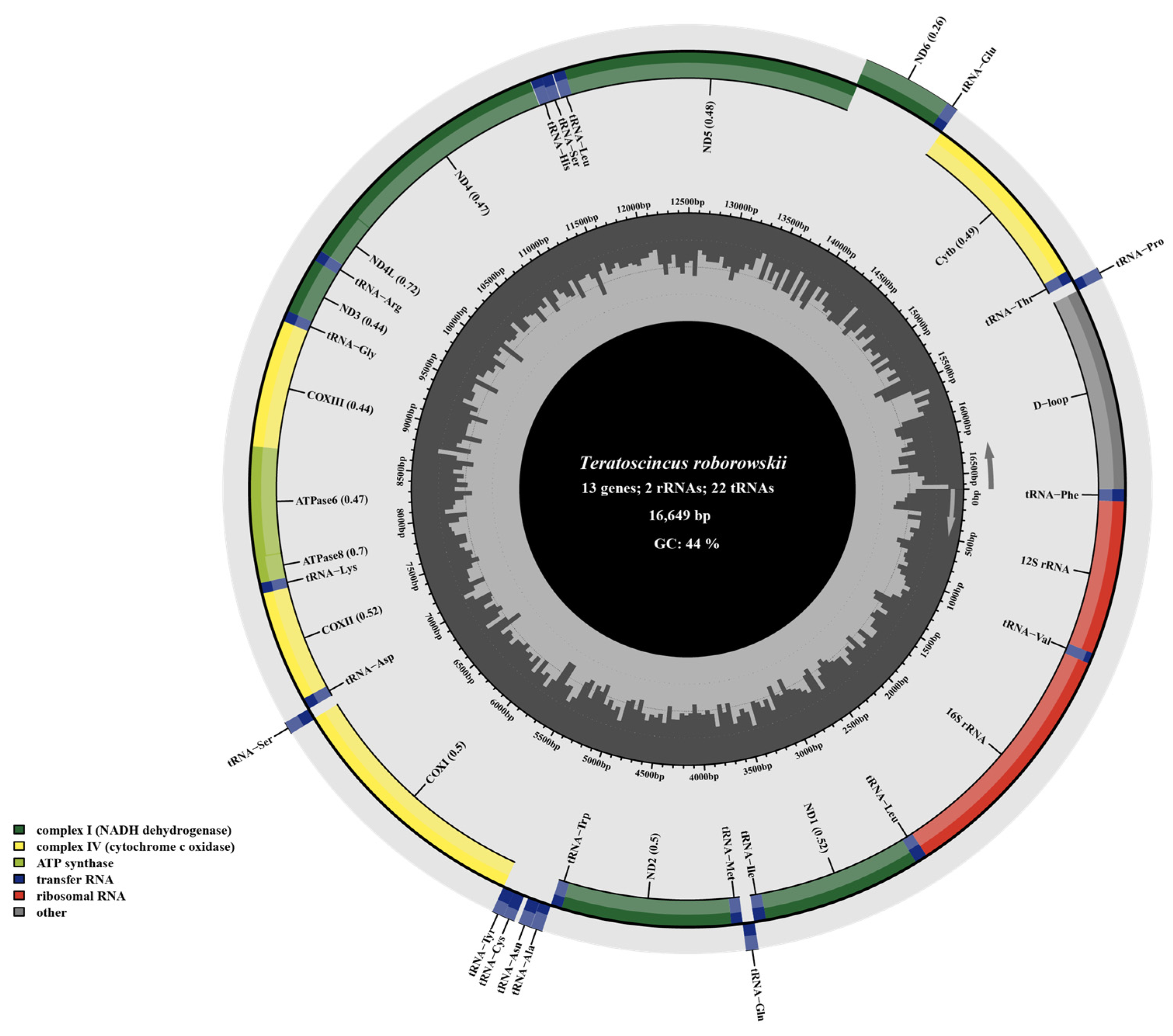
3.1. Mitogenome Organization and Nucleotide Composition
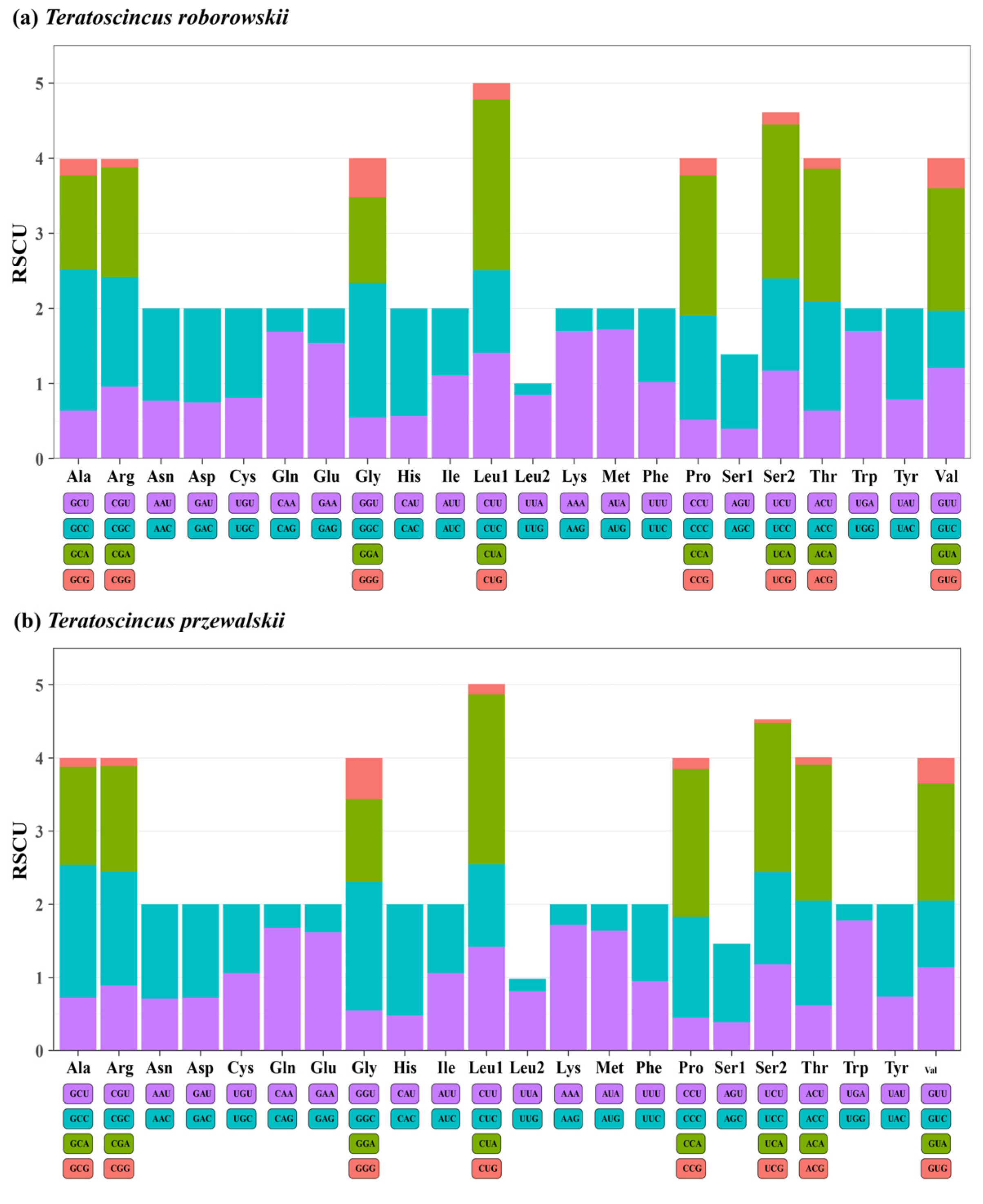
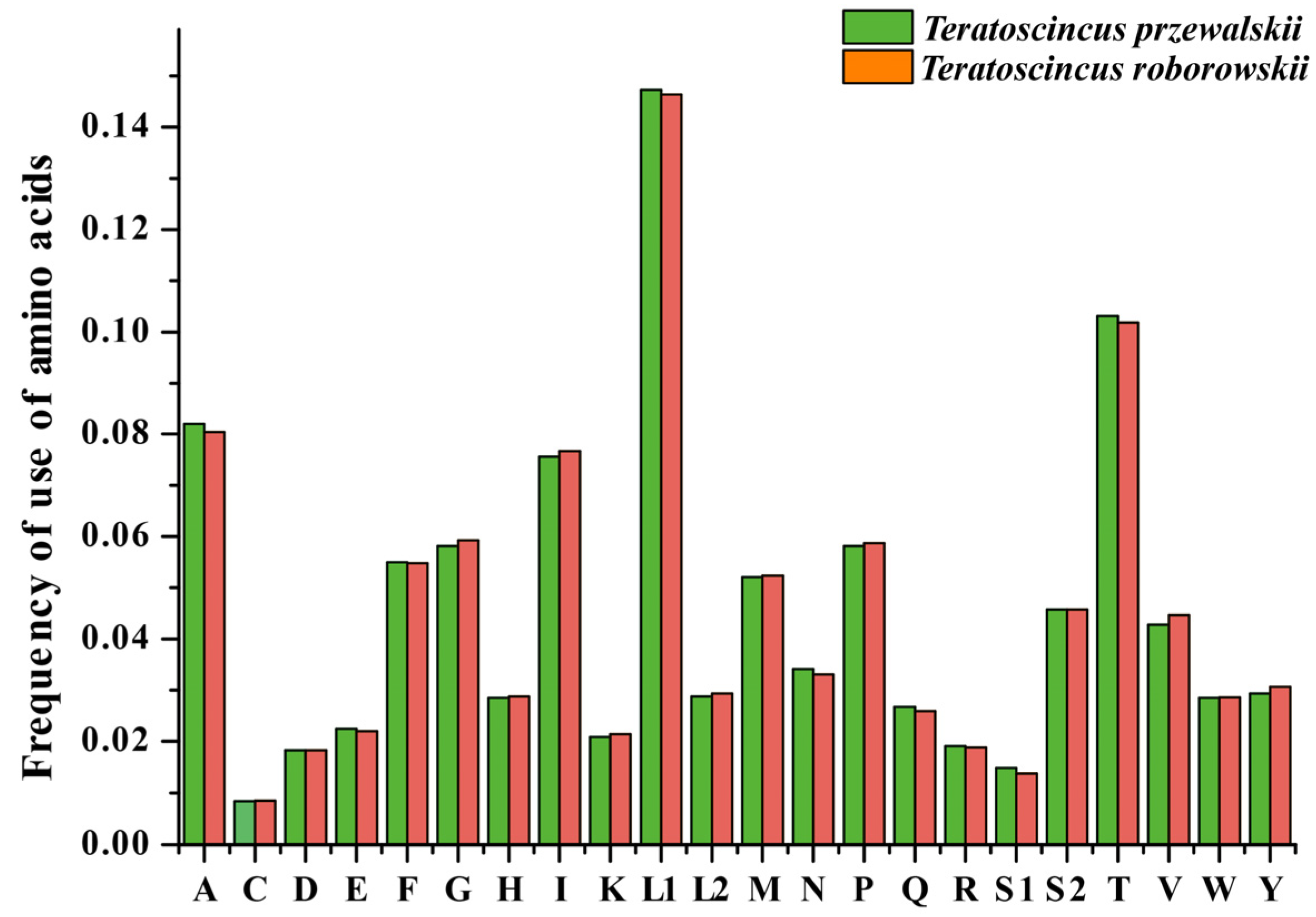
3.2. PCGs and Codon Usage
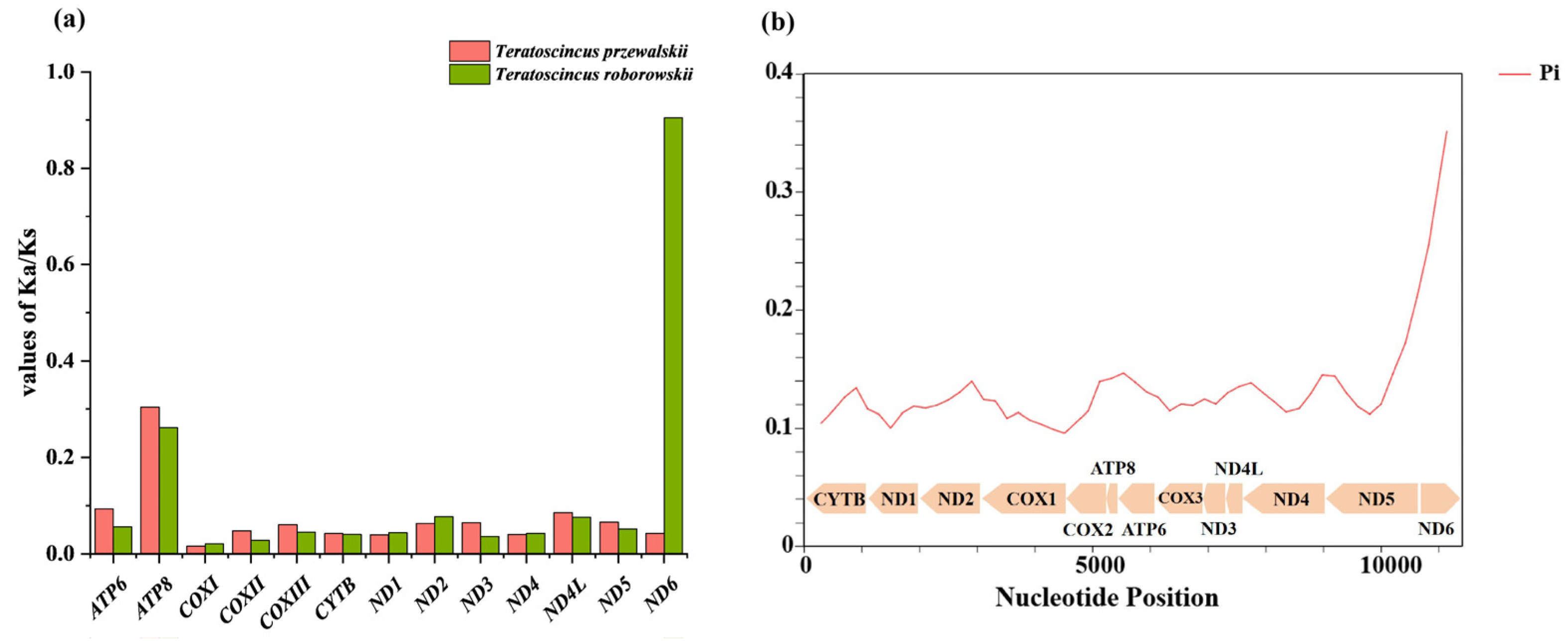
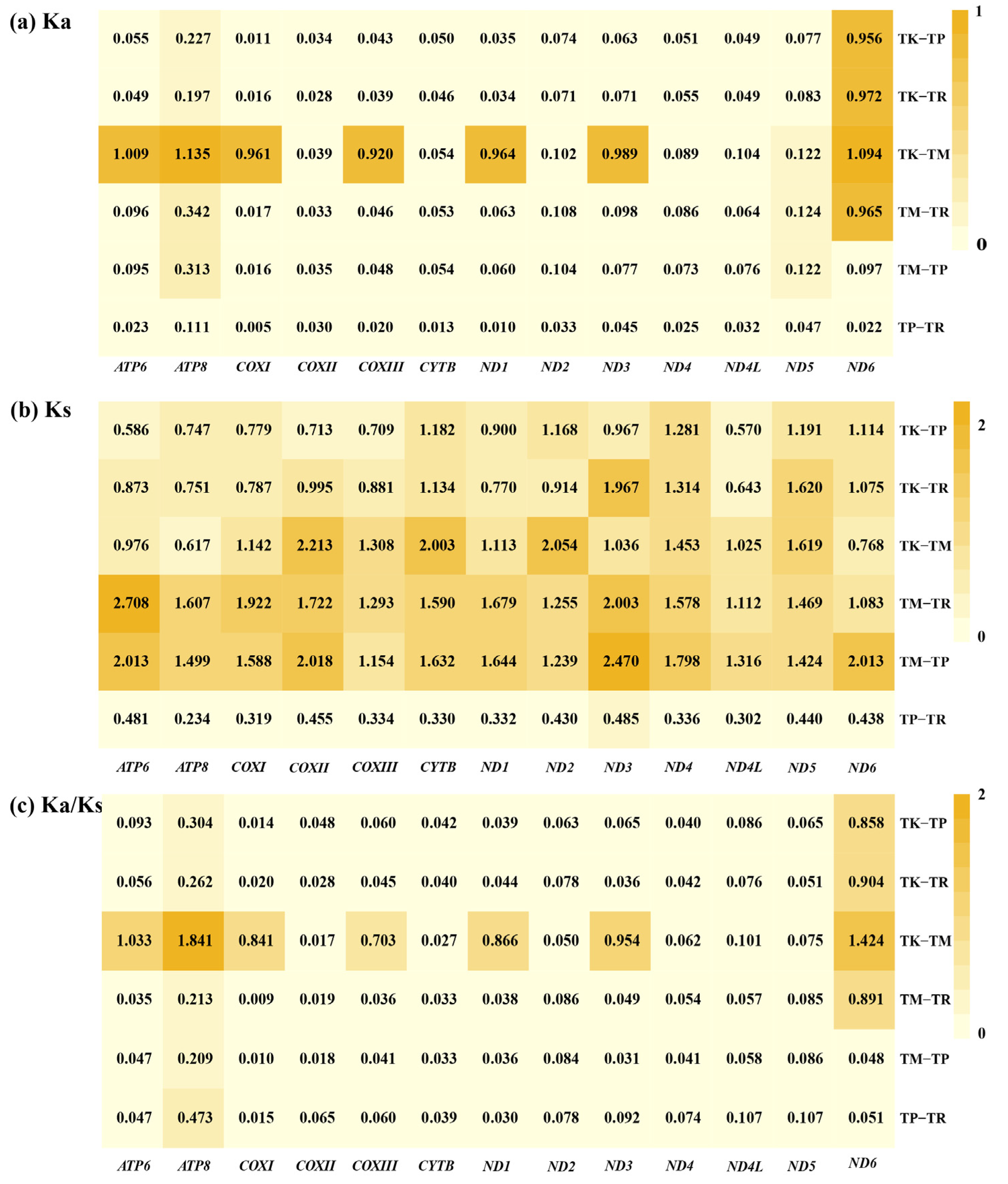
3.3. Comparative Analysis of Evolutionary Selection in Teratoscincus Species
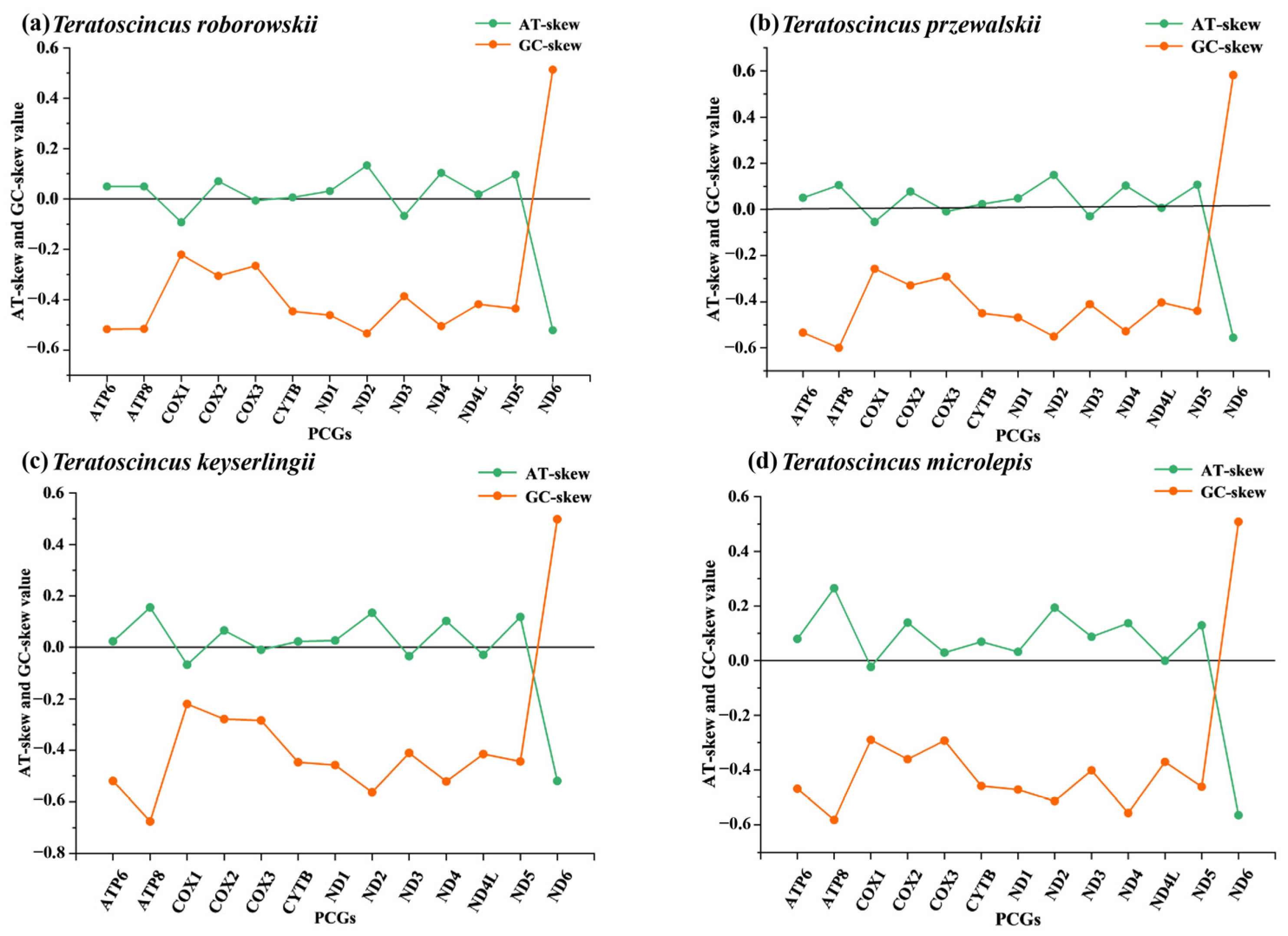
3.4. AT/GC-Skew Analysis in Teratoscincus Species
3.5. Driver of Codon Usage Bias in Teratoscincus Species
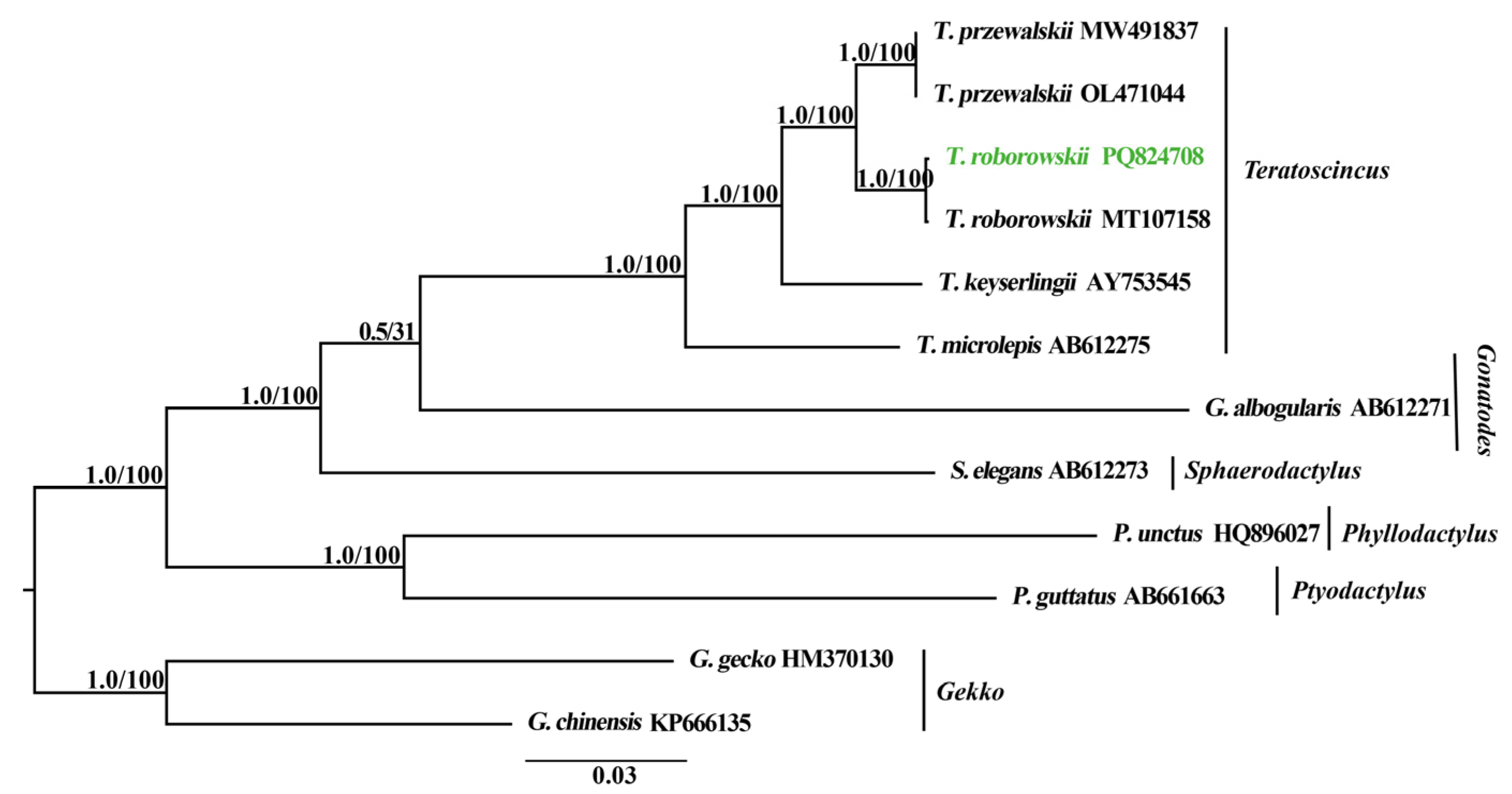
3.6. Phylogenetic Relationships
4. Discussion
4.1. Mitochondrial Genome Organization and Composition
4.2. Selection Pressure on PCGs
4.3. Evolutionary Dynamics of Mitochondrial Genes in Teratoscincus
4.4. Phylogeny of Teratoscincus
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Macey, J.R.; Wang, Y.; Ananjeva, N.B.; Larson, A.; Papenfuss, T.J. Vicariant patterns of fragmentation among gekkonid lizards of the genus Teratoscincus produced by the Indian collision: A molecular phylogenetic perspective and an area cladogram for Central Asia. Mol. Phylogenet. Evol. 1999, 12, 320–332. [Google Scholar] [CrossRef][Green Version]
- Macey, J.R.; Fong, J.J.; Kuehl, J.V.; Shafiei, S.; Ananjeva, N.B.; Papenfuss, T.J.; Boore, J.L. The complete mitochondrial genome of a gecko and the phylogenetic position of the Middle Eastern Teratoscincus keyserlingii. Mol. Phylogenet. Evol. 2005, 36, 188–193. [Google Scholar] [CrossRef][Green Version]
- Gamble, T.; Bauer, A.M.; Greenbaum, E.; Jackman, T.R. Evidence for Gondwanan vicariance in an ancient clade of gecko lizards. J. Biogeogr. 2008, 35, 88–104. [Google Scholar] [CrossRef]
- Sindaco, R.; Jeremcenko, V.K. The Reptiles of the Western Palearctic. Annotated Checklist and Distributional Atlas of the Turtles, Crocodiles, Amphisbaenians and Lizards of Europe, North Africa, Middle East and Central Asia; Edizioni Belvedere: Latina, Italy, 2008. [Google Scholar]
- Akbarpour, M.; Shafiei, S.; Sehhatisabet, M.E.; Damadi, E. A new species of frog-eyed gecko, genus Teratoscincus Strauch, 1863 (Squamata: Sphaerodactylidae), from southeastern Iran. Zool. Middle East 2017, 63, 296–302. [Google Scholar] [CrossRef]
- Wang, G.; Fan, Y.; Zhi, R.X.; Ar, L.P.; Li, S.Z.; Chen, X.R. A new species of Teratoscincus from Xinjiang China (Lacertilia: Gekkonidae). J. August 1st Agric. Coll. 1989, 4, 10–13, (In Chinese with English abstract). [Google Scholar]
- Uetz, P.; Freed, P.; Hosek, J. The Reptile Database. Available online: http://www.reptile-database.org (accessed on 20 December 2024).
- Shi, L.; Zhou, Y.H.; Yuan, H. On the dividing of the zoogeographical region of reptiles of Xinjiang Uygur Autonomous Region. Sichuan J. Zool. 2002, 21, 152–157. [Google Scholar]
- Li, H.M.; She, Y.; Hou, L.X.; Zhang, Y.; Guo, D.N.; Qin, X.M. The complete mitochondrial genome of Teratoscincus roborowskii (Squamata: Gekkonidae). Mitochondrial DNA Part A 2016, 27, 1916–1917. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.C.; Liu, Y.; Lin, Y.Y.; Liang, T.; Shi, L. Burrow characteristics and microhabitat Use of the Turpan Wonder Gecko Teratoscincus roborowskii (Squamata, Gekkonidae). Stud. Asian Amphib. Reptiles 2017, 18, 61–69. [Google Scholar]
- Semenov, D.V.; Borkin, L.J. On the ecology of Przewalsky’s Gecko (Teratoscincus przewalskii) in the Transaltai Gobi, Mongolia. Asiat. Herpetol. Res. 1992, 4, 99–112. [Google Scholar]
- Yu, H.; Liu, Y.; Liu, Y.; Yang, J.; Li, S.; Bi, J.; Zhang, R. Complete mitochondrial genome of Teratoscincus przewalskii (Reptilia, Squamata, Sphaerodactylidae) and phylogenetic analysis. Mitochondrial DNA Part B 2021, 6, 3166–3168. [Google Scholar] [CrossRef]
- Seligmann, H.; Anderson, S.C.; Autumn, K.; Bouskila, A.; Saf, R.; Tuniyev, B.S.; Werner, Y.L. Analysis of the locomotor activity of a nocturnal desert lizard (Reptilia: Gekkonidae: Teratoscincus scincus) under varying moonlight. Zoology 2007, 110, 104–117. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhou, S.; Wu, X.; Wei, Q.; Shang, Y.; Sun, G.; Mei, X.; Dong, Y.; Sha, W.; Zhang, H. High-altitude adaptation in vertebrates as revealed by mitochondrial genome analyses. Ecol. Evol. 2021, 11, 15077–15084. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.H.; Hu, S.Y.; Li, M.; Liu, M.; Sun, H.; Zhao, J.R.; Chen, W.T.; Yuan, M.L. Comparative mitogenomic analyses of darkling beetles (Coleoptera: Tenebrionidae) provide evolutionary insights into tRNA-like sequences. Genes 2023, 14, 1738. [Google Scholar] [CrossRef]
- Rehman, A.; Huo, Q.B.; Du, Y.Z. The first complete mitochondrial Genome of genus Isocapnia (Plecoptera: Capniidae) and phylogenetic assignment of superfamily Nemouroidea. Genes 2023, 14, 965. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef]
- Wang, I.C.; Lin, H.D.; Liang, C.M.; Huang, C.C.; Wang, R.D.; Yang, J.Q.; Wang, W.K. Complete mitochondrial genome of the freshwater fish Onychostoma lepturum (Teleostei, Cyprinidae): Genome characterization and phylogenetic analysis. Zookeys 2020, 1005, 57–72. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [PubMed]
- Böhme, M.U.; Fritzsch, G.; Tippmann, A.; Schlegel, M.; Berendonk, T.U. The complete mitochondrial genome of the green lizard Lacerta viridis viridis (Reptilia: Lacertidae) and its phylogenetic position within squamate reptiles. Gene 2007, 394, 69–77. [Google Scholar] [CrossRef]
- Tian, L.; Yang, W.; Si, C.; Guo, X.; Zhang, B. Complete mitogenome analysis of five leafhopper species of Idiocerini (Hemiptera: Cicadellidae). Genes 2022, 13, 2000. [Google Scholar] [CrossRef]
- Shan, W.; Tursun, M.; Zhou, S.; Zhang, Y.; Dai, H. Complete mitochondrial genome sequence of Lepus yarkandensis Günther, 1875 (Lagomorpha, Leporidae): Characterization and phylogenetic analysis. Zookeys 2021, 1012, 135–150. [Google Scholar] [CrossRef]
- Li, J.; Guo, X.G. A complete mitogenome of the Przewalski’s Wonder Gecko (Teratoscincus przewalskii) from the Junggar Basin in Northwest China with its phylogenetic implications. Mitochondrial DNA Part B 2023, 8, 398–401. [Google Scholar] [CrossRef] [PubMed]
- Tarroso, P.; Simó-Riudalbas, M.; Carranza, S. The complete mitochondrial genome of Pristurusru pestrisrupestris. Mitochondrial DNA Part B 2017, 2, 802–803. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.L.; Dai, X.Y.; Xu, X.D.; Guan, J.Y.; Zhang, Y.P.; Zhang, J.Y.; Yu, D.N. The complete mitochondrial genome of Teratoscincus roborowskii (Squamata: Gekkonidae) and its phylogeny. Mitochondrial DNA Part B 2020, 5, 1575–1577. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, W.; Fukunaga, T.; Isagozawa, R.; Yamada, K.; Maeda, Y.; Sato, T.P.; Sado, T.; Mabuchi, K.; Takeshima, H.; Miya, M.; et al. MitoFish and MitoAnnotator: A mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol. Biol. Evol. 2013, 30, 2531–2540. [Google Scholar] [CrossRef]
- Zhu, T.; Sato, Y.; Sado, T.; Miya, M.; Iwasaki, W. MitoFish, MitoAnnotator, and MiFishpipeline: Updates in ten years. Mol. Biol. Evol. 2023, 40, msad035. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Zhang, Z. KaKs_Calculator 3.0: Calculating selective pressure on coding and non-coding sequences. Genom. Proteom. Bioinform. 2022, 20, 536–540. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Racine, J.S. RStudio: A platform-independent IDE for R and Sweave. J. Appl. Econom. 2012, 27, 167–172. [Google Scholar]
- Hao, S.; Ping, J.; Zhang, Y. Complete mitochondrial genome of Gekko chinensis (Squamata, Gekkonidae). Mitochondrial DNA Part A 2016, 27, 4226–4227. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Yang, Z.; Bielawski, J.P. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 2000, 15, 496–503. [Google Scholar] [CrossRef]
- Pyron, R.A.; Burbrink, F.T.; Wiens, J.J. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Ecol. Evol. 2013, 13, 93. [Google Scholar] [CrossRef]
- Tamar, K.; Els, J.; Kornilios, P.; Soorae, P.; Tarroso, P.; Thanou, E.; Pereira, J.; Shah, J.N.; Elhassan, E.E.M.; Aguhob, J.C.; et al. The demise of a wonder: Evolutionary history and conservation assessments of the Wonder Gecko Teratoscincus keyserlingii (Gekkota, Sphaerodactylidae) in Arabia. PLoS ONE 2021, 16, e0244150. [Google Scholar] [CrossRef]
- Macey, J.R.; Larson, A.; Ananjeva, N.B.; Fang, Z.; Papenfuss, T.J. Two novel gene orders and the role of light-strand replication in rearrangement of the vertebrate mitochondrial genome. Mol. Biol. Evol. 1997, 14, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Liu, J.; Wang, S.; Guo, X. Comparative analysis of mitochondrial genomes in two subspecies of the sunwatcher toadheaded agama (Phrynocephalus helioscopus): Prevalent intraspecific gene rearrangements in Phrynocephalus. Genes 2022, 13, 203. [Google Scholar] [CrossRef] [PubMed]
- Lavrov, D.V.; Boore, J.L.; Brown, W.M. Complete mtDNA sequences of two millipedes suggest a new model for mitochondrial gene rearrangements: Duplication and nonrandom loss. Mol. Biol. Evol. 2002, 19, 163–169. [Google Scholar] [CrossRef]
- Pääbo, S.; Thomas, W.K.; Whitfield, K.M.; Kumazawa, Y.; Wilson, A.C. Rearrangements of mitochondrial transfer RNA genes in marsupials. J. Mol. Evol. 1991, 33, 426–430. [Google Scholar] [CrossRef]
- Okajima, Y.; Kumazawa, Y. Mitochondrial genomes of acrodont lizards: Timing of gene rearrangements and phylogenetic and biogeographic implications. BMC Evol. Biol. 2010, 10, 141. [Google Scholar] [CrossRef]
- Chen, M.; Liu, J.; Chen, D.; Guo, X. Complete mitochondrial genome of a blue-tailed skink Plestiodoncapito (Reptilia, Squamata, Scincidae) and comparison with other Scincidae lizards. Genetica 2020, 148, 229–241. [Google Scholar] [CrossRef]
- Hassanin, A.; Leger, N.; Deutsch, J. Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of Metazoa, and consequences for phylogenetic inferences. Syst. Biol. 2005, 54, 277–298. [Google Scholar] [CrossRef] [PubMed]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA procession in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Castellana, S.; Vicario, S.; Saccone, C. Evolutionary patterns of the mitochondrial genome in Metazoa: Exploring the role of mutation and selection in mitochondrial protein coding genes. Genome Biol. Evol. 2011, 3, 1067–1079. [Google Scholar] [CrossRef]
- Muse, S.V. Examining rates and patterns of nucleotide substitution in plants. Plant Mol. Biol. 2000, 42, 25–43. [Google Scholar] [CrossRef]
- Castoe, T.A.; de Koning, A.J.; Kim, H.M.; Gu, W.; Noonan, B.P.; Naylor, G.; Jiang, Z.J.; Parkinson, C.L.; Pollock, D.D. Evidence for an ancient adaptive episode of convergent molecular evolution. Proc. Natl. Acad. Sci. USA 2009, 106, 8986–8991. [Google Scholar] [CrossRef]
- Yang, Z.; Nielsen, R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 2002, 19, 908–917. [Google Scholar] [CrossRef] [PubMed]
- Gharib, W.H.; Robinson-Rechavi, M. The branch-site test of positive selection is surprisingly robust but lacks power under synonymous substitution saturation and variation in GC. Mol. Biol. Evol. 2013, 30, 1675–1686. [Google Scholar] [CrossRef]
- Jin, Y.; Wo, Y.; Tong, H.; Song, S.; Zhang, L.; Brown, R.P. Evolutionary analysis of mitochondrially encoded proteins of toad-headed lizards, Phrynocephalus, along an altitudinal gradient. BMC Genom. 2018, 19, 185. [Google Scholar] [CrossRef] [PubMed]
- Mei, Q.; Deng, Y.; Zhao, D.; Jiang, D.; Liao, Y.; Yu, X.; Liu, P.; Jiang, L. Characterization of the Complete Mitogenome of the Ring-Necked Pheasant Phasianus colchicus (Galliformes: Phasianidae) and Systematic Implications for Phasianinae Phylogenetics. Genes 2024, 15, 1569. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.H.; Lu, C.H. Comparative Analysis and Phylogenetic Study of Dawkinsia filamentosa and Pethia nigrofasciata Mitochondrial Genomes. Int. J. Mol. Sci. 2024, 25, 3004. [Google Scholar] [CrossRef]
- Jeffares, D.C.; Tomiczek, B.; Sojo, V.; Reis, M. A beginners guide to estimating the non-synonymous to synonymous rate ratio of all protein-coding genes in a genome. Parasite Genom. Protoc. 2015, 1201, 65–90. [Google Scholar]
- Roy, A.; Mukhopadhyay, S.; Sarkar, I.; Sen, A. Comparative investigation of the various determinants that influence the codon and amino acid usage patterns in the genus Bifidobacterium. World J. Microbiol. Biotechnol. 2015, 31, 959–981. [Google Scholar] [CrossRef]
- Smith, R.D. Enhanced effective codon numbers to understand codon usage bias. Biosystems 2022, 220, 104734. [Google Scholar] [CrossRef]
- Xu, X.D.; Guan, J.Y.; Zhang, Z.Y.; Cao, Y.R.; Cai, Y.Y.; Storey, K.B.; Yu, D.N.; Zhang, J.Y. Insight into the phylogenetic relationships among three subfamilies within Heptageniidae (Insecta: Ephemeroptera) along with low-temperature selection pressure analyses using mitogenomes. Insects 2021, 12, 656. [Google Scholar] [CrossRef]
- Ballard, J.W.; Melvin, R.G.; Katewa, S.D.; Maas, K. Mitochondrial DNA variation is associated with measurable differences in life-history traits and mitochondrial metabolism in Drosophila simulans. Evolution 2007, 61, 1735–1747. [Google Scholar] [CrossRef] [PubMed]
- Domrös, M.; Peng, G.; Zhang, S. The characteristics of the desert climate at Turpan, China. Erdkunde 1992, 46, 217–233. [Google Scholar] [CrossRef]
- Wei, Q.; Wang, X.; Dong, Y.; Shang, Y.; Sun, G.; Wu, X.; Zhao, X.; Sha, W.; Yang, G.; Zhang, H. Analysis of the Complete Mitochondrial Genome of Pteronura brasiliensis and Lontra canadensis. Animals 2023, 13, 3165. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; He, W.; Huang, H.; Ou, D.; Wang, L.; Li, J.; Lou, S. A comprehensive analysis of the Fowleria variegata (Valenciennes, 1832) mitochondrial genome and its phylogenetic implications within the family Apogonidae. Genes 2023, 14, 1612. [Google Scholar] [CrossRef]

| Region | Size (bp) | A + T Content (%) | G + C Content (%) | AT-Skew | GC-Skew | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| TP | TR | TP | TR | TP | TR | TP | TR | TP | TR | |
| Whole genome | 17,184 | 16649 | 55.8 | 56.2 | 44.2 | 43.8 | 0.103 | 0.089 | 0.371 | 0.356 |
| PCGs | 11,367 | 11340 | 56.4 | 56.5 | 43.6 | 43.5 | 0.026 | 0.012 | 0.380 | 0.366 |
| rRNA genes | 2493 | 2503 | 54.2 | 54.6 | 45.7 | 45.4 | 0.240 | 0.224 | 0.225 | 0.207 |
| tRNA genes | 1531 | 1531 | 56.3 | 56.2 | 43.7 | 43.8 | 0.058 | 0.06 | 0.001 | 0.004 |
| CR | 1774 | 1247 | 54.1 | 56.8 | 45.9 | 43.2 | 0.083 | 0.067 | 0.315 | 0.343 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, D.; Ma, R.; Guo, X.; Li, J. Comparative Mitogenomics of Wonder Geckos (Sphaerodactylidae: Teratoscincus Strauch, 1863): Uncovering Evolutionary Insights into Protein-Coding Genes. Genes 2025, 16, 531. https://doi.org/10.3390/genes16050531
Zheng D, Ma R, Guo X, Li J. Comparative Mitogenomics of Wonder Geckos (Sphaerodactylidae: Teratoscincus Strauch, 1863): Uncovering Evolutionary Insights into Protein-Coding Genes. Genes. 2025; 16(5):531. https://doi.org/10.3390/genes16050531
Chicago/Turabian StyleZheng, Dongqing, Rongrong Ma, Xianguang Guo, and Jun Li. 2025. "Comparative Mitogenomics of Wonder Geckos (Sphaerodactylidae: Teratoscincus Strauch, 1863): Uncovering Evolutionary Insights into Protein-Coding Genes" Genes 16, no. 5: 531. https://doi.org/10.3390/genes16050531
APA StyleZheng, D., Ma, R., Guo, X., & Li, J. (2025). Comparative Mitogenomics of Wonder Geckos (Sphaerodactylidae: Teratoscincus Strauch, 1863): Uncovering Evolutionary Insights into Protein-Coding Genes. Genes, 16(5), 531. https://doi.org/10.3390/genes16050531







