Analysis of the DYNC1H1 Gene Polymorphic Variants’ Association with ASD Occurrence and Clinical Phenotype of Affected Children
Highlights
- The DYNC1H1 rs3818188 polymorphism is associated with ASD in a subgroup of girls.
- rs2403015 is associated with a transient increase in muscle tone during infancy.
- This variant affects gene expression in skeletal muscle (based on in silico analysis).
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Materials
2.3. Genetic Analysis
2.4. Statistical Analysis
3. Results
3.1. Analysis of the Association Between DYNC1H1 Gene Polymorphism and ASD Occurrence
3.2. Analysis of the Association Between DYNC1H1 Gene Polymorphism and Clinical Phenotype
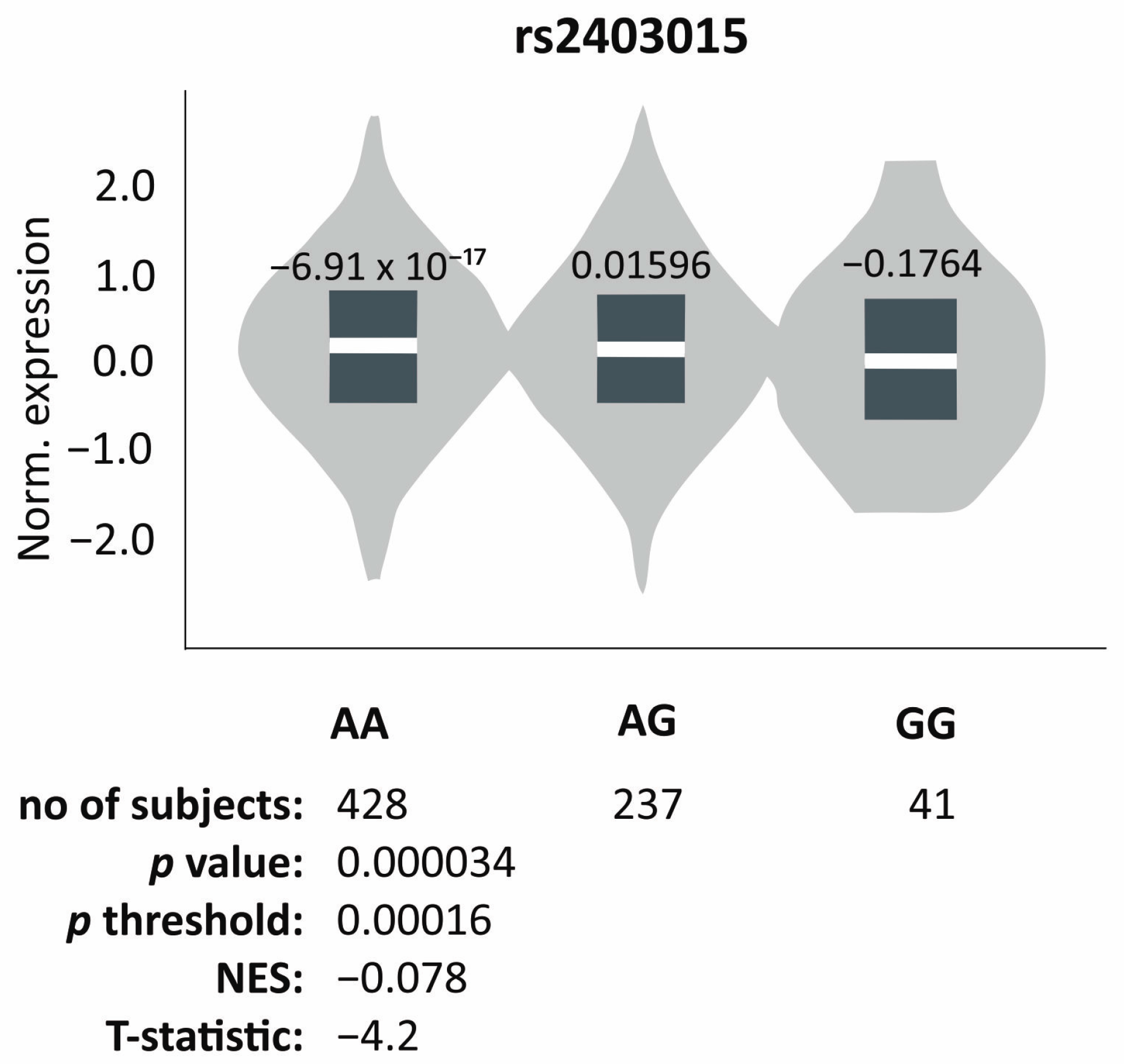
3.3. In Silico Analysis of the Influence of Polymorphic Variants on Gene Expression
4. Discussion
Study Limitations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DYNC1H1 | Dynein cytoplasmic 1 heavy chain 1 |
| ASD | Autism spectrum disorder |
| TDT | Transmission disequilibrium test |
References
- Kereszturi, É. Diversity and Classification of Genetic Variations in Autism Spectrum Disorder. Int. J. Mol. Sci. 2023, 24, 16768. [Google Scholar] [CrossRef] [PubMed]
- Schiavo, G.; Greensmith, L.; Hafezparast, M.; Fisher, E.M. Cytoplasmic dynein heavy chain: The servant of many masters. Trends Neurosci. 2013, 36, 641–651. [Google Scholar] [CrossRef] [PubMed]
- Viollet, L.M.; Swoboda, K.J.; Mao, R.; Best, H.; Ha, Y.; Toutain, A.; Guyant-Marechal, L.; Laroche-Raynaud, C.; Ghorab, K.; Barthez, M.A.; et al. A novel pathogenic variant in DYNC1H1 causes various upper and lower motor neuron anomalies. Eur. J. Med. Genet. 2020, 63, 104063. [Google Scholar] [CrossRef] [PubMed]
- Becker, L.L.; Dafsari, H.S.; Schallner, J.; Abdin, D.; Seifert, M.; Petit, F.; Smol, T.; Bok, L.; Rodan, L.; Krapels, I.; et al. The clinical-phenotype continuum in DYNC1H1-related disorders-genomic profiling and proposal for a novel classification. J. Hum. Genet. 2020, 65, 1003–1017. [Google Scholar] [CrossRef] [PubMed]
- Hoang, H.T.; Schlager, M.A.; Carter, A.P.; Bullock, S.L. DYNC1H1 mutations associated with neurological diseases compromise processivity of dynein-dynactin-cargo adaptor complexes. Proc. Natl. Acad. Sci. USA 2017, 114, E1597–E1606. [Google Scholar] [CrossRef] [PubMed]
- Yuen, R.K.; Merico, D.; Bookman, M.; Howe, J.; Thiruvahindrapuram, B.; Patel, R.V.; Whitney, J.; Deflaux, N.; Bingham, J.; Wang, Z.; et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat. Neurosci. 2017, 20, 602–611. [Google Scholar] [CrossRef] [PubMed]
- Abrahams, B.S.; Arking, D.E.; Campbell, D.B.; Mefford, H.C.; Morrow, E.M.; Weiss, L.A.; Menashe, I.; Wadkins, T.; Banerjee-Basu, S.; Packer, A. SFARI Gene 2.0: A community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol. Autism 2013, 4, 36. [Google Scholar] [CrossRef] [PubMed]
- Amabile, S.; Jeffries, L.; McGrath, J.M.; Ji, W.; Spencer-Manzon, M.; Zhang, H.; Lakhani, S.A. DYNC1H1-related disorders: A description of four new unrelated patients and a comprehensive review of previously reported variants. Am. J. Med. Genet. 2020, 182, 2049–2057. [Google Scholar] [CrossRef] [PubMed]
- Harrison, P.W.; Amode, M.R.; Austine-Orimoloye, O.; Azov, A.G.; Barba, M.; Barnes, I.; Becker, A.; Bennett, R.; Berry, A.; Bhai, J.; et al. Ensembl 2024. Nucleic Acids Res. 2024, 52, D891–D899. [Google Scholar] [CrossRef] [PubMed]
- Jankowska, K.; Meyer, D.; Holcomb, D.D.; Kames, J.; Hamasaki-Katagiri, N.; Katneni, U.K.; Hunt, R.C.; Ibla, J.C.; Kimchi-Sarfaty, C. Synonymous ADAMTS13 variants impact molecular characteristics and contribute to variability in active protein abundance. Blood Adv. 2022, 6, 5364–5378. [Google Scholar] [CrossRef] [PubMed]
- Stachelek, K.; Harutyunyan, N.; Lee, S.; Beck, A.; Kim, J.; Xu, L.; Berry, J.L.; Nagiel, A.; Reynolds, C.P.; Murphree, A.L.; et al. Non-synonymous, synonymous, and non-coding nucleotide variants contribute to recurrently altered biological processes during retinoblastoma progression. Genes Chromosomes Cancer 2023, 62, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Huang, K.; Wang, Y.; Hu, E.; Wei, B.; Song, Z.; Zou, Y.; Ge, L.; Chen, L.; Li, W. Integration of SNP Disease Association, eQTL, and Enrichment Analyses to Identify Risk SNPs and Susceptibility Genes in Chronic Obstructive Pulmonary Disease. BioMed Res. Int. 2020, 2020, 3854196. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Zhang, T.; Wang, Y.; Wang, L.; Yan, Z.; Teng, Y.; Li, Z.; Lou, Q.; Liu, S.; Cai, J.; et al. Association of DYNC1H1 gene SNP/CNV with disease susceptibility, GCs efficacy, HRQOL, anxiety, and depression in Chinese SLE patients. J. Clin. Lab Anal. 2021, 35, e23892. [Google Scholar] [CrossRef] [PubMed]
- Kanne, S.M.; Randolph, J.K.; Farmer, J.E. Diagnostic and assessment findings: A bridge to academic planning for children with autism spectrum disorders. Neuropsychol. Rev. 2008, 18, 367–384. [Google Scholar] [CrossRef] [PubMed]
- GTEx Portal. Available online: https://gtexportal.org/home/testyourown (accessed on 26 September 2024).
- Menyhart, O.; Weltz, B.; Győrffy, B. MultipleTesting.com: A tool for life science researchers for multiple hypothesis testing correction. PLoS ONE 2021, 16, e0245824. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, S.B.; Schaffner, S.F.; Nguyen, H.; Moore, J.M.; Roy, J.; Blumenstiel, B.; Higgins, J.; DeFelice, M.; Lochner, A.; Faggart, M.; et al. The structure of haplotype blocks in the human genome. Science 2002, 296, 2225–2229. [Google Scholar] [CrossRef] [PubMed]
- Asemota, S.; Effah, W.; Young, K.L.; Holt, J.; Cripe, L.; Ponnusamy, S.; Thiyagarajan, T.; Hwang, D.J.; He, Y.; Mcnamara, K.; et al. Identification of a targetable JAK-STAT enriched androgen receptor and androgen receptor splice variant positive triple-negative breast cancer subtype. Cell Rep. 2023, 42, 113461. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Han, J.; Zhou, H.; Ai, S.; Wan, D. A Prognosis Marker Dynein Cytoplasmic 1 Heavy Chain 1 Correlates with EMT and Immune Signature in Liver Hepatocellular Carcinoma by Bioinformatics and Experimental Analysis. Dis. Markers 2022, 2022, 6304859. [Google Scholar] [CrossRef] [PubMed]
| Genotype | Genotype Distribution, n (%) | |||
|---|---|---|---|---|
| Children | Mothers | Fathers | ||
| rs3818188 | n | 205 | 205 | 205 |
| GG | 132 (64.4) | 135 (65.8) | 118 (57.6) | |
| GA | 66 (32.2) | 60 (29.3) | 79 (38.5) | |
| AA | 7 (3.4) | 10 (4.9) | 8 (3.9) | |
| rs941793 | n | 205 | 205 | 205 |
| AA | 141 (68.8) | 137 (66.8) | 142 (69.3) | |
| AG | 55 (26.8) | 59 (28.8) | 54 (26.3) | |
| GG | 9 (4.4) | 9 (4.4) | 9 (4.4) | |
| rs2403015 | n | 203 | 202 | 203 |
| AA | 153 (75.4) | 144 (71.3) | 154 (75.9) | |
| AG | 44 (21.7) | 55 (27.2) | 42 (20.7) | |
| GG | 6 (2.9) | 3 (1.5) | 7 (3.4) | |
| Allele | Transmitted n (%) | Not Transmitted n (%) | χ2; p |
|---|---|---|---|
| rs3818188 | n informative trios = 115 | ||
| G A | 77 (55.4) 62 (44.6) | 62 (44.6) 77 (55.4) | 1.619, 0.203 |
| rs941793 | n informative trios = 99 | ||
| A G | 57 (50.4) 56 (49.6) | 56 (49.6) 57 (50.4) | 0.009, 0.925 |
| rs2403015 | n informative trios = 86 | ||
| A G | 50 (52.1) 46 (47.9) | 46 (47.9) 50 (52.1) | 0.167, 0.683 |
| Allele | Transmitted n (%) | Not Transmitted n (%) | χ2; p |
|---|---|---|---|
| rs3818188 | Boys n informative trios = 86 | ||
| G A | 54 (50.5) 53 (49.5) | 53 (49.5) 54 (50.5) | 0.019; 0.891 |
| rs3818188 | Girls n informative trios = 29 | ||
| G A | 23 (71.9) 9 (28.1) | 9 (28.1) 23 (71.9) | 6.125; 0.013 * |
| Characteristics | n (%) | n Available Data * |
|---|---|---|
| Abnormal motor development | 79 (41.6) | 190 |
| Transient increase in muscle tone (infant period) | 37 (19.3) | 192 |
| Decrease in muscle tone | 67 (35.3) | 190 |
| Asymmetric position | 45 (23.2) | 194 |
| SI impairments | 172 (89.6) | 192 |
| Need for rehabilitation | 64 (32.3) | 198 |
| Hearing impairments | 32 (16.6) | 193 |
| Vision impairments | 54 (28.1) | 192 |
| Regression in communication | 77 (39.5) | 195 |
| Impairment in eye contact | 135 (74.6) | 181 |
| Bye-bye gesture | 119 (68.0) | 175 |
| Reaction to name | 131 (67.5) | 194 |
| Pretend play | 59 (30.4) | 194 |
| Compulsive, ritualistic behaviour | 141 (75.0) | 188 |
| Self-aggressive behaviour | 78 (39.8) | 196 |
| Genotype Frequency of rs3818188 Polymorphism and Skill of Pretend Play (Recessive/Dominant Model) | ||||
| Yes (n, %) | No (n, %) | χ2; p | ||
| AA + GA GG | 28 (47.5) 31 (52.5) | 43 (31.9) 92 (68.1) | vs. GG | 4.31; 0.038 1 |
| Genotype frequency of rs941793 polymorphism and bye-bye gesture (recessive/dominant model) | ||||
| Yes (n, %) | No (n, %) | χ2; p | ||
| GG + AG AA | 29 (24.4) 90 (75.6) | 22 (39.3) 34 (60.7) | vs. AA | 4.10; 0.043 1 |
| Genotype frequency of rs2403015 polymorphism and transient increase in muscle tone during infancy (recessive/dominant model) | ||||
| Yes (n, %) | No (n, %) | χ2; p | ||
| GG + AG AA | 14 (37.8) 23 (62.2) | 30 (19.4) 125 (80.6) | vs. AA | 5.78; 0.016 1,2 |
| Genotype frequency of rs2403015 polymorphism and hearing impairments (additive model) | ||||
| Yes (n, %) | No (n, %) | χ2; p | ||
| AA AG GG | 27 (84.4) 3 (9.4) 2 (6.2) | 121 (75.2) 38 (23.6) 2 (1.2) | 6.07; 0.048 1 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balcerzyk-Matić, A.; Iwanicki, T.; Jarosz, A.; Nowak, T.; Emich-Widera, E.; Kazek, B.; Kapinos-Gorczyca, A.; Kapinos, M.; Iwanicka, J.; Gawron, K.; et al. Analysis of the DYNC1H1 Gene Polymorphic Variants’ Association with ASD Occurrence and Clinical Phenotype of Affected Children. Genes 2025, 16, 510. https://doi.org/10.3390/genes16050510
Balcerzyk-Matić A, Iwanicki T, Jarosz A, Nowak T, Emich-Widera E, Kazek B, Kapinos-Gorczyca A, Kapinos M, Iwanicka J, Gawron K, et al. Analysis of the DYNC1H1 Gene Polymorphic Variants’ Association with ASD Occurrence and Clinical Phenotype of Affected Children. Genes. 2025; 16(5):510. https://doi.org/10.3390/genes16050510
Chicago/Turabian StyleBalcerzyk-Matić, Anna, Tomasz Iwanicki, Alicja Jarosz, Tomasz Nowak, Ewa Emich-Widera, Beata Kazek, Agnieszka Kapinos-Gorczyca, Maciej Kapinos, Joanna Iwanicka, Katarzyna Gawron, and et al. 2025. "Analysis of the DYNC1H1 Gene Polymorphic Variants’ Association with ASD Occurrence and Clinical Phenotype of Affected Children" Genes 16, no. 5: 510. https://doi.org/10.3390/genes16050510
APA StyleBalcerzyk-Matić, A., Iwanicki, T., Jarosz, A., Nowak, T., Emich-Widera, E., Kazek, B., Kapinos-Gorczyca, A., Kapinos, M., Iwanicka, J., Gawron, K., Likus, W., & Niemiec, P. (2025). Analysis of the DYNC1H1 Gene Polymorphic Variants’ Association with ASD Occurrence and Clinical Phenotype of Affected Children. Genes, 16(5), 510. https://doi.org/10.3390/genes16050510






