Phylogenetic Analyses of Bostrichiformia and Characterization of the Mitogenome of Gibbium aequinoctiale (Bostrichiformia Ptinidae)
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimen Sampling and DNA Isolation
2.2. Mitogenome Amplification and Sequencing
2.3. Mitogenome Assembly, Annotation, and Analyses
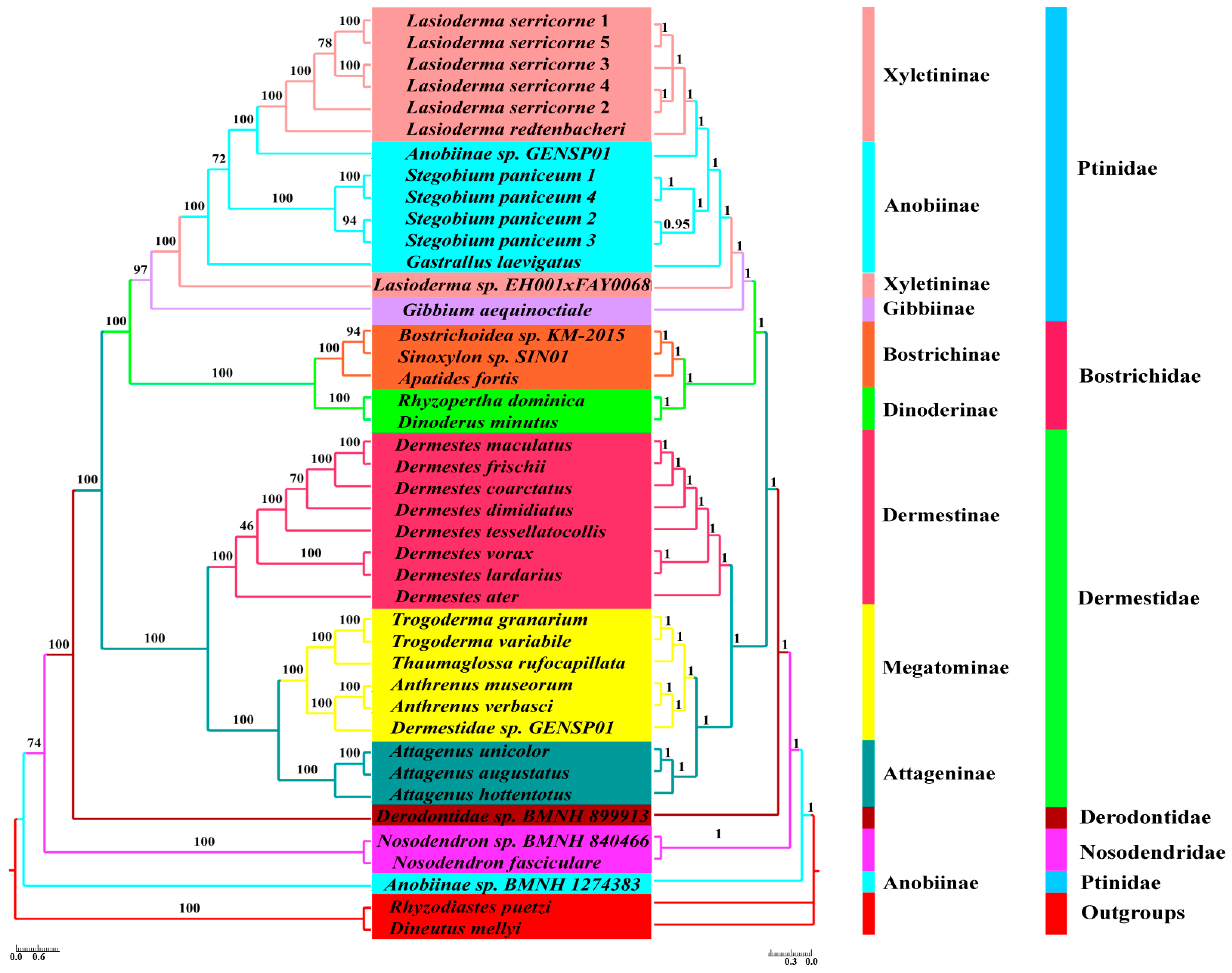
2.4. Phylogeny Reconstruction
| Family | Subfamily | Species | ACC.NO | Integrity | Lentgh (bp) | References |
|---|---|---|---|---|---|---|
| Bostrichidae | Dinoderinae | Rhyzopertha dominica | NC_042820 | Circular | 15,859 | [35] |
| Bostrichidae | Dinoderinae | Dinoderus minutus | KX087284 | Circular | 15,230 | unpublished |
| Bostrichidae | Bostrichinae | Apatides fortis | NC_013582 | Circular | 16,171 | [8] |
| Bostrichidae | Bostrichinae | Sinoxylon sp. SIN01 | JX412742 | Linear | 14,275 | unpublished |
| Bostrichidae | Bostrichoidea sp. KM-2015 | KX035220 | Circular | 15,462 | unpublished | |
| Dermestidae | Dermestinae | Dermestes dimidiatus | NC_067497 | Circular | 16,073 | [36] |
| Dermestidae | Dermestinae | Dermestes vorax | NC_086760 | Circular | 15,775 | unpublished |
| Dermestidae | Dermestinae | Dermestes ater | NC_053877 | Circular | 15,812 | [37] |
| Dermestidae | Dermestinae | Dermestes lardarius | NC_053876 | Circular | 15,772 | [37] |
| Dermestidae | Dermestinae | Dermestes maculatus | NC_037200 | Circular | 16,390 | [38] |
| Dermestidae | Dermestinae | Dermestes coarctatus | NC_044851 | Circular | 15,873 | [39] |
| Dermestidae | Dermestinae | Dermestes frischii | NC_044850 | Circular | 15,873 | [39] |
| Dermestidae | Dermestinae | Dermestes tessellatocollis | NC_044849 | Circular | 16,218 | [39] |
| Dermestidae | Megatominae | Thaumaglossa rufocapillata | NC_084110 | Circular | 16,026 | [40] |
| Dermestidae | Megatominae | Anthrenus verbasci | KX087239 | Linear | 15,818 | unpublished |
| Dermestidae | Megatominae | Anthrenus museorum | NC_063826 | Circular | 15,555 | unpublished |
| Dermestidae | Megatominae | Trogoderma granarium | NC_053875 | Circular | 15,505 | [37] |
| Dermestidae | Megatominae | Trogoderma variabile | MG011537 | Linear | 15,592 | [41] |
| Dermestidae | Dermestidae sp. GENSP01 | JX412839 | Linear | 12,521 | unpublished | |
| Dermestidae | Attageninae | Attagenus unicolor japonicus | OP235946 | Circular | 15,462 | [37] |
| Dermestidae | Attageninae | Attagenus augustatus augustatus | MT113339 | Circular | 15,538 | [37] |
| Dermestidae | Attageninae | Attagenus hottentotus | JX412837 | Linear | 14,309 | unpublished |
| Derodontidae | Derodontidae sp. BMNH 899913 | KX035159 | Circular | 16,711 | unpublished | |
| Nosodendridae | Nosodendron sp. BMNH 840466 | KX035152 | Circular | 16,704 | unpublished | |
| Nosodendridae | Nosodendron fasciculare | KX087322 | Linear | 15,896 | unpublished | |
| Ptinidae | Xyletininae | Lasioderma sp. EH001xFAY0068 | OQ716351 | Linear | 15,344 | unpublished |
| Ptinidae | Xyletininae | Lasioderma serricorne 1 | NC_038197 | Circular | 14,476 | [42] |
| Ptinidae | Xyletininae | Lasioderma serricorne 2 | MF417629 | Circular | 15,958 | [43] |
| Ptinidae | Xyletininae | Lasioderma serricorne 3 | MT254408 | Circular | 15,009 | unpublished |
| Ptinidae | Xyletininae | Lasioderma serricorne 4 | MH817138 | Circular | 14,550 | [35] |
| Ptinidae | Xyletininae | Lasioderma serricorne 5 | MG592705 | Circular | 14,476 | [42] |
| Ptinidae | Xyletininae | Lasioderma redtenbacheri | KX087303 | Linear | 15,042 | unpublished |
| Ptinidae | Ptininae | Gibbium aequinoctiale | KY549398 | Circular | 17,020 | this study |
| Ptinidae | Anobiinae | Anobiinae sp. BMNH 1274383 | KT696225 | Circular | 18,471 | unpublished |
| Ptinidae | Anobiinae | Gastrallus laevigatus | KX087292 | Linear | 15,361 | unpublished |
| Ptinidae | Anobiinae | Anobiinae sp. GENSP01 | JX412730 | Linear | 12,786 | unpublished |
| Ptinidae | Anobiinae | Stegobium paniceum 1 | NC_036678 | Circular | 15,271 | [44] |
| Ptinidae | Anobiinae | Stegobium paniceum 2 | MK947052 | Circular | 15,474 | unpublished |
| Ptinidae | Anobiinae | Stegobium paniceum 3 | MH817140 | Linear | 14,101 | [35] |
| Ptinidae | Anobiinae | Stegobium paniceum 4 | KX819317 | Circular | 15,271 | [44] |
| Outgroups | Rhyzodiastes puetzi | NC_067745 | Circular | 16,793 | unpublished | |
| Dineutus mellyi | NC_054236 | Circular | 16,123 | [45] | ||
3. Results
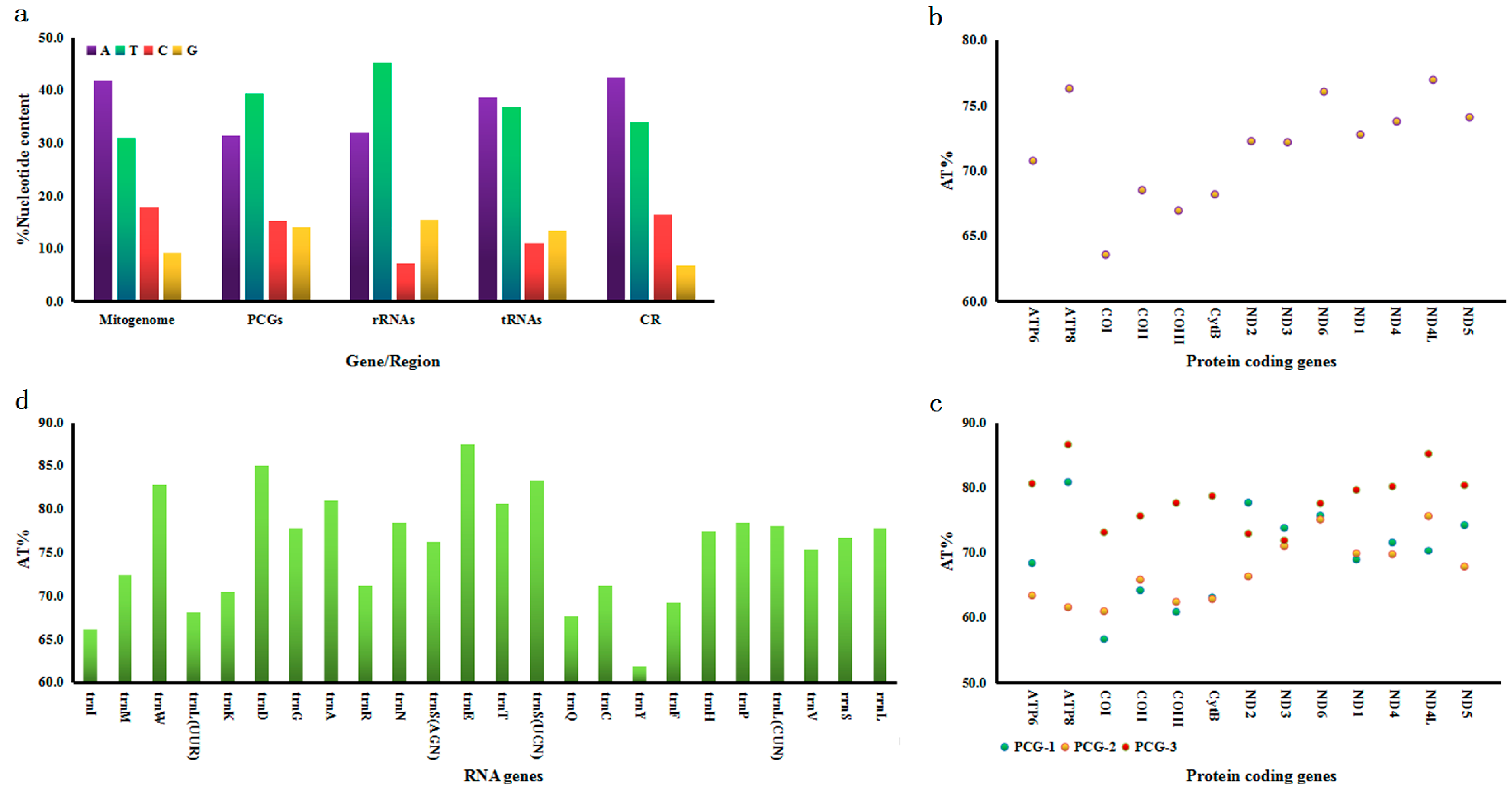
3.1. Mitogenome Description
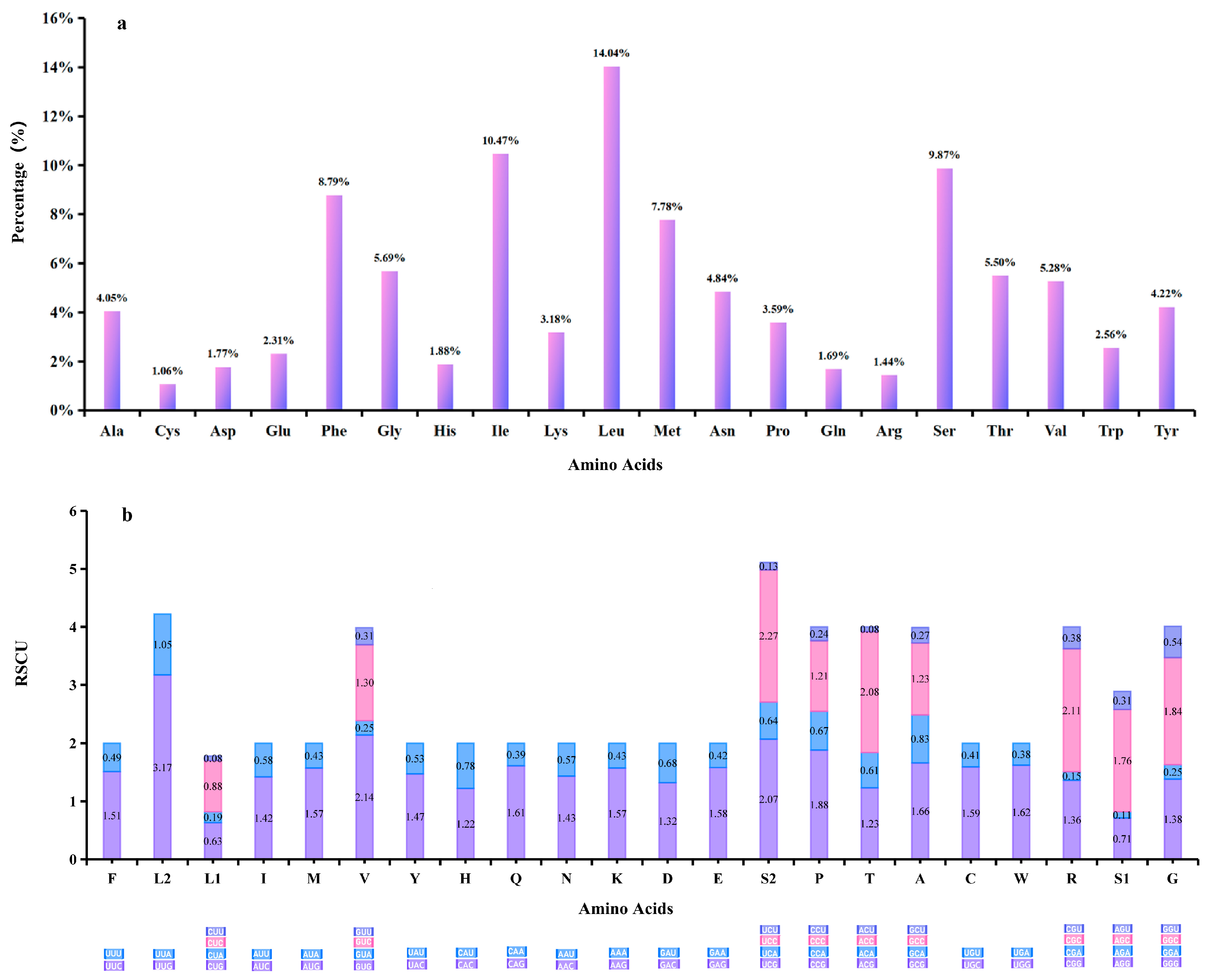
3.2. Protein-Coding Genes (PCGs)
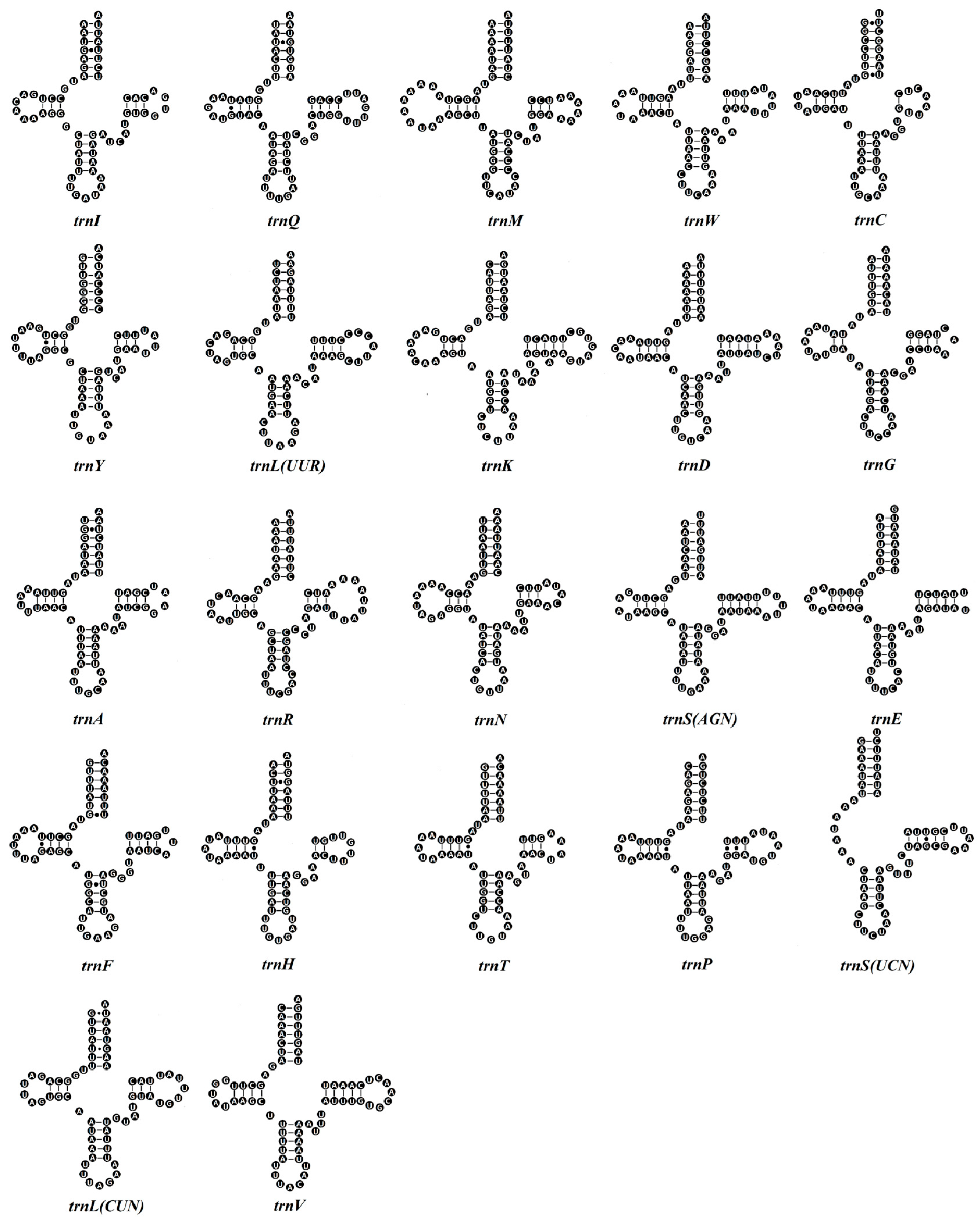
3.3. tRNA and rRNA Genes
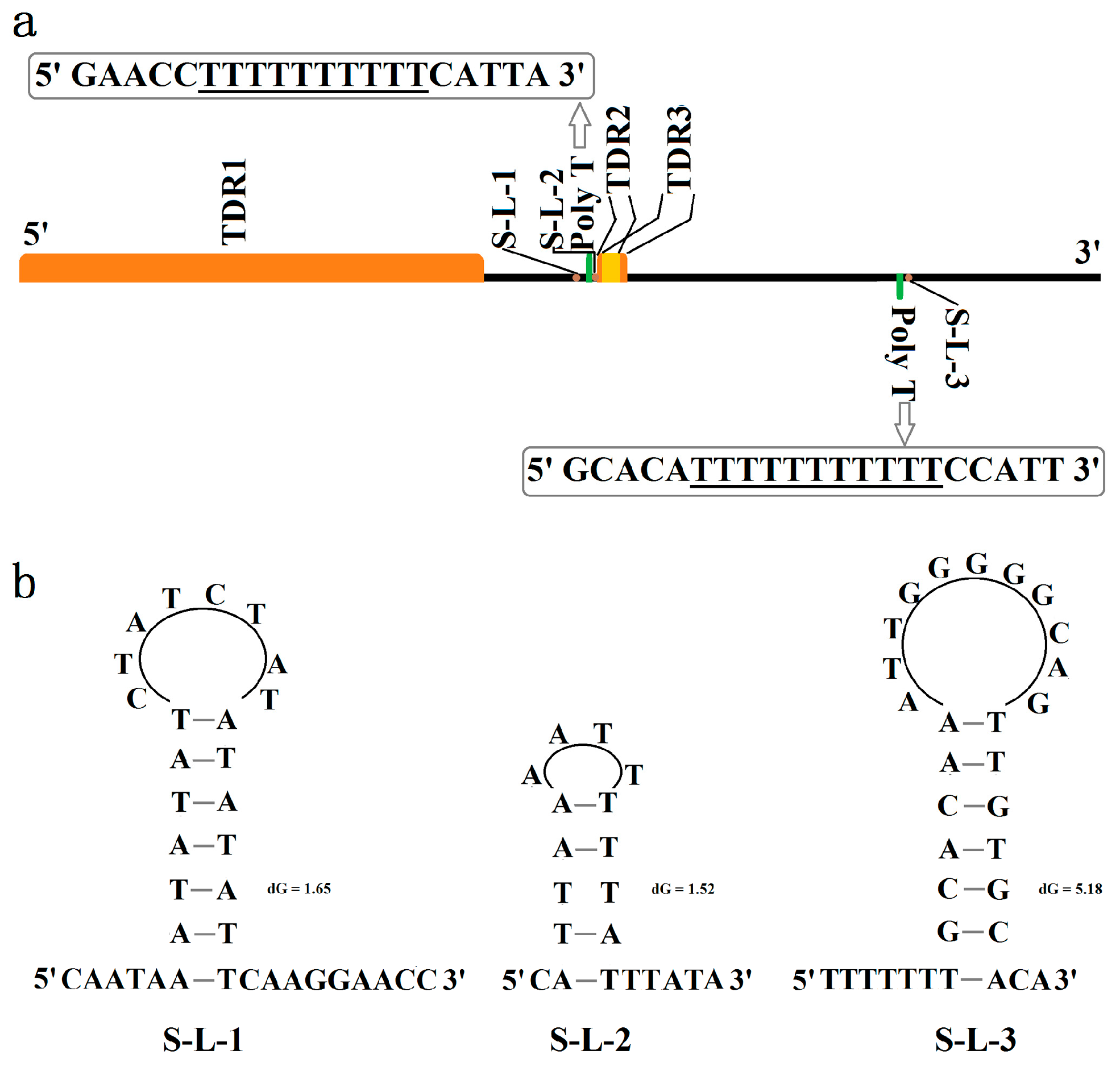
3.4. Control Region (CR)
3.5. Phylogenetic Analyses
4. Discussion
4.1. Mitogenome Characteristics
4.2. Non-Coding Region Analyses
4.3. Phylogenetic Relationship for Bostrichiformia and Ptinidae
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Mitogenome | mitochondrial genome |
| PCGs | protein-coding genes |
| tRNAs | transfer RNA genes |
| rRNAs | ribosomal RNA genes |
| CR | control region |
| TDRs | tandem repeats |
| PCR | polymerase chain reaction |
| rrnS | small rRNA subunits |
| rrnL | large rRNA subunits |
| PCG | 13 PCGs |
| mtDNA | 13 PCGs and two rRNAs |
| PCG12 | 13 PCGs excluding the 3rd codon position |
| mtDNA12 | PCG12 and two rRNAs |
| AIC | Akaike information criterion |
| BIC | Bayesian information criterion |
| BI | Bayesian inference |
| ML | maximum likelihood |
| ATP6 and ATP8 | ATP synthase subunits 6 and 8 |
| COI–III | cytochrome oxidase subunits 1–3 |
| CytB | cytochrome b |
| ND1–6 and ND4L | NADH dehydrogenase subunits 1–6 and 4L |
| RSCUs | relative synonymous codon usages |
| DHU arm | dihydrouridine arm |
| Poly-T | polythymine stretch |
| S–L | stem–loop |
| ID | identification |
References
- Lawrence, J.F. Anobiidae (Bostrichoidea). In Immature Insects; Stehr, F.W., Ed.; Kendall/Hunt Publishing Co.: Dubuque, IA, USA, 1991; pp. 441–444. [Google Scholar]
- Lawrence, J.F.; Viedma, M.G. Ptinidae (Bostrichidae). In Immature Insects; Stehr, F.W., Ed.; Kendall/Hunt Publishing Co.: Dubuque, IA, USA, 1991; pp. 444–445. [Google Scholar]
- Philips, T.K. Phylogenetic analysis of the New World Ptininae (Coleoptera: Bostrichoidea). Syst. Entomol. 2000, 25, 235–262. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhu, X.P.; Liu, Y.P.; Luo, S.L.; Zhangsun, D.T. Mitogenome Characterization of Four Conus Species and Comparative Analysis. Int. J. Mol. Sci. 2023, 24, 9411. [Google Scholar] [CrossRef] [PubMed]
- Avise, J.C.; Arnold, J.; Ball, R.M.; Bermingham, E.; Lamb, T.; Neigel, J.E.; Reeb, C.A.; Saunders, N.C. Intraspecific phylogeography: The mitochondrial-DNA bridge between population-genetics and systematics. Annu. Rev. Ecol. Syst. 1987, 18, 489–522. [Google Scholar] [CrossRef]
- Moritz, C.; Dowling, T.E.; Brown, W.M. Evolution of animal mitochondrial DNA: Relevance for population biology and systematics. Annu. Rev. Ecol. Syst. 1987, 18, 269–292. [Google Scholar] [CrossRef]
- Sheffield, N.C.; Song, H.; Cameron, S.L.; Whiting, M.F. Nonstationary evolution and compositional heterogeneity in beetle mitochondrial phylogenomics. Syst. Biol. 2009, 58, 381–394. [Google Scholar] [CrossRef]
- Kim, K.G.; Hong, M.Y.; Kim, M.J.; Im, H.H.; Kim, M.I.; Bae, C.H.; Seo, S.J.; Lee, S.H.; Kim, I. Complete mitochondrial genome sequence of the yellow-spotted longhorned beetle Psacothea hilaris (Coleoptera: Cerambycidae) and phylogenetic analysis among coleopteran insects. Mol. Cells. 2009, 27, 429–441. [Google Scholar] [CrossRef]
- Pons, J.; Ribera, I.; Bertranpetit, J.; Balke, M. Nucleotide substitution rates for the full set of mitochondrial protein-coding genes in Coleoptera. Mol. Phylogenet. Evol. 2010, 56, 796–807. [Google Scholar] [CrossRef]
- Coates, B.S. Assembly and annotation of full mitochondrial genomes for the corn rootworm species, Diabrotica virgifera virgifera and Diabrotica barberi (Insecta: Coleoptera: Chrysomelidae), using Next Generation Sequence data. Gene 2014, 542, 190–197. [Google Scholar] [CrossRef]
- Timmermans, M.J.T.N.; Barton, C.; Haran, J.; Dirk, A.; Culverwell, C.L.; Ollikainen, A.; Dodsworth, S.; Foster, P.G.; Bocak, L.; Vogler, A.P. Family-level sampling of mitochondrial genomes in Coleoptera: Compositional heterogeneity and phylogenetics. Genome Biol. Evol. 2015, 8, 161–175. [Google Scholar] [CrossRef]
- Zhang, H.L.; Liu, N.; Han, Z.P.; Liu, J.X. Phylogenetic analyses and evolutionary timescale of Coleoptera based on mitochondrial sequence. Biochem. Syst. Ecol. 2016, 66, 229–238. [Google Scholar] [CrossRef]
- Sun, H.Q.; Zhao, W.X.; Lin, R.Z.; Zhou, Z.F.; Huai, W.X.; Yao, Y.X. The conserved mitochondrial genome of the jewel beetle (Coleoptera: Buprestidae) and its phylogenetic implications for the suborder Polyphaga. Genomics 2020, 112, 3713–3721. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yang, Y.H.; Yan, Z.H.; Wang, H.S.; Bai, M.; Shi, C.M.; Li, J. Analysis of the Mitogenomes of Two Helotid Species Provides New Insights into the Phylogenetic Relationship of the Basal Cucujoidea (Insecta: Coleoptera). Biology 2023, 12, 135. [Google Scholar] [CrossRef]
- Gao, R.R.; Lei, Q.L.; Jin, X.; Zafar, I.; Yang, X.K.; Su, C.Y.; Hao, J.S.; Nie, R.E. Characterization of Four Complete Mitogenomes of Monolepta Species and Their Related Phylogenetic Implications. Insects 2024, 15, 50. [Google Scholar] [CrossRef]
- Yi, C.H.; Shu, X.; Wang, L.M.; Yin, J.; Wang, Y.H.; Wang, Y.C.; Zhang, H.H.; He, Q.J.; Zhao, M. The first report of complete mitogenomes of two endangered species of genus Propomacrus (Coleoptera: Scarabaeidae: Euchirinae) and phylogenetic implications. PLoS ONE 2024, 19, e0310559. [Google Scholar] [CrossRef] [PubMed]
- Long, T.T.; Zhu, W.L.; Yang, L.; Long, J.K.; Chang, Z.M.; Chen, X.S. First report of the complete mitochondrial genome of 3 beetles (Coleoptera: Scarabaeidae) harming Gastrodia elata (Asparagales: Orchidaceae). J. Insect. Sci. 2024, 24, 1–10. [Google Scholar] [CrossRef]
- Bouchard, P.; Bousquet, Y.; Davies, A.E.; Alonso-Zarazaga, M.A.; Lawrence, J.F.; Lyal, C.H.C.; Newton, A.F.; Reid, C.A.M.; Schmitt, M.; Slipinski, S.A.; et al. Family-group names in Coleoptera (Insecta). ZooKeys 2011, 88, 1–972. [Google Scholar] [CrossRef]
- Zahradník, P.; Háva, J. Catalogue of the world genera and subgenera of the superfamilies Derodontoidea and Bostrichoidea (Coleoptera: Derodontiformia, Bostrichiformia). Zootaxa 2014, 3754, 301–352. [Google Scholar] [CrossRef][Green Version]
- Simon, C.; Buckley, T.R.; Frati, F.; Stewart, J.B.; Beckenbach, A.T. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 2006, 37, 545–579. [Google Scholar] [CrossRef]
- Staden, R.; Beal, K.F.; Bonfield, J.K. The Staden package, 1998. Methods. Mol. Biol. 2000, 132, 115–130. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Stothard, P.; Wishart, D.S. Circular genome visualization and exploration using CGView. Bioinformatics 2005, 21, 537–539. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL X Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Stöver, B.C.; Müller, K.F. TreeGraph 2: Combining and visualizing evidence from different Phylogenetic analyses. BMC Bioinform. 2010, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.L.; Zhang, Q.L.; Zhang, L.; Guo, Z.L.; Liu, Y.J.; Shen, Y.Y.; Shao, R.F. High-level phylogeny of the Coleoptera inferred with mitochondrial genome sequences. Mol. Phylogenet. Evol. 2016, 104, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.N.; Zhang, Y.; Wei, S.H.; Ullah, F.M.; Pang, Y.T.; Ma, C.C.; Li, Z.H. The first complete mitochondrial genome of Dermestes dimidiatus ab. rosea Kusnezova and its phylogenetic implications for the superfamily Bostrichoidea. Mitochondrial DNA B 2020, 5, 3805–3807. [Google Scholar] [CrossRef]
- Zeng, L.Y.; Pang, Y.T.; Feng, S.Q.; Wang, Y.N.; Stejskal, V.; Aulicky, R.; Zhang, S.F.; Li, Z.H. Comparative mitochondrial genomics of five Dermestid beetles (Coleoptera: Dermestidae) and its implications for phylogeny. Genomics 2021, 113, 927–934. [Google Scholar] [CrossRef]
- Karagozlu, M.Z.; Park, S.H.; Shin, S.E.; Kim, C.B. Complete mitogenome and phylogenetic analysis of hide beetle Dermestes maculatus (Insecta, Coleoptera, Dermestidae). Mitochondrial DNA B 2017, 2, 827–828. [Google Scholar] [CrossRef]
- Karagozlu, M.Z.; An, H.; Park, S.H.; Shin, S.E.; Kim, C.B. Comparative analyses of the three complete mitochondrial genomes from forensic important beetle genus Dermestes with phylogenetic relationships. Gene 2019, 706, 146–153. [Google Scholar] [CrossRef]
- Chang, G.D.; Shim, J.; Ji, S.M.; Song, J.H. Discovery of Thaumaglossa rufocapillata Redtenbacher (Dermestidae, Coleoptera) from Mantis oothecae in Korea and its complete mitochondrial genome. Int. J. Indust. Entomol. 2023, 47, 115–125. [Google Scholar] [CrossRef]
- Pang, Y.T.; Feng, S.Q.; Wu, Y.; Zhang, S.F.; Li, Z.H. Sequencing and analysis of the mitochondrial genome of Trogoderma variabile (Coleoptera: Dermestidae). Plant Quar. 2019, 33, 8–13. [Google Scholar]
- Liu, Q.; Jiang, X.; Hou, X.; Cai, R.; Tan, J.; Chen, W. The complete mitochondrial genome of Lasioderma serricorne (Coleoptera, Anobiidae). Mitochondrial DNA B 2018, 3, 64–65. [Google Scholar] [CrossRef]
- Yang, W.J.; Xu, K.K.; Zhu, X.Y.; Chen, S.C.; Cai, X.Y.; Cao, Y.; Meng, Y.L.; Yang, H.; Li, C. The complete mitochondrial genome of the cigarette beetle, Lasioderma serricorne (Coleoptera: Anobiidae). Mitochondrial DNA B 2017, 2, 430–431. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.J.; Cai, X.Y.; Xu, K.K.; Cao, Y.; Meng, Y.L.; Li, C. The complete mitochondrial genome of Stegobium paniceum. (Coleoptera: Anobiidae). Mitochondrial DNA B 2016, 1, 815–816. [Google Scholar] [CrossRef]
- Tian, T.; Yuan, H.; Chen, B. Phylogeny of hydradephagan water beetles (Coleoptera: Adephaga) inferred with mitochondrial genome sequences. Kun. Chong Xue Bao 2020, 63, 1016–1027. [Google Scholar]
- Wolstenholme, D.R. Animal mitochondrial DNA: Structure and evolution. Int. Rev. Cytol. 1992, 141, 173–216. [Google Scholar] [CrossRef]
- Yang, F.; Du, Y.Z.; Wang, L.P.; Cao, J.M.; Yu, W.W. The complete mitochondrial genome of the leafminer Liriomyza sativae (Diptera: Agromyzidae): Great difference in the A+T-rich region compared to Liriomyza trifolii. Gene 2011, 485, 7–15. [Google Scholar] [CrossRef]
- Cheng, K.C.; Cahill, D.S.; Kasai, H.; Nishimura, S.; Loeb, L.A. 8- Hydroxyguanine, an abundant form of oxidative DNA damage, causes G→T and A→C substitutions. J. Biol. Chem. 1992, 267, 166–172. [Google Scholar] [CrossRef]
- Francino, M.P.; Ochman, H. Strand asymmetries in DNA evolution. Trends Genet. 1997, 13, 240–245. [Google Scholar] [CrossRef]
- Sheffield, N.C.; Song, H.; Cameron, S.L.; Whiting, M.F.A. Comparative analysis of mitochondrial genomes in Coleoptera (Arthropoda: Insecta) and genome descriptions of six new beetles. Mol. Biol. Evol. 2008, 25, 2499–2509. [Google Scholar] [CrossRef]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Taanman, J.W. The mitochondrial genome: Structure, transcription, translation and replication. Biochim. Biophys. Acta. 1999, 1410, 103–123. [Google Scholar] [CrossRef]
- Buroker, N.E.; Brown, J.R.; Gilbert, T.A.; O’Hara, P.J.; Beckenbach, A.T.; Thomas, W.K.; Smith, M.J. Length heteroplasmy of sturgeon mitochondrial DNA: An illegitimate elongation model. Genetics 1990, 124, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.X.; Hewitt, G.M. Insect mitochondrial control region: A review of its structure, evolution and usefulness in evolutionary studies. Biochem. Syst. Ecol. 1997, 25, 99–210. [Google Scholar] [CrossRef]
- Bell, K.L.; Philips, T.K. Molecular systematics and evolution of the Ptinidae (Coleoptera: Bostrichoidea) and related families. Zool. J. Linn. Soc. 2012, 165, 88–108. [Google Scholar] [CrossRef][Green Version]
- Tang, P.A.; Feng, R.Q.; Zhang, L.; Wang, J.; Wang, X.T.; Zhang, L.J.; Yuan, M.L. Mitochondrial genomes of three Bostrichiformia species and phylogeneticanalysis of Polyphaga (Insecta, Coleoptera). Genomics 2020, 112, 2970–2977. [Google Scholar] [CrossRef]

| Gene (Region) | Coding Strand | Position | Intergenic Gaps | Start Codon | Stop Codon | Anticodon | Length (bp) | AT% |
|---|---|---|---|---|---|---|---|---|
| trnI | + | 1–62 | GAT | 62 | 66.1 | |||
| trnQ | − | 60–127 | 3 | TTG | 68 | 67.6 | ||
| trnM | + | 131–199 | CAT | 69 | 72.5 | |||
| ND2 | + | 194–1204 | ATT | TAA | 1011 | 72.2 | ||
| trnW | + | 1203–1266 | TCA | 64 | 82.8 | |||
| trnC | − | 1259–1317 | GCA | 59 | 71.2 | |||
| trnY | − | 1318–1380 | 1 | GTA | 63 | 61.9 | ||
| COI | + | 1382–2917 | AAT | TAA | 1536 | 63.5 | ||
| trnLUUR | + | 2913–2978 | TAA | 66 | 68.2 | |||
| COII | + | 2978–3653 | ATG | T | 676 | 68.5 | ||
| trnK | + | 3651–3721 | CTT | 71 | 70.4 | |||
| trnD | + | 3721–3787 | GTC | 67 | 85.1 | |||
| ATP8 | + | 3788–3943 | ATT | TAA | 156 | 76.3 | ||
| ATP6 | + | 3940–4602 | ATA | TAA | 663 | 70.7 | ||
| COIII | + | 4602–5390 | ATG | TAA | 789 | 66.9 | ||
| trnG | + | 5390–5452 | TCC | 63 | 77.8 | |||
| ND3 | + | 5453–5804 | ATC | T | 352 | 72.2 | ||
| trnA | + | 5805–5867 | 3 | TGC | 63 | 81.0 | ||
| trnR | + | 5871–5936 | TCG | 66 | 71.2 | |||
| trnN | + | 5934–5998 | GTT | 65 | 78.5 | |||
| trnSAGN | + | 5999–6057 | AGT | 59 | 76.3 | |||
| trnE | + | 6058–6121 | TTC | 64 | 87.5 | |||
| trnF | − | 6120–6184 | GAA | 65 | 69.2 | |||
| ND5 | − | 6185–7889 | ATT | T | 1705 | 74.1 | ||
| trnH | − | 7890–7951 | 3 | GTG | 62 | 77.4 | ||
| ND4 | − | 7955–9280 | ATG | TAA | 1326 | 73.8 | ||
| ND4L | − | 9274–9555 | 2 | ATG | TAA | 282 | 77.0 | |
| trnT | + | 9558–9619 | TGT | 62 | 80.6 | |||
| trnP | − | 9620–9684 | 2 | TGG | 65 | 78.5 | ||
| ND6 | + | 9687–10,166 | ATT | TAA | 480 | 76.0 | ||
| CytB | + | 10,166–11,302 | ATG | TAA | 1137 | 68.2 | ||
| trnSUCN | + | 11,301–11,366 | 17 | TGA | 66 | 83.3 | ||
| ND1 | − | 11,384–12,337 | ATA | TAG | 954 | 72.7 | ||
| trnLCUN | − | 12,335–12,398 | TAG | 64 | 78.1 | |||
| rrnL | − | 12,399–13,668 | 1270 | 77.9 | ||||
| trnV | − | 13,669–13,737 | TAC | 69 | 75.4 | |||
| rrnS | − | 13,738–14,530 | 793 | 76.7 | ||||
| Control region | + | 14,531–17,020 | 2490 | 76.6 |
| ID of TDR | Repeat Region in CR | Period Size | Copy Number | Percent Matches | AT% |
|---|---|---|---|---|---|
| TDR1 | 1–1074 | 473 | 2.3 | 96 | 75.8 |
| TDR2 | 1339–1388 | 23 | 2.1 | 89 | 100 |
| TDR3 | 1350–1405 | 21 | 2.5 | 80 | 95.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Han, Z.; Zhang, R.; Zhang, Y.; Wu, J.; Wang, Z. Phylogenetic Analyses of Bostrichiformia and Characterization of the Mitogenome of Gibbium aequinoctiale (Bostrichiformia Ptinidae). Genes 2025, 16, 509. https://doi.org/10.3390/genes16050509
Zhang H, Han Z, Zhang R, Zhang Y, Wu J, Wang Z. Phylogenetic Analyses of Bostrichiformia and Characterization of the Mitogenome of Gibbium aequinoctiale (Bostrichiformia Ptinidae). Genes. 2025; 16(5):509. https://doi.org/10.3390/genes16050509
Chicago/Turabian StyleZhang, Hongli, Zhiping Han, Rui Zhang, Yongfang Zhang, Juan Wu, and Zhichao Wang. 2025. "Phylogenetic Analyses of Bostrichiformia and Characterization of the Mitogenome of Gibbium aequinoctiale (Bostrichiformia Ptinidae)" Genes 16, no. 5: 509. https://doi.org/10.3390/genes16050509
APA StyleZhang, H., Han, Z., Zhang, R., Zhang, Y., Wu, J., & Wang, Z. (2025). Phylogenetic Analyses of Bostrichiformia and Characterization of the Mitogenome of Gibbium aequinoctiale (Bostrichiformia Ptinidae). Genes, 16(5), 509. https://doi.org/10.3390/genes16050509





