Recombination and Genetic Diversity Analysis of Porcine Reproductive and Respiratory Syndrome 1 Nonstructural Protein 2 Genes in China
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain Collection
2.2. Sequence Analysis of the PRRSV-1 NSP2
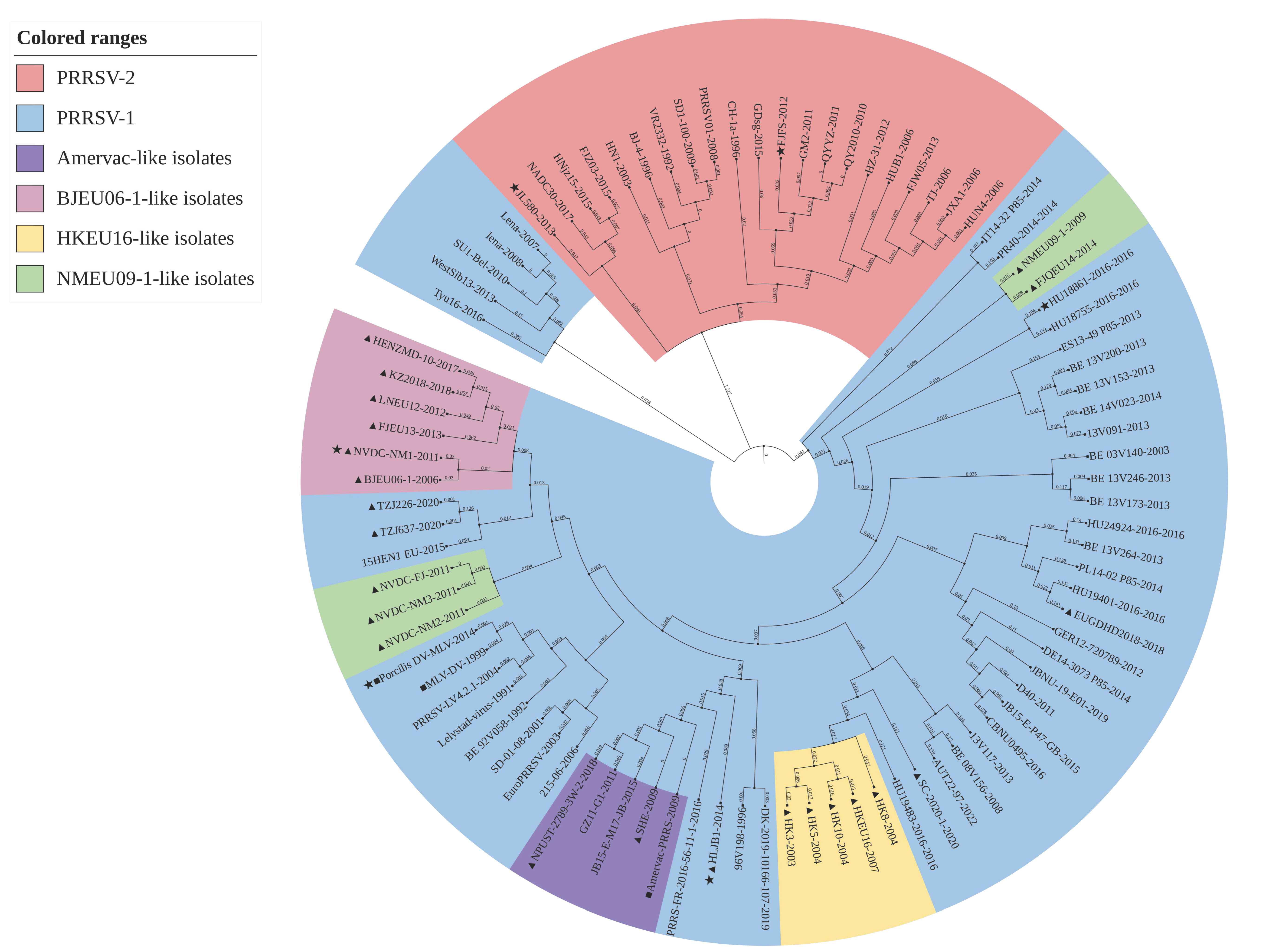
2.3. Phylogenetic Analysis
2.4. Recombination Analysis
3. Results
3.1. Analysis of Nucleotide and Amino Acid Similarity of the PRRSV-1 NSP2
3.2. Amino Acid Sequence Alignment
3.3. Analysis of Phylogenetic Analysis
3.4. Recombinant Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brashishkite, D.A.; Galetsky, S.A.; Kuliffay, P.; Lizonová, A.; Grófová, M. BrdU-induced changes in the provirus of PR-RSV in mammalian cells. Neoplasma 1988, 35, 643–650. [Google Scholar] [PubMed]
- Benfield, D.A.; Nelson, E.; Collins, J.E.; Harris, L.; Goyal, S.M.; Robison, D.; Christianson, W.T.; Morrison, R.B.; Gorcyca, D.; Chladek, D. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332). J. Vet. Diagn. Investig. 1992, 4, 127–133. [Google Scholar] [CrossRef]
- Wensvoort, G.; Terpstra, C.; Pol, J.M.; ter Laak, E.A.; Bloemraad, M.; de Kluyver, E.P.; Kragten, C.; van Buiten, L.; den Besten, A.; Wagenaar, F.; et al. Mystery swine disease in The Netherlands: The isolation of Lelystad virus. Vet. Q. 1991, 13, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Kappes, M.A.; Faaberg, K.S. PRRSV structure, replication and recombination: Origin of phenotype and genotype diversity. Virology 2015, 479–480, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Snijder, E.J.; Meulenberg, J.J. The molecular biology of arteriviruses. J. Gen. Virol. 1998, 79 Pt 5, 961–979. [Google Scholar] [CrossRef]
- Chen, J.; Liu, T.; Zhu, C.G.; Jin, Y.F.; Zhang, Y.Z. Genetic Variation of Chinese PRRSV Strains Based on ORF5 Sequence. Biochem. Genet. 2006, 44, 421–431. [Google Scholar] [CrossRef]
- Franzo, G.; Cecchinato, M.; Martini, M.; Ceglie, L.; Gigli, A.; Drigo, M. Observation of high recombination occurrence of Porcine Reproductive and Respiratory Syndrome Virus in field condition. Virus Res. 2014, 194, 159–166. [Google Scholar] [CrossRef]
- Sun, Q.; Xu, H.; Li, C.; Gong, B.; Li, Z.; Tian, Z.J.; Zhang, H. Emergence of a novel PRRSV-1 strain in mainland China: A recombinant strain derived from the two commercial modified live viruses Amervac and DV. Front. Vet. Sci. 2022, 9, 974743. [Google Scholar] [CrossRef]
- Fang, Y.; Treffers, E.E.; Li, Y.H.; Tas, A.; Sun, Z.; van der Meer, Y.; de Ru, A.H.; van Veelen, P.A.; Atkins, J.F.; Snijder, E.J.; et al. Efficient-2 frameshifting by mammalian ribosomes to synthesize an additional arterivirus protein. Proc. Natl. Acad. Sci. USA 2012, 109, E2920–E2928. [Google Scholar] [CrossRef]
- den Boon, J.A.; Faaberg, K.S.; Meulenberg, J.J.; Wassenaar, A.L.; Plagemann, P.G.; Gorbalenya, A.E.; Snijder, E.J. Processing and evolution of the N-terminal region of the arterivirus replicase ORF1a protein: Identification of two papainlike cysteine proteases. J. Virol. 1995, 69, 4500–4505. [Google Scholar] [CrossRef]
- Fang, Y.; Kim, D.Y.; Ropp, S.; Steen, P.; Christopher-Hennings, J.; Nelson, E.A.; Rowland, R.R. Heterogeneity in Nsp2 of European-like porcine reproductive and respiratory syndrome viruses isolated in the United States. Virus Res. 2004, 100, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Snijder, E.J. The PRRSV replicase: Exploring the multifunctionality of an intriguing set of nonstructural proteins. Virus Res. 2010, 154, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Rutherford, M.S.; Faaberg, K.S. The porcine reproductive and respiratory syndrome virus nsp2 cysteine protease domain possesses both trans- and cis-cleavage activities. J. Virol. 2009, 83, 9449–9463. [Google Scholar] [CrossRef]
- Snijder, E.J.; Wassenaar, A.L.; Spaan, W.J.; Gorbalenya, A.E. The arterivirus Nsp2 protease. An unusual cysteine protease with primary structure similarities to both papain-like and chymotrypsin-like proteases. J. Biol. Chem. 1995, 270, 16671–16676. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Wu, W.; Gao, J.; Smith, N.; Burkard, C.; Xia, D.; Zhang, M.; Wang, C.; Archibald, A.; Digard, P.; et al. Characterization of the Interactome of the Porcine Reproductive and Respiratory Syndrome Virus Nonstructural Protein 2 Reveals the Hyper Variable Region as a Binding Platform for Association with 14-3-3 Proteins. J. Proteome Res. 2016, 15, 1388–1401. [Google Scholar] [CrossRef]
- Ziebuhr, J.; Snijder, E.J.; Gorbalenya, A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000, 81 Pt 4, 853–879. [Google Scholar] [CrossRef]
- Guo, R.; Yan, X.; Li, Y.; Cui, J.; Misra, S.; Firth, A.E.; Snijder, E.J.; Fang, Y. A swine arterivirus deubiquitinase stabilizes two major envelope proteins and promotes production of viral progeny. PLoS Pathog. 2021, 17, e1009403. [Google Scholar] [CrossRef]
- Liu, B.; Luo, L.; Shi, Z.; Ju, H.; Yu, L.; Li, G.; Cui, J. Research Progress of Porcine Reproductive and Respiratory Syndrome Virus NSP2 Protein. Viruses 2023, 15, 2310. [Google Scholar] [CrossRef]
- Han, J.; Wang, Y.; Faaberg, K.S. Complete genome analysis of RFLP 184 isolates of porcine reproductive and respiratory syndrome virus. Virus Res. 2006, 122, 175–182. [Google Scholar] [CrossRef]
- Shi, M.; Lam, T.Y.; Hon, C.C.; Hui, K.H.; Faaberg, K.S.; Wennblom, T.; Murtaugh, M.P.; Stadejek, T.; Leung, C.C. Molecular epidemiology of PRRSV: A phylogenetic perspective. Virus Res. 2010, 154, 7–17. [Google Scholar] [CrossRef]
- Chen, N.; Liu, Q.; Qiao, M.; Deng, X.; Chen, X.; Sun, M. Whole genome characterization of a novel porcine reproductive and respiratory syndrome virus 1 isolate: Genetic evidence for recombination between Amervac vaccine and circulating strains in mainland China. Infect. Genet. Evol. 2017, 54, 308–313. [Google Scholar] [CrossRef] [PubMed]
- Hsueh, F.C.; Kuo, K.L.; Hsu, F.Y.; Wang, S.Y.; Chiu, H.J.; Wu, M.T.; Lin, C.F.; Huang, Y.H.; Chiou, M.T.; Lin, C.N. Molecular Characteristics and Pathogenicity of Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) 1 in Taiwan during 2019-2020. Life 2023, 13, 843. [Google Scholar] [CrossRef]
- Wang, X.; Bai, X.; Wang, Y.; Wang, L.; Wei, L.; Tan, F.; Zhou, Z.; Tian, K. Pathogenicity characterization of PRRSV-1 181187-2 isolated in China. Microb. Pathog. 2023, 180, 106158. [Google Scholar] [CrossRef]
- Gauger, P.C.; Faaberg, K.S.; Guo, B.; Kappes, M.A.; Opriessnig, T. Genetic and phenotypic characterization of a 2006 United States porcine reproductive and respiratory virus isolate associated with high morbidity and mortality in the field. Virus Res. 2012, 163, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Ming, S.; Yongying, M.; Bohua, L.; Huiying, L.; Xiaoyu, D.; Qiaorong, L.; Mingming, Q.; Xi, C.; Xinyan, Y.; Xizhao, C. Pathogenic Characterization of European Genotype Porcine Reproductive and Respiratory Syndrome Virus Recently Isolated in Mainland China. Open Virol. J. 2017, 11, 83–89. [Google Scholar] [CrossRef]
- Chen, N.; Xiao, Y.; Ye, M.; Li, X.; Zhu, J. High genetic diversity of Chinese porcine reproductive and respiratory syndrome viruses from 2016 to 2019. Res. Vet. Sci. 2020, 131, 38–42. [Google Scholar] [CrossRef]
- Dewey, C.; Charbonneau, G.; Carman, S.; Hamel, A.; Nayar, G.; Friendship, R.; Eernisse, K.; Swenson, S. Lelystad-like strain of porcine reproductive and respiratory syndrome virus (PRRSV) identified in Canadian swine. Can. Vet. J. 2000, 41, 493–494. [Google Scholar] [PubMed]
- Renson, P.; Touzain, F.; Lebret, A.; Le Dimna, M.; Quenault, H.; Normand, V.; Claude, J.B.; Pez, F.; Rose, N.; Blanchard, Y.; et al. Complete Genome Sequence of a Recombinant Porcine Reproductive and Respiratory Syndrome Virus Strain from Two Genotype 1 Modified Live Virus Vaccine Strains. Genome Announc. 2017, 5, e00454-17. [Google Scholar] [CrossRef]
- Yu, F.; Liu, L.; Tian, X.; Chen, L.; Huang, X.; Sun, Y.; Yan, Y.; Tian, Z.; Cai, X.; Liu, D.; et al. Genomic Analysis of Porcine Reproductive and Respiratory Syndrome Virus 1 Revealed Extensive Recombination and Potential Introduction Events in China. Vet. Sci. 2022, 9, 450. [Google Scholar] [CrossRef]
- Li, C.; Xu, H.; Zhao, J.; Gong, B.; Sun, Q.; Xiang, L.; Li, W.; Guo, Z.; Li, J.; Tang, Y.D.; et al. Epidemiological investigation and genetic evolutionary analysis of PRRSV-1 on a pig farm in China. Front. Microbiol. 2022, 13, 1067173. [Google Scholar] [CrossRef]
- Zhang, H.; Sha, H.; Qin, L.; Wang, N.; Kong, W.; Huang, L.; Zhao, M. Research Progress in Porcine Reproductive and Respiratory Syndrome Virus–Host Protein Interactions. Animals 2022, 12, 1381. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.X.; Wen, Y.J.; Yang, B.C.; Liu, Z.; Shi, X.C.; Leng, X.; Song, N.; Wu, H.; Chen, L.Z.; Cheng, S.P. Role of non-structural protein 2 in the regulation of the replication of the porcine reproductive and respiratory syndrome virus in MARC-145 cells: Effect of gene silencing and over expression. Vet. Microbiol. 2012, 161, 58–65. [Google Scholar] [CrossRef]
- Yu, X.; Chen, N.; Wang, L.; Wu, J.; Zhou, Z.; Ni, J.; Li, X.; Zhai, X.; Shi, J.; Tian, K. New genomic characteristics of highly pathogenic porcine reproductive and respiratory syndrome viruses do not lead to significant changes in pathogenicity. Vet. Microbiol. 2012, 158, 291–299. [Google Scholar] [CrossRef]
- Kim, D.Y.; Calvert, J.G.; Chang, K.O.; Horlen, K.; Kerrigan, M.; Rowland, R.R. Expression and stability of foreign tags inserted into nsp2 of porcine reproductive and respiratory syndrome virus (PRRSV). Virus Res. 2007, 128, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Bai, J.; Wang, H.; Fan, B.; Li, Y.; Jiang, P. Effect of amino acids residues 323-433 and 628-747 in Nsp2 of representative porcine reproductive and respiratory syndrome virus strains on inflammatory response in vitro. Virus Res. 2015, 208, 13–21. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, F.X.; Wen, Y.J.; Li, Z.G.; Liu, X.; Sun, N.; Yang, Y.; Zhang, S.Q.; Zhu, H.W.; Cheng, S.P.; et al. Effect of Nonstructural Protein 2 Hypervariable Regions in the Replication of Porcine Reproductive and Respiratory Syndrome Virus in Marc-145 Cells. Intervirology 2015, 58, 288–296. [Google Scholar] [CrossRef] [PubMed]
- Ling-Xue, Y.; Xin, W.; Hai, Y.; Yi-Feng, J.; Fei, G.; Wu, T.; Li-Wei, L.; Hui-Chun, L.; Shen, Y.; Peng-Fei, C. The emergence of a highly pathogenic porcine reproductive and respiratory syndrome virus with additional 120aa deletion in Nsp2 region in Jiangxi, China. Transbound. Emerg. Dis. 2018, 65, 1740–1748. [Google Scholar] [CrossRef]
- Zhang, Q.; Song, Z.; Yu, Y.; Huang, J.; Jiang, P.; Shan, H. Genetic analysis of a porcine reproductive and respiratory syndrome virus 1 strain in China with new patterns of amino acid deletions in nsp2, GP3 and GP4. Microb. Pathog. 2020, 149, 104531. [Google Scholar] [CrossRef]
- Shen, S.; Kwang, J.; Liu, W.; Liu, D.X. Determination of the complete nucleotide sequence of a vaccine strain of porcine reproductive and respiratory syndrome virus and identification of the Nsp2 gene with a unique insertion. Arch. Virol. 2000, 145, 871–883. [Google Scholar] [CrossRef]
- Nam, E.; Park, C.K.; Kim, S.H.; Joo, Y.S.; Yeo, S.G.; Lee, C. Complete genomic characterization of a European type 1 porcine reproductive and respiratory syndrome virus isolate in Korea. Arch. Virol. 2009, 154, 629–638. [Google Scholar] [CrossRef]
- Zhang, H.; Luo, Q.; Zheng, Y.; Sha, H.; Li, G.; Kong, W.; Huang, L.; Zhao, M. Genetic Variability and Recombination of the NSP2 Gene of PRRSV-2 Strains in China from 1996 to 2021. Vet. Sci. 2023, 10, 325. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wang, K.G.; Zhou, B.J.; Luo, X.F.; Wen, M.; An, T.Q.; Liu, C.G.; Cheng, Z.T.; Lu, X.L.; Chen, J. Cloning and sequence analysis of the Nsp2 gene of porcine reproductive and respiratory syndrome virus isolated in Guizhou area. Chin. J. Prev. Vet. Med. 2010, 32, 924–928. (In Chinese) [Google Scholar] [CrossRef]
- Shi, X.; Fan, X.; Nie, S.; Kou, L.; Zhang, X.; Liu, H.; Ji, S.; Deng, R.; Wang, A.; Zhang, G. Identification of a linear B-cell epitope on glycoprotein (GP) 2a of porcine reproductive and respiratory syndrome virus (PRRSV). Int. J. Biol. Macromol. 2019, 139, 1288–1294. [Google Scholar] [CrossRef]
- Lin, C.C.; Lee, C.C.; Lin, S.H.; Huang, P.J.; Li, H.P.; Chang, Y.S.; Tang, P.; Chao, M. RNA recombination in Hepatitis delta virus: Identification of a novel naturally occurring recombinant. J. Microbiol. Immunol. Infect. 2017, 50, 771–780. [Google Scholar] [CrossRef]
- Simon-Loriere, E.; Holmes, E.C. Why do RNA viruses recombine? Nat. Rev. Microbiol. 2011, 9, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Kvisgaard, L.K.; Kristensen, C.S.; Ryt-Hansen, P.; Pedersen, K.; Stadejek, T.; Trebbien, R.; Andresen, L.O.; Larsen, L.E. A recombination between two Type 1 Porcine Reproductive and Respiratory Syndrome Virus (PRRSV-1) vaccine strains has caused severe outbreaks in Danish pigs. Transbound. Emerg. Dis. 2020, 67, 1786–1796. [Google Scholar] [CrossRef]
- Chen, N.; Cao, Z.; Yu, X.; Deng, X.; Zhao, T.; Wang, L.; Liu, Q.; Li, X.; Tian, K. Emergence of novel European genotype porcine reproductive and respiratory syndrome virus in mainland China. J. Gen. Virol. 2011, 92 Pt 4, 880–892. [Google Scholar] [CrossRef]
- Nan, Y.; Wu, C.; Gu, G.; Sun, W.; Zhang, Y.J.; Zhou, E.M. Improved Vaccine against PRRSV: Current Progress and Future Perspective. Front. Microbiol. 2017, 8, 1635. [Google Scholar] [CrossRef]




| Year | Aera | Strain | GenBank Accession Number | Type |
|---|---|---|---|---|
| 1991 | Netherlands | Lelystad virus | M96262 | PRRSV-1 |
| 1992 | Belgium | BE_92V058 | MW448197 | PRRSV-1 |
| 1996 | Belgium | 96V198 | MK876228 | PRRSV-1 |
| 1999 | Netherlands | MLV-DV | KJ127878 | PRRSV-1 |
| 2001 | USA | SD-01-08 | DQ489311 | PRRSV-1 |
| 2003 | China | HK3 | KF287129 | PRRSV-1 |
| 2003 | USA | EuroPRRSV | AY366525 | PRRSV-1 |
| 2003 | Belgium | BE_03V140 | MW053394 | PRRSV-1 |
| 2004 | China | HK5 | KF287130 | PRRSV-1 |
| 2004 | China | HK8 | KF287128 | PRRSV-1 |
| 2004 | China | HK10 | KF287131 | PRRSV-1 |
| 2004 | Netherlands | PRRSV LV4.2.1 | AY588319 | PRRSV-1 |
| 2006 | China | BJEU06-1 | GU047344 | PRRSV-1 |
| 2006 | United Kingdom | 215-06 | OP047897 | PRRSV-1 |
| 2007 | China | HKEU16 | EU076704 | PRRSV-1 |
| 2007 | Belarus | Lena | JF802085 | PRRSV-1 |
| 2008 | Belgium | BE_08V156 | MW053397 | PRRSV-1 |
| 2008 | Belarus | lena | JF802085 | PRRSV-1 |
| 2009 | China | SHE | GQ461593 | PRRSV-1 |
| 2009 | China | NMEU09-1 | GU047345 | PRRSV-1 |
| 2009 | Spain | Amervac PRRS | GU067771 | PRRSV-1 |
| 2010 | Belarus | SU1-Bel | KP889243 | PRRSV-1 |
| 2011 | China | GZ11-G1 | KF001144 | PRRSV-1 |
| 2011 | China | NVDC-FJ | KC492506 | PRRSV-1 |
| 2011 | China | NVDC-NM1 | JX187609 | PRRSV-1 |
| 2011 | China | NVDC-NM2 | KC492504 | PRRSV-1 |
| 2011 | China | NVDC-NM3 | KC492505 | PRRSV-1 |
| 2011 | South Korea | D40 | MZ287330 | PRRSV-1 |
| 2012 | China | LNEU12 | KM196101 | PRRSV-1 |
| 2012 | Germany | GER12-720789 | OP529852 | PRRSV-1 |
| 2013 | China | FJEU13 | KP860912 | PRRSV-1 |
| 2013 | Russia | WestSib13 | KX668221 | PRRSV-1 |
| 2013 | Belgium | 13V117 | KT159249 | PRRSV-1 |
| 2013 | Belgium | BE_13V246 | MW053396 | PRRSV-1 |
| 2013 | Belgium | BE_13V173 | MW053395 | PRRSV-1 |
| 2013 | Belgium | 13V091 | KT159248 | PRRSV-1 |
| 2013 | Belgium | BE_13V200 | MW053399 | PRRSV-1 |
| 2013 | Belgium | BE_13V153 | MW053398 | PRRSV-1 |
| 2013 | Spain | ES13-49_P85 | MK024325 | PRRSV-1 |
| 2013 | Belgium | BE_13V264 | MW053400 | PRRSV-1 |
| 2014 | China | FJQEU14 | KP860913 | PRRSV-1 |
| 2014 | China | HLJB1 | KT224385 | PRRSV-1 |
| 2014 | Germany | DE14-3073_P85 | MK024324 | PRRSV-1 |
| 2014 | Denmark | Porcilis_DV-MLV | MT311646 | PRRSV-1 |
| 2014 | Belgium | BE_14V023 | MW053401 | PRRSV-1 |
| 2014 | Poland | PL14-02_P85 | MK024327 | PRRSV-1 |
| 2014 | Italy | PR40/2014 | MF346695 | PRRSV-1 |
| 2014 | Italy | IT14-32_P85 | MK024326 | PRRSV-1 |
| 2015 | China | 15HEN1_EU | KX967492 | PRRSV-1 |
| 2015 | South Korea | JB15-E-M17-JB | MZ287329 | PRRSV-1 |
| 2015 | South Korea | JB15-E-P47-GB | MZ287328 | PRRSV-1 |
| 2016 | South Korea | CBNU0495 | MZ287327 | PRRSV-1 |
| 2016 | France | PRRS-FR-2016-56-11-1 | MH018883 | PRRSV-1 |
| 2016 | Hungary | HU19401/2016 | MH463457 | PRRSV-1 |
| 2016 | Hungary | HU24924/2016 | MH463459 | PRRSV-1 |
| 2016 | Hungary | HU19483/2016 | MH463458 | PRRSV-1 |
| 2016 | Hungary | HU18861/2016 | MH463456 | PRRSV-1 |
| 2016 | Hungary | HU18755/2016 | MH463455 | PRRSV-1 |
| 2016 | Russia | Tyu16 | MT008024 | PRRSV-1 |
| 2017 | China | HENZMD-10 | KY363382 | PRRSV-1 |
| 2018 | China | KZ2018 | MN550991 | PRRSV-1 |
| 2018 | China | EUGDHD2018 | MK639926 | PRRSV-1 |
| 2018 | China | NPUST-2789-3W-2 | MN242825 | PRRSV-1 |
| 2019 | South Korea | JBNU-19-E01 | MW847781 | PRRSV-1 |
| 2019 | Denmark | DK-2019-10166-107 | MN603982 | PRRSV-1 |
| 2020 | China | SC-2020-1 | MW115431 | PRRSV-1 |
| 2020 | China | TZJ226 | OP566682 | PRRSV-1 |
| 2020 | China | TZJ637 | OP566683 | PRRSV-1 |
| 2022 | Austria | AUT22-97 | OP627116 | PRRSV-1 |
| 1992 | USA | VR2332 | EF536003.1 | PRRSV-2 |
| 1996 | China | BJ-4 | AF331831 | PRRSV-2 |
| 1996 | China | CH-1a | AY032626 | PRRSV-2 |
| 2003 | China | HN1 | AY457635.1 | PRRSV-2 |
| 2006 | China | TJ | EU860248 | PRRSV-2 |
| 2006 | China | HUB1 | EF075945 | PRRSV-2 |
| 2006 | China | JXA1 | EF112445 | PRRSV-2 |
| 2006 | China | HUN4 | EF635006.1 | PRRSV-2 |
| 2008 | China | PRRSV01 | FJ175687 | PRRSV-2 |
| 2009 | China | SD1-100 | GQ914997 | PRRSV-2 |
| 2010 | China | QY2010 | JQ743666 | PRRSV-2 |
| 2011 | China | GM2 | JN662424 | PRRSV-2 |
| 2011 | China | QYYZ | JQ308798 | PRRSV-2 |
| 2012 | China | HZ-31 | KC445138 | PRRSV-2 |
| 2012 | China | FJFS | KP998476 | PRRSV-2 |
| 2013 | China | JL580 | KR706343.1 | PRRSV-2 |
| 2013 | China | FJW05 | KP860911 | PRRSV-2 |
| 2015 | China | GDsg | KX621003 | PRRSV-2 |
| 2015 | China | HNjZ15 | KT945017 | PRRSV-2 |
| 2015 | China | FJZ03 | KP860909 | PRRSV-2 |
| 2017 | China | NADC30 | MH500776.1 | PRRSV-2 |
| Tyu16 | Lena | |
|---|---|---|
| 15HEN1_EU | a 64.3 (aa) | |
| lena | b 100 (nt, aa) | |
| BE_08V156 | a 67.3 (nt) |
| Recombination Event | Recombinant Strain | Recombinant Breakpoint | Recombination Analysis Method (p-Value) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Main Parental Strain | Minor Parental Strain | RDP | GENECONV | BootScan | MaxChi | Chimaera | SiScan | 3seq | ||
| 1 | JL580 | 2134–2205 (3328–3386) | 1.509 × 10−64 | 1.123 × 10−54 | NS | 4.298 × 10−27 | 2.053 × 10−29 | 1.630 × 10−38 | 1.020 × 10−11 | |
| FJZ03 | HUB1 | |||||||||
| 2 | NVDC-NM1 | 3718–66 (1204–1301) | 4.846 × 10−30 | 5.298 × 10−26 | 8.873 × 10−26 | 1.700 × 10−20 | 4.974 × 10−16 | 6.034 × 10−34 | 1.020 × 10−11 | |
| LNEU12 | BJEU06-1-2006 | |||||||||
| 3 | HU18861-2016 | 3641–35 (713–884) | 6.887 × 10−30 | 9.225 × 10−29 | 5.293 × 10−32 | 1.287 × 10−6 | 1.224 × 1012 | 1.179 × 10−22 | 1.020 × 10−11 | |
| JB15-E-M17-JB | HU18755-2016 | |||||||||
| 4 | Porcilis_DV-MLV | 881–940 (1090–1198) | 2.414 × 10−27 | 1.319 × 10−27 | 3.401 × 10−12 | 1.545 × 10−10 | 7.047 × 10−10 | 7.332 × 10−8 | 1.020 × 10−11 | |
| PRRSV-LV4.2.1 | D40 | |||||||||
| 5 | FJFS | 746–945 (3590–14) | 5.124 × 10−5 | 6.492 × 10−20 | 2.196 × 10−23 | 1.345 × 10−13 | 1.414 × 10−8 | 1.543 × 10−45 | 3.633 × 10−25 | |
| HUN4 | QYYZ | |||||||||
| 6 | HLJB1 | 2783–3002 (3678–33) | 2.268 × 10−10 | 5.446 × 10−6 | 1.729 × 10−8 | 5.043 × 10−7 | 3.227 × 10−4 | 4.590 × 10−10 | 5.884 × 10−3 | |
| BJEU06-1 | JB15-E-M17-JB | |||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lv, C.; Guan, B.; Pang, J.; Kong, W.; Wang, R.; Wang, L.; Zhao, M.; Zhang, H. Recombination and Genetic Diversity Analysis of Porcine Reproductive and Respiratory Syndrome 1 Nonstructural Protein 2 Genes in China. Genes 2025, 16, 507. https://doi.org/10.3390/genes16050507
Lv C, Guan B, Pang J, Kong W, Wang R, Wang L, Zhao M, Zhang H. Recombination and Genetic Diversity Analysis of Porcine Reproductive and Respiratory Syndrome 1 Nonstructural Protein 2 Genes in China. Genes. 2025; 16(5):507. https://doi.org/10.3390/genes16050507
Chicago/Turabian StyleLv, Chen, Baoyi Guan, Jiankun Pang, Weili Kong, Ruining Wang, Lin Wang, Mengmeng Zhao, and Hang Zhang. 2025. "Recombination and Genetic Diversity Analysis of Porcine Reproductive and Respiratory Syndrome 1 Nonstructural Protein 2 Genes in China" Genes 16, no. 5: 507. https://doi.org/10.3390/genes16050507
APA StyleLv, C., Guan, B., Pang, J., Kong, W., Wang, R., Wang, L., Zhao, M., & Zhang, H. (2025). Recombination and Genetic Diversity Analysis of Porcine Reproductive and Respiratory Syndrome 1 Nonstructural Protein 2 Genes in China. Genes, 16(5), 507. https://doi.org/10.3390/genes16050507







