Identification and Characterization of LINE and SINE Retrotransposons in the African Hedgehog (Atelerix albiventris, Erinaceidae) and Their Association with 3D Genome Organization and Gene Expression
Abstract
1. Introduction
2. Methods
2.1. Extraction of LINEs and SINEs
- (i)
- Reference database: RepeatModeler v2.0.215 was used to perform ab initio repeat family prediction from the A. albiventris genome [46]. Unknown classification of the ab initio prediction was performed using TEclass v2.1.3d [47]. The predicted repeats from ab initio, Dfam database, and the Repbase library were merged and clustered using cd-hit-est (parameters: -n 5, -aL 0.99, -c 0.8 and -s 0.8) to create a new repetitive sequence library [48].
- (ii)
- (TE annotation: Repeats in the A. albiventris were detected and classified by searches for similarity to sequences in the reference repeat database with RepeatMasker v4.0.7 (http://www.repeatmasker.org, accessed on 11 September 2024) using default settings. We extracted LINE and SINE annotation information from the RepeatMasker output by filtering based on the classification qualifier.
- (iii)
- TE consensus sequence: To obtain the consensus sequence for LINE and SINE families, we followed the pipeline proposed by Goubert et al. (2022) [49]. First, each raw sequence of LINE and SINE from the output was mapped onto the genome using BLAST v2.15 [50], applying a 95% similarity threshold, which has been commonly used in repeat annotation pipelines to ensure that only closely related copies are considered. Next, the putative copies of the query sequence were used as input for the multiple alignments using mafft v7.520 software with default parameters [51]. Gaps, rare insertions, and highly divergent sequences were removed using the t-coffee tool [52]. Finally, the cons function from the EMBOSS package (http://emboss.open-bio.org/rel/dev/apps/cons.html, accessed on 15 July 2024) was used to generate a consensus sequence that serves as a representative model for each subfamily, not a specific genomic locus.
2.2. Genome Feature of LINEs and SINEs
2.3. Insert Age Estimation
2.4. Phylogenetic Tree and Family Classification for LINEs and SINEs
2.5. Hi-C Analysis and 3D Chromatin Architecture
2.6. DNA Methylation
2.7. Transcriptomic Analysis of LINEs and SINEs
2.8. Statistical Analysis and Data Visualization
3. Results
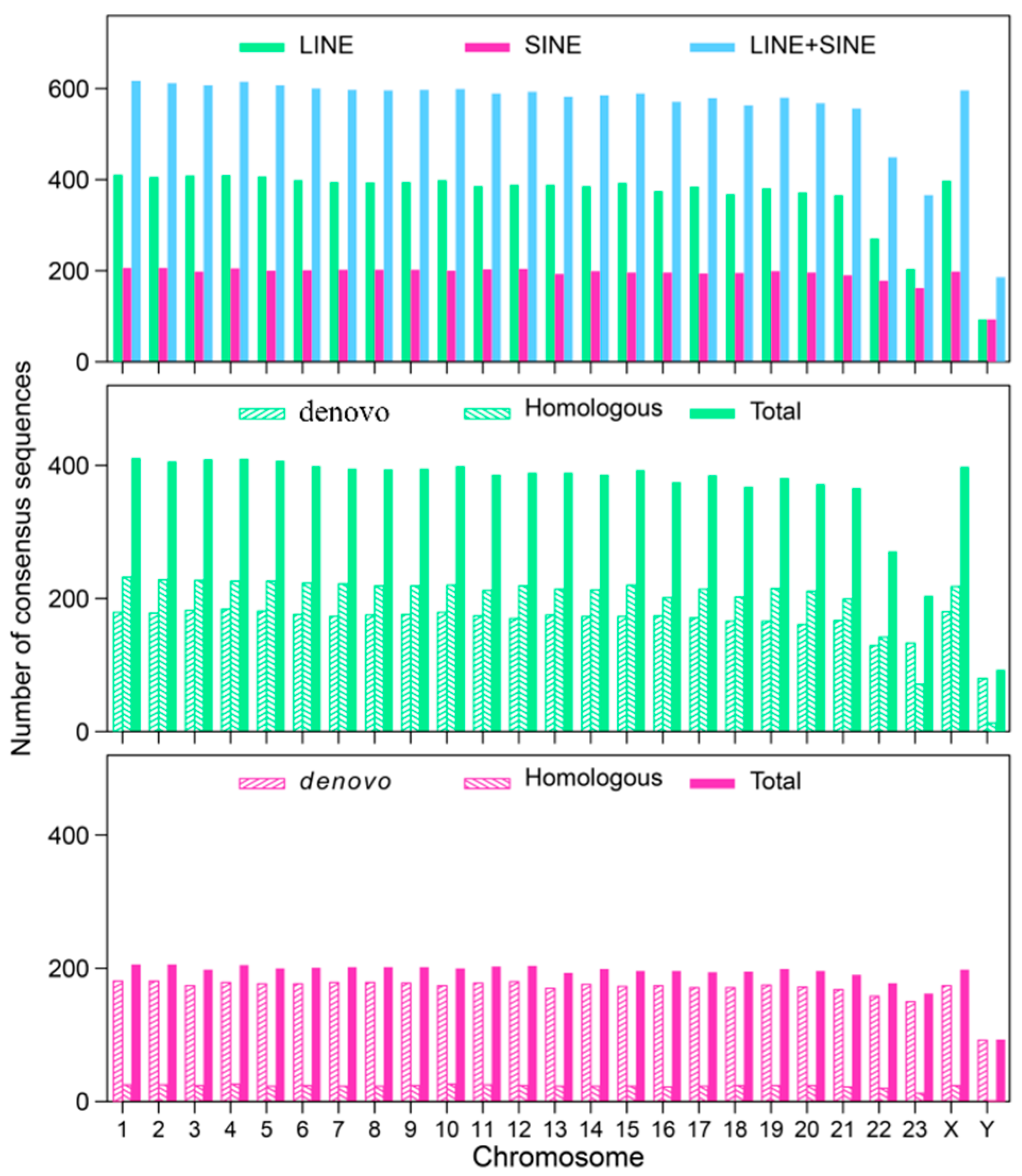
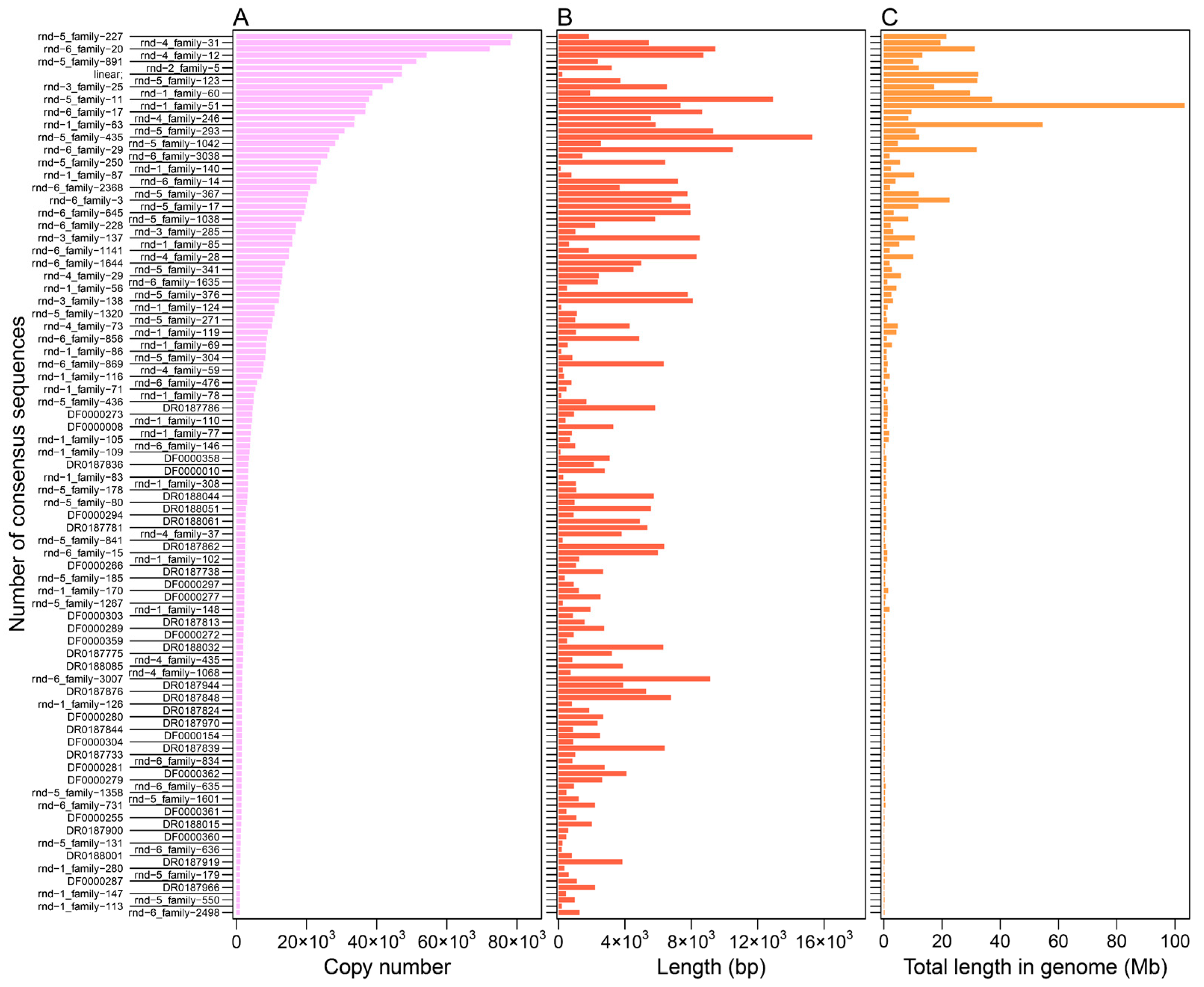
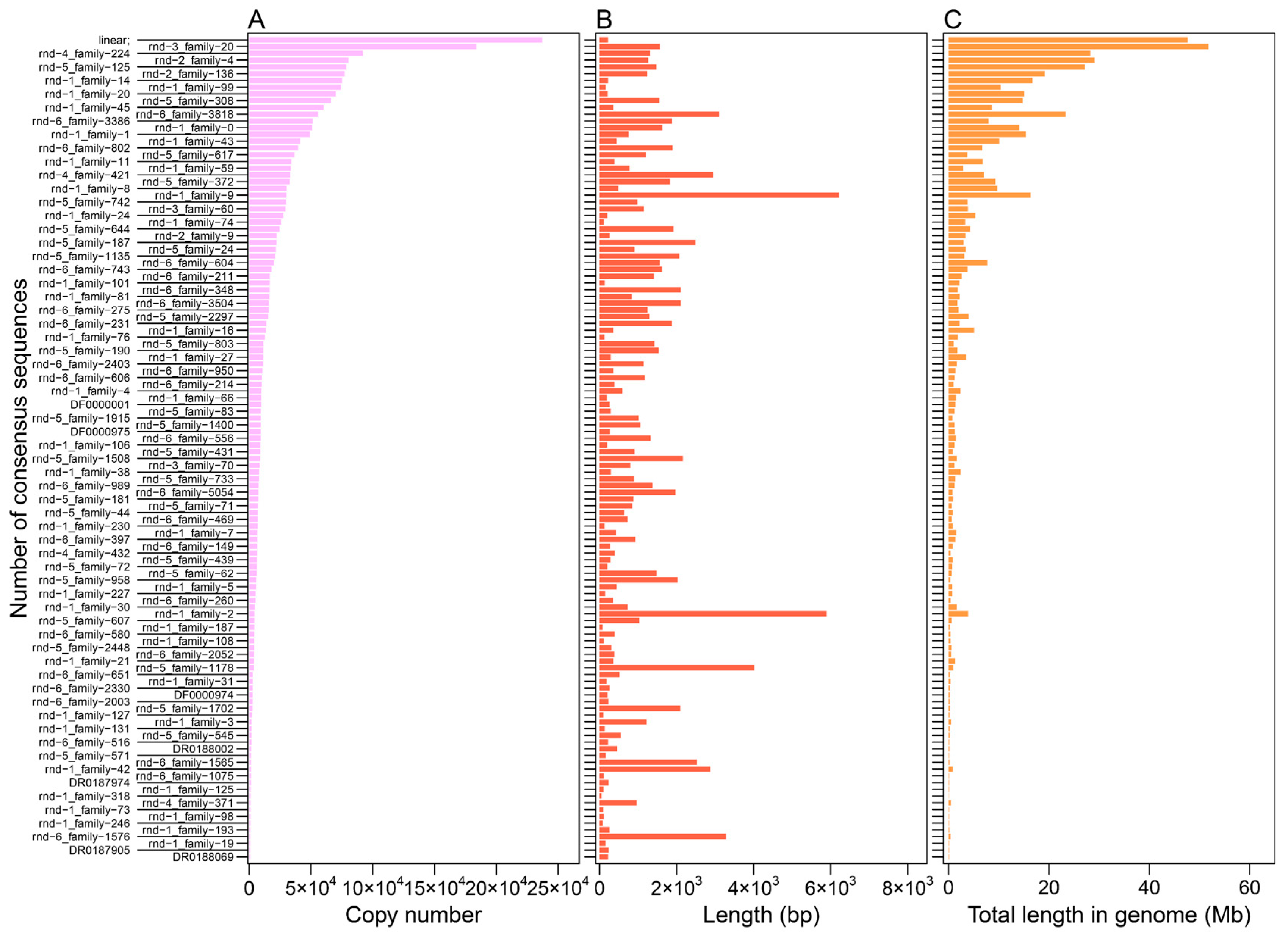
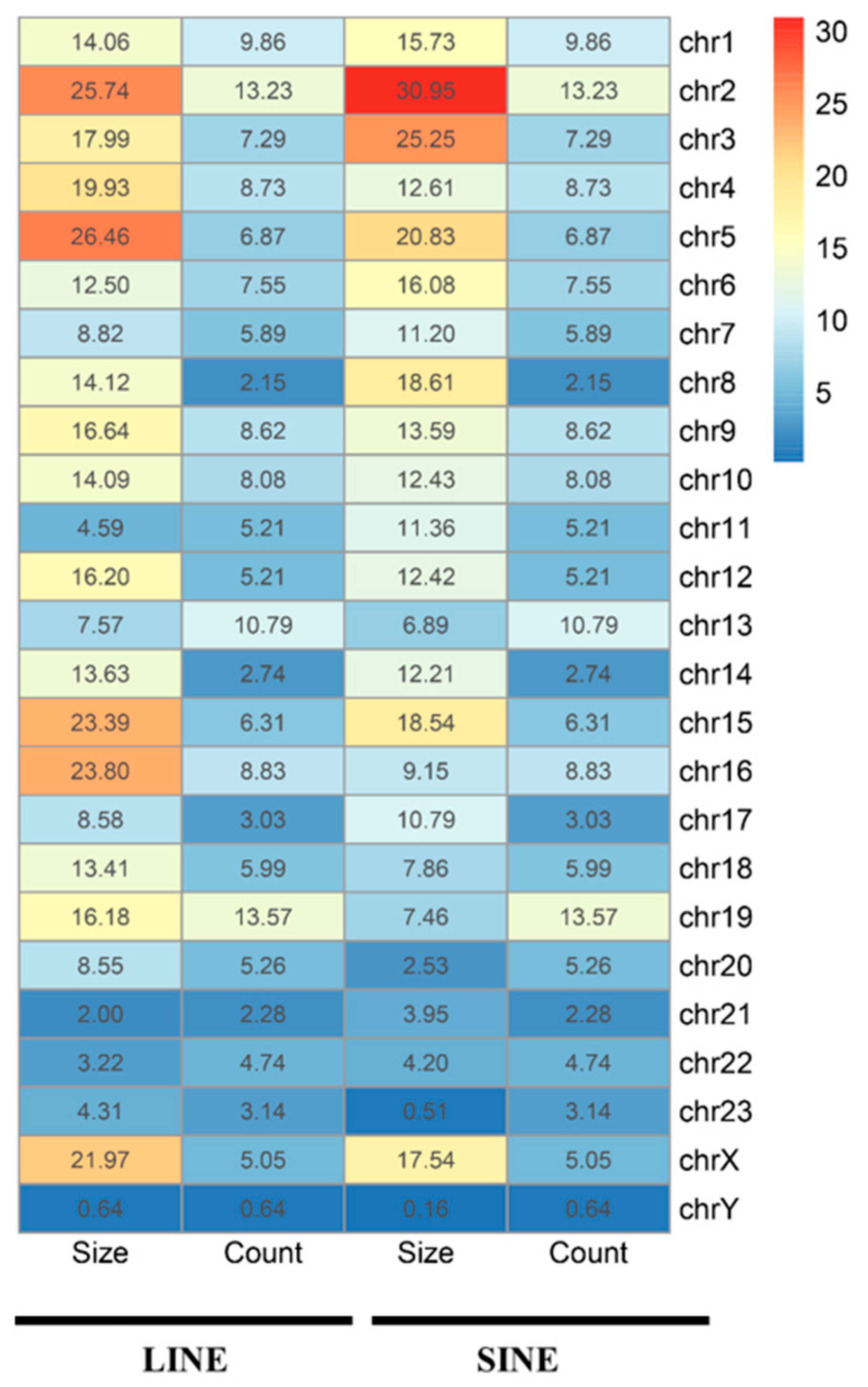
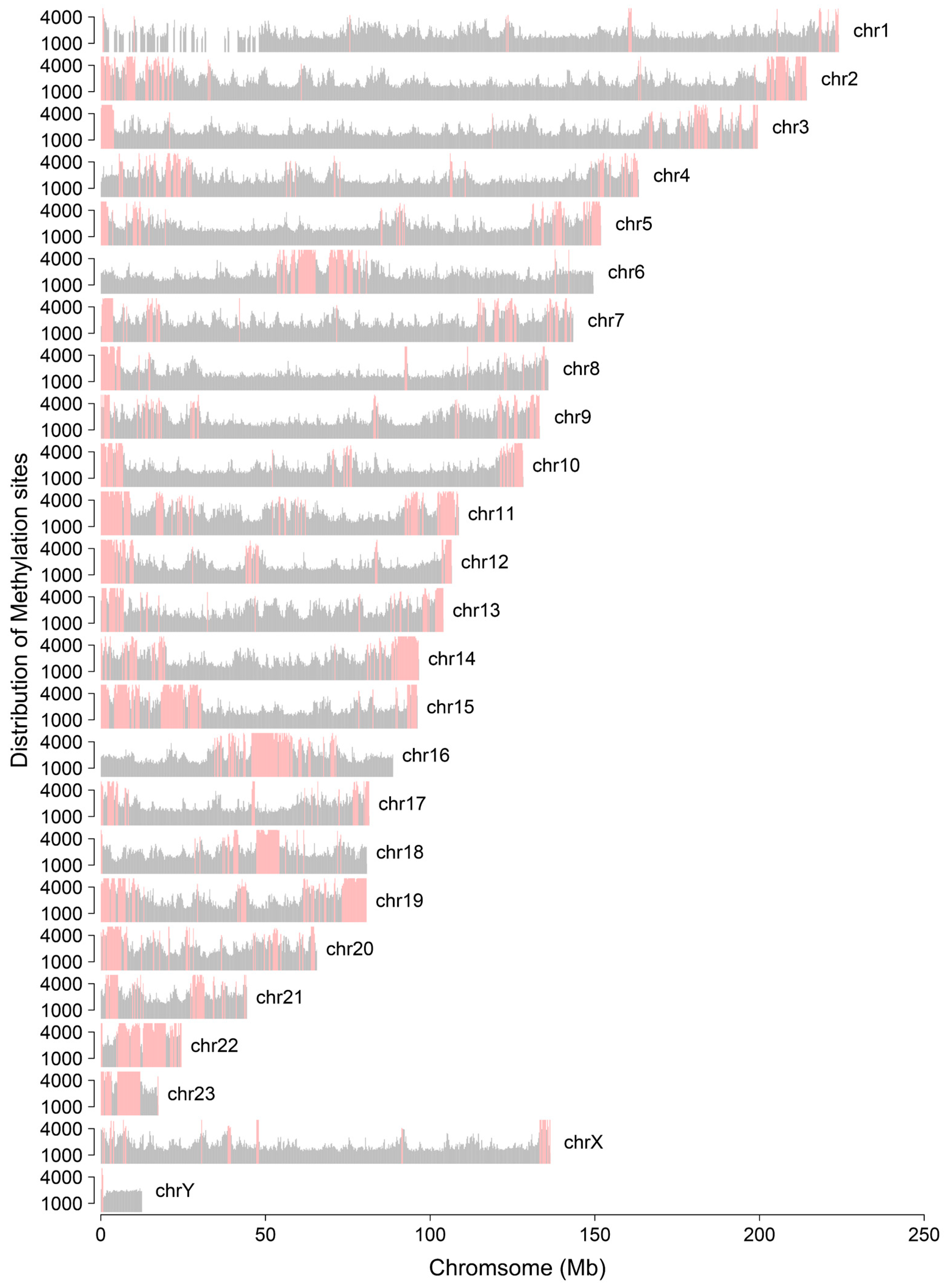
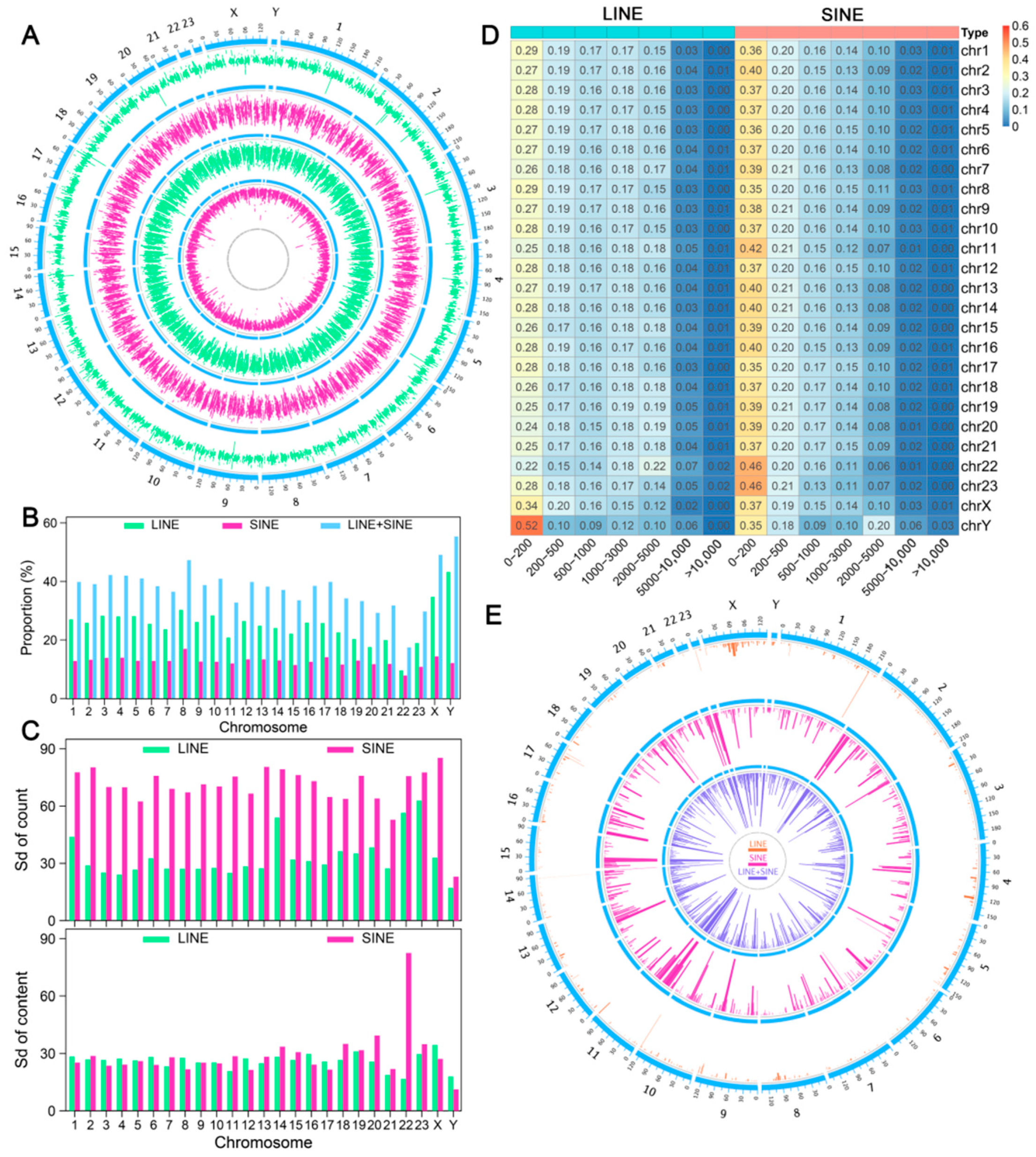
3.1. Distribution Feature of LINE and SINE Elements in the A. albiventris Genome
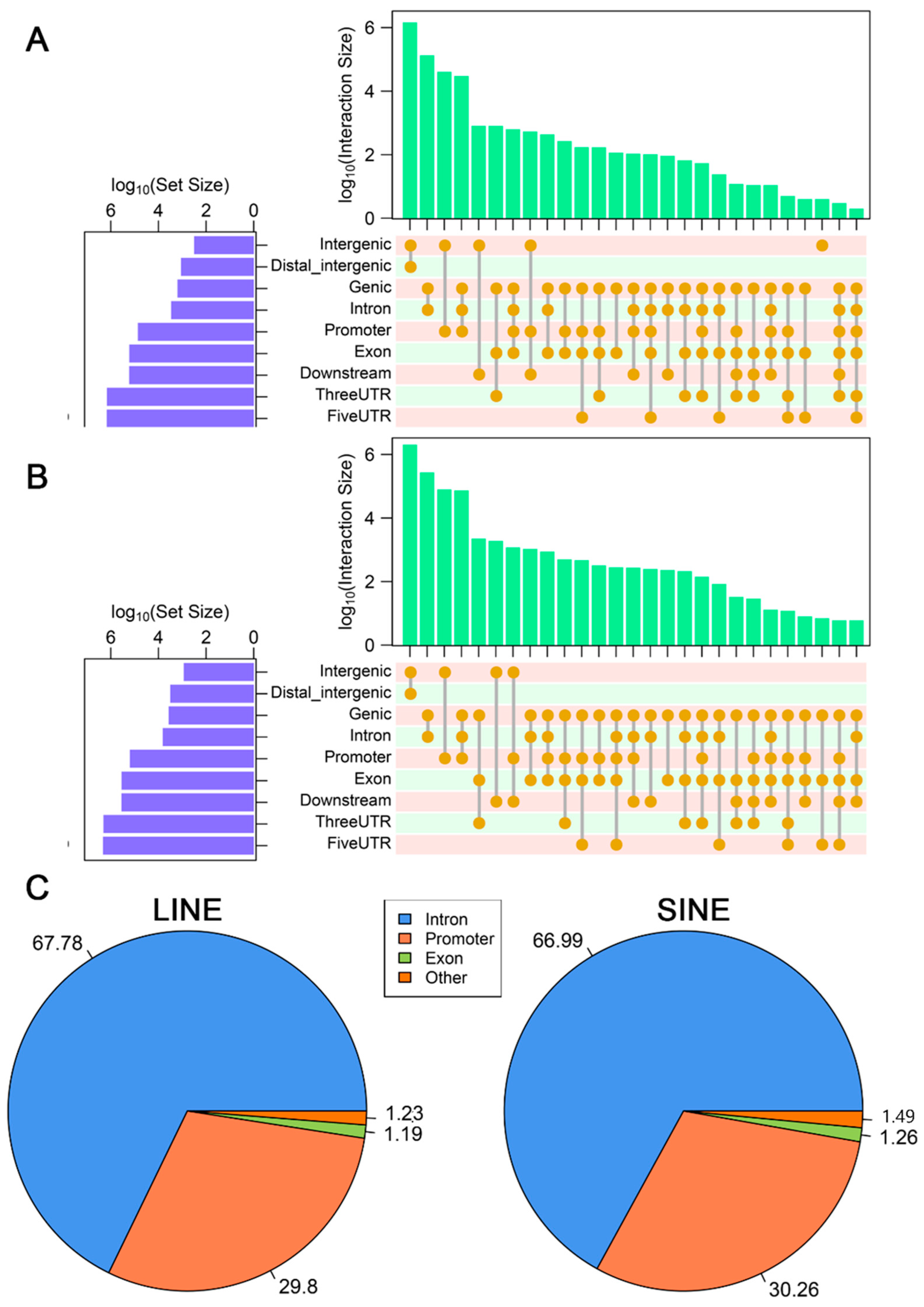
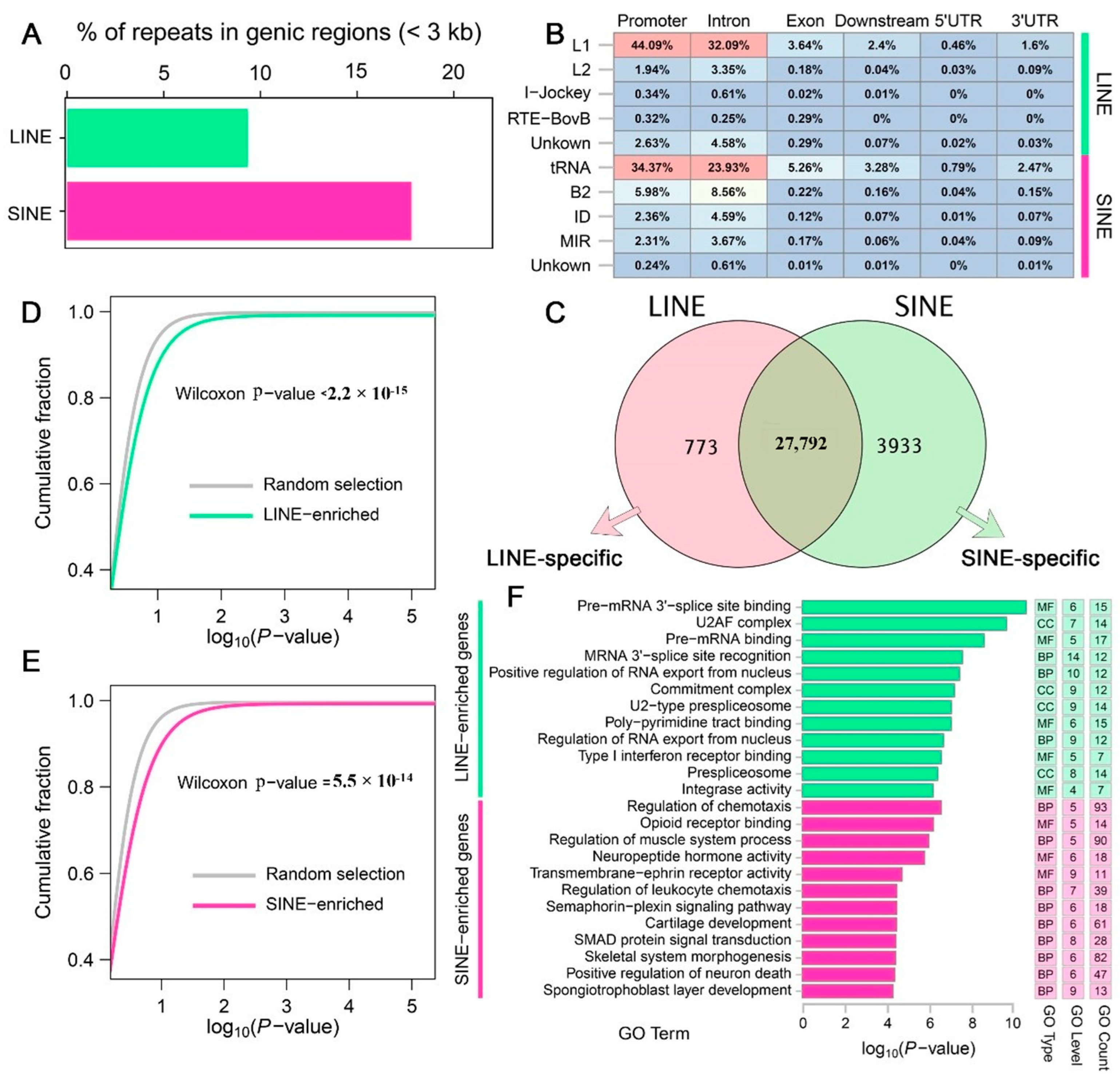
3.2. Impact of LINEs and SINEs on Gene Function in A. albiventris
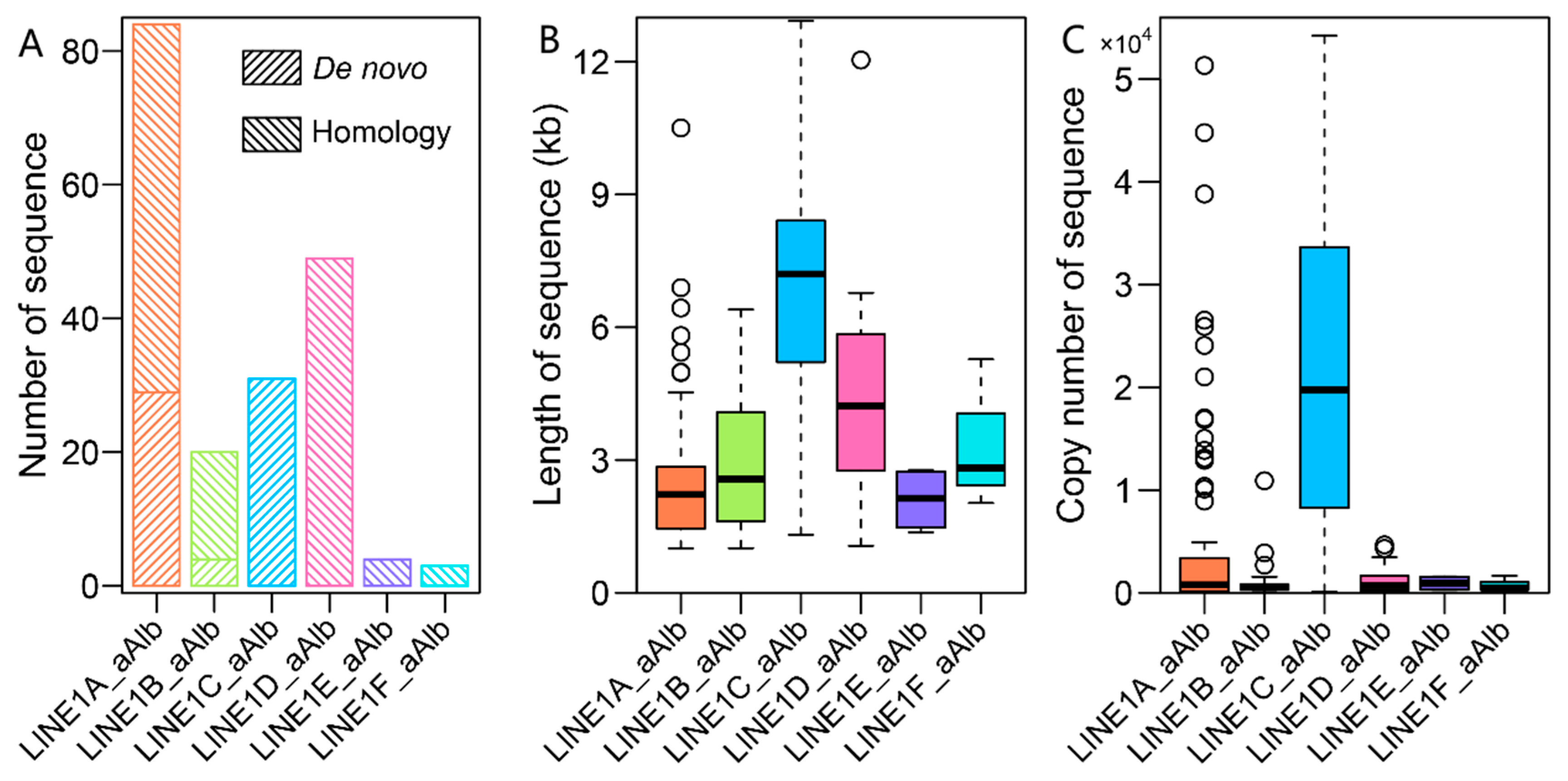
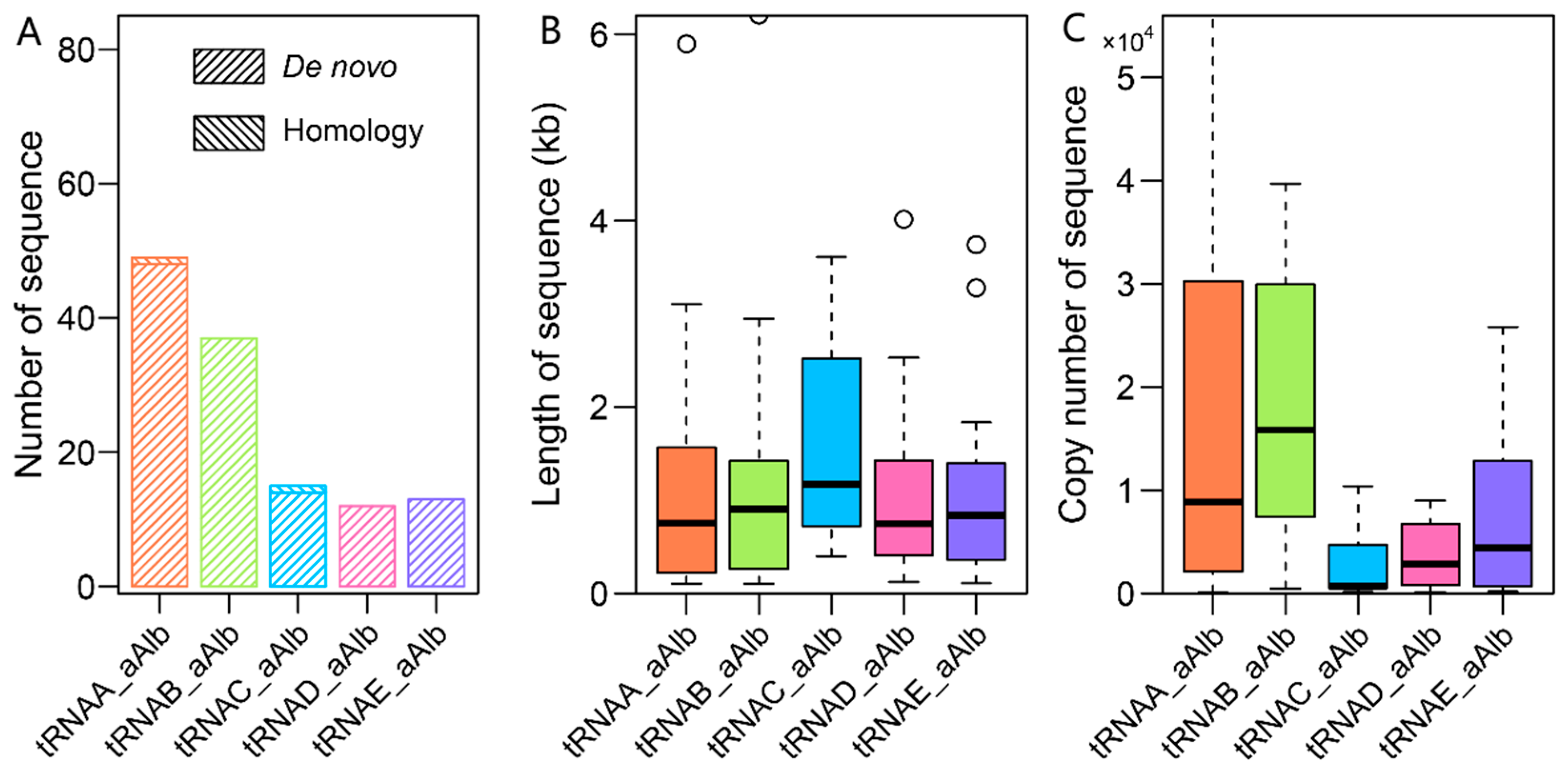
3.3. Evolution of LINEs and SINEs in the A. albiventris Genome
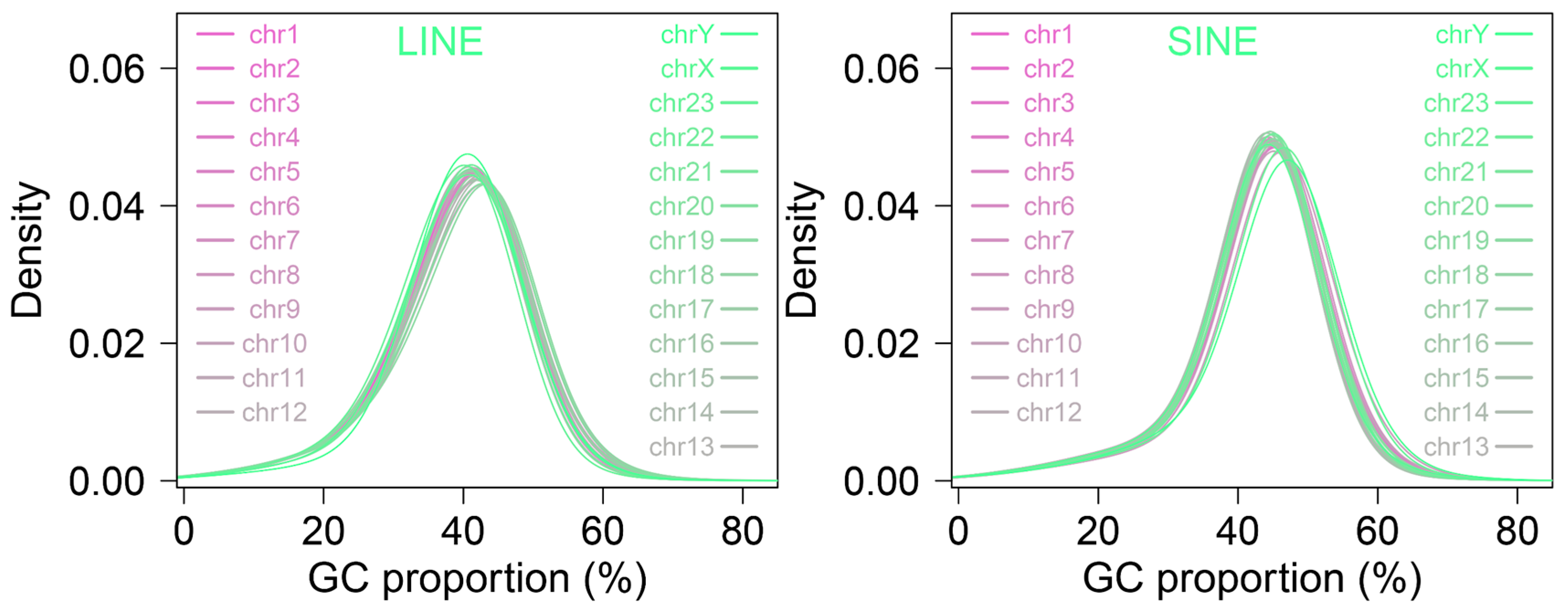
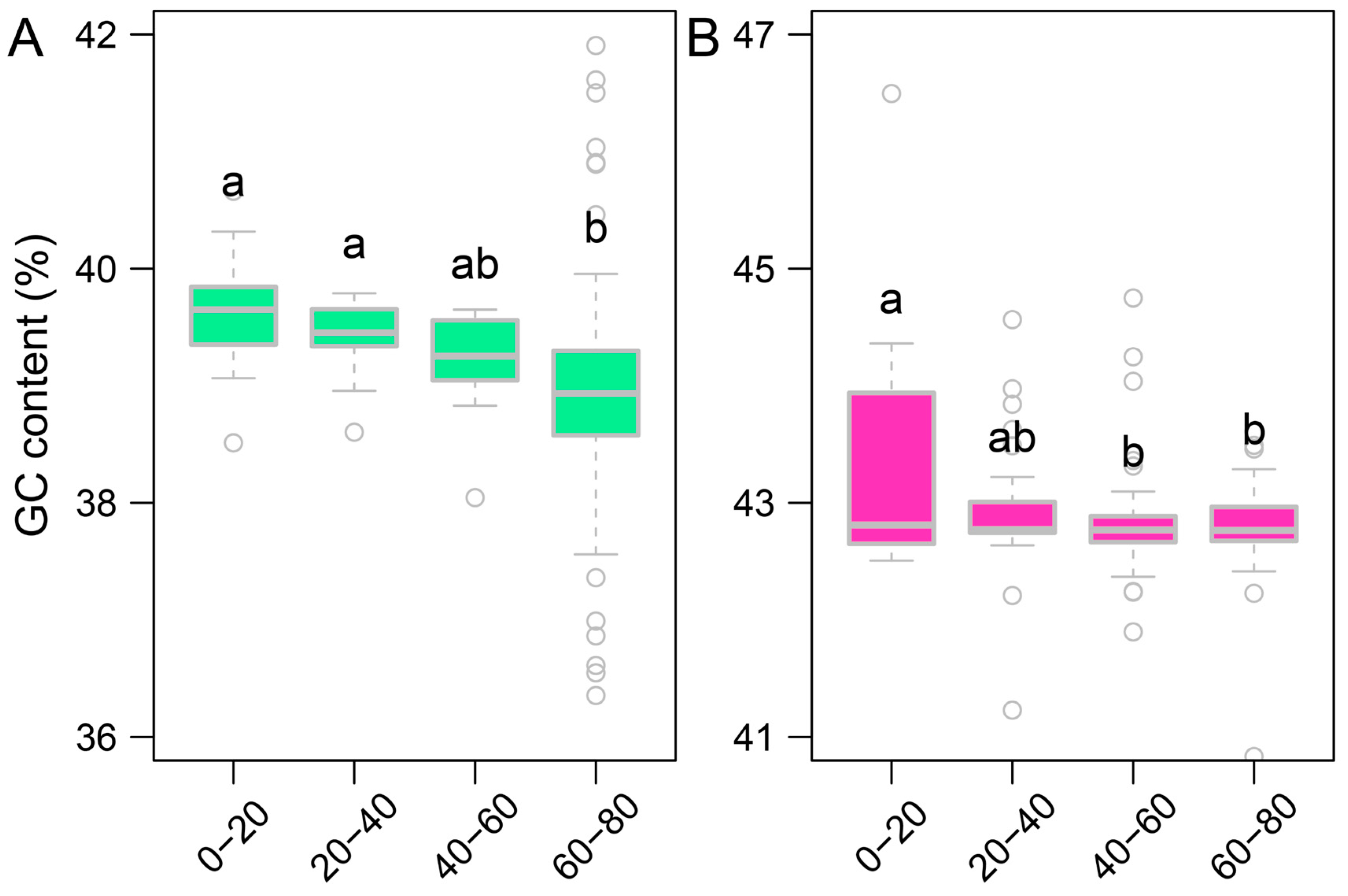
3.4. GC Content Associated with LINEs and SINEs Insertions
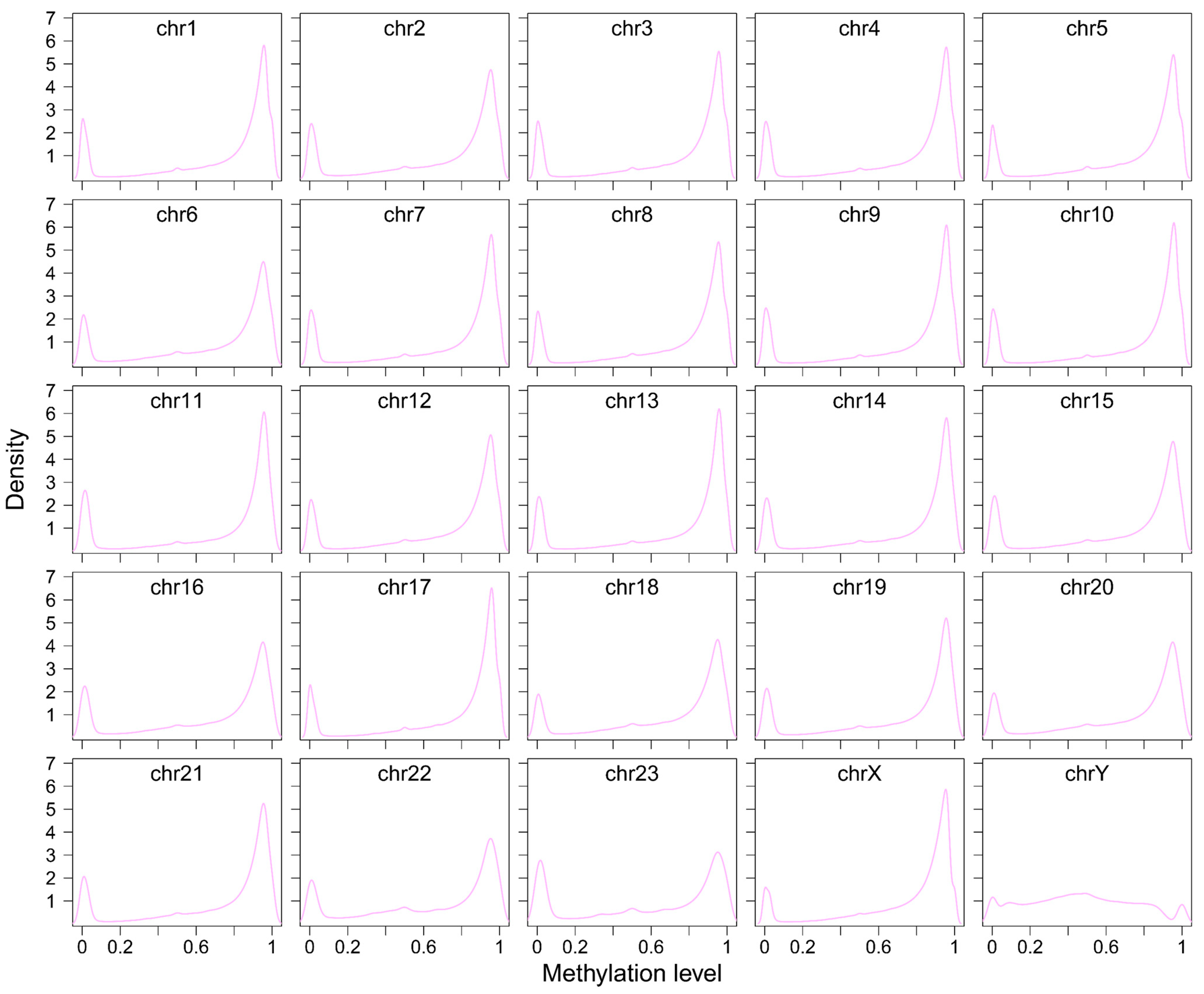
3.5. DNA Methylation Patterns of LINEs and SINEs in the A. albiventris Genome
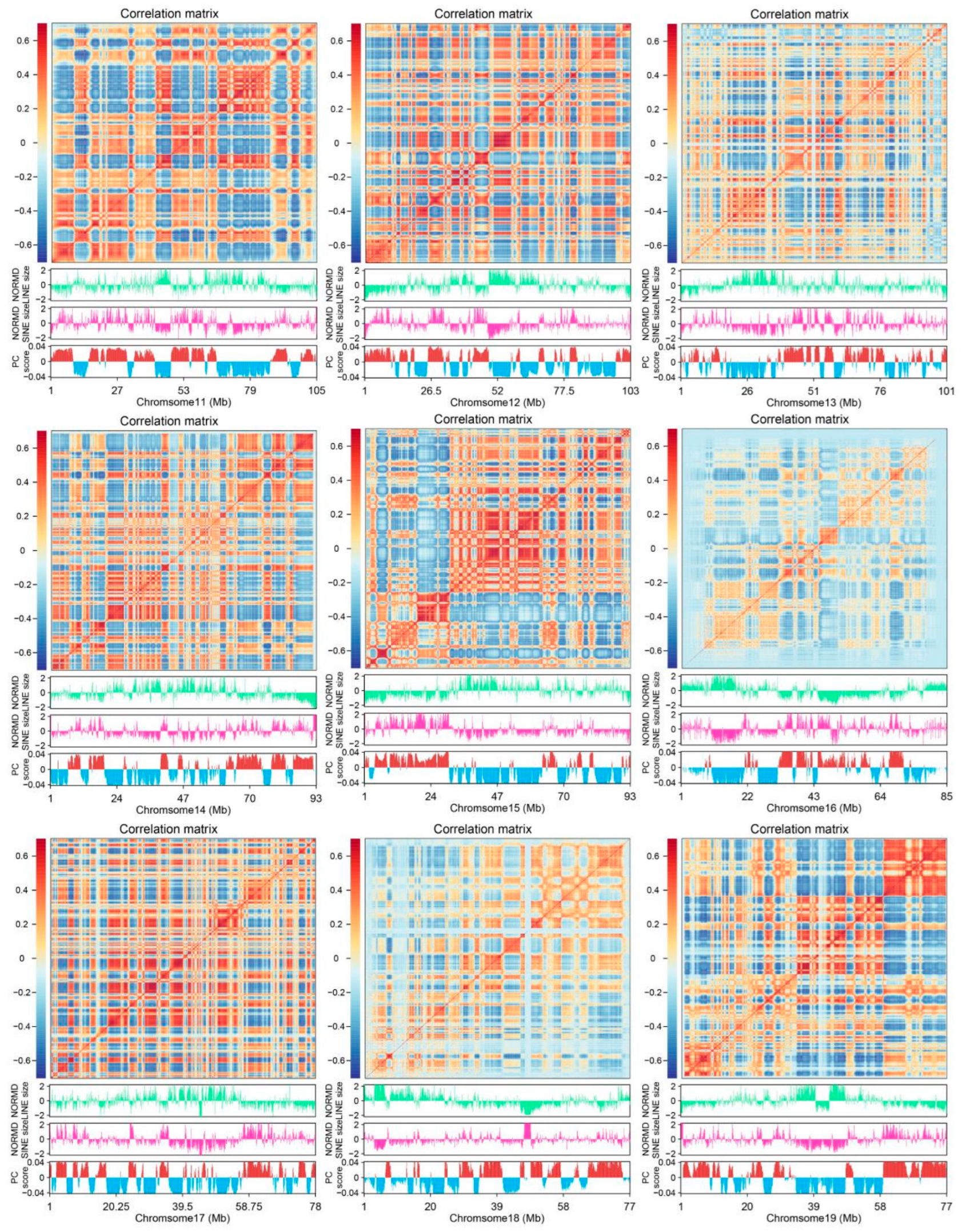
3.6. LINEs and SINEs Distributions Correlate with Global Compartmentalization
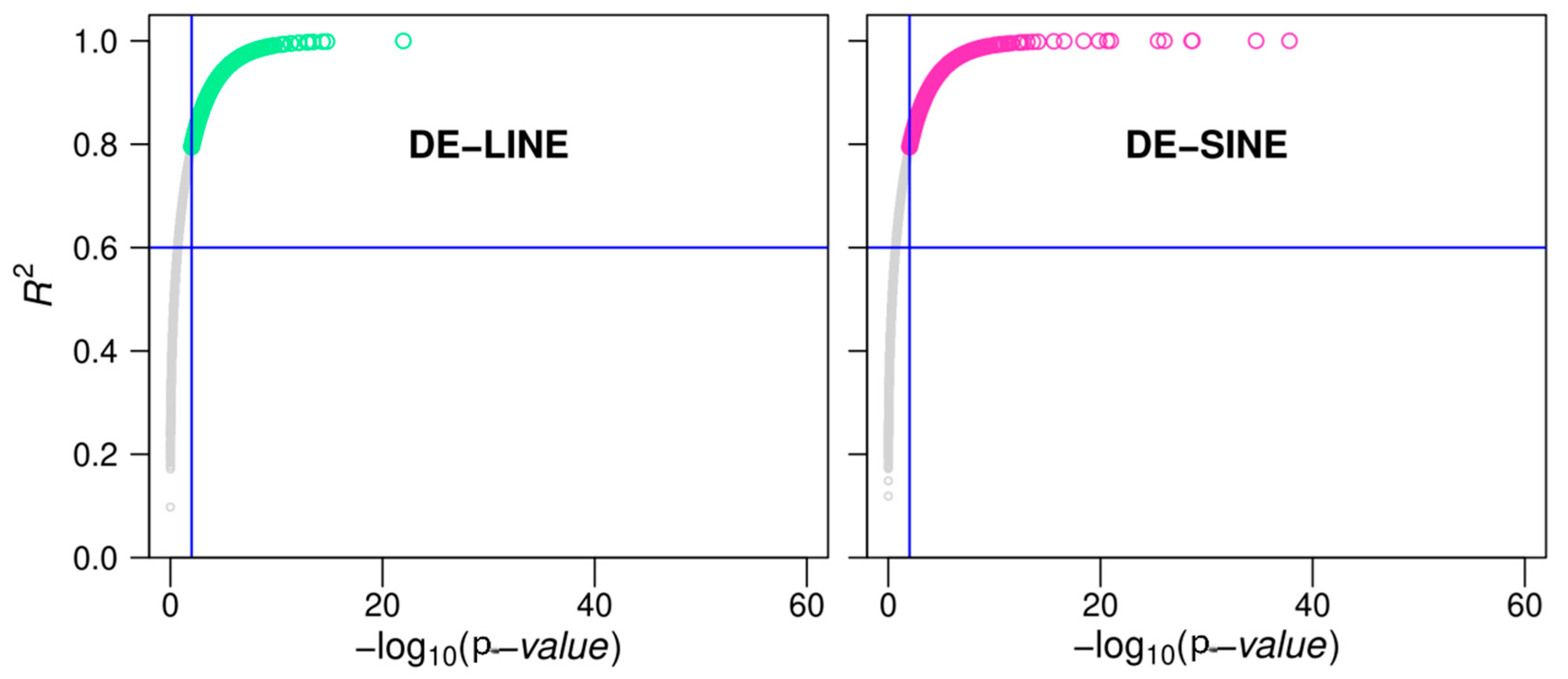
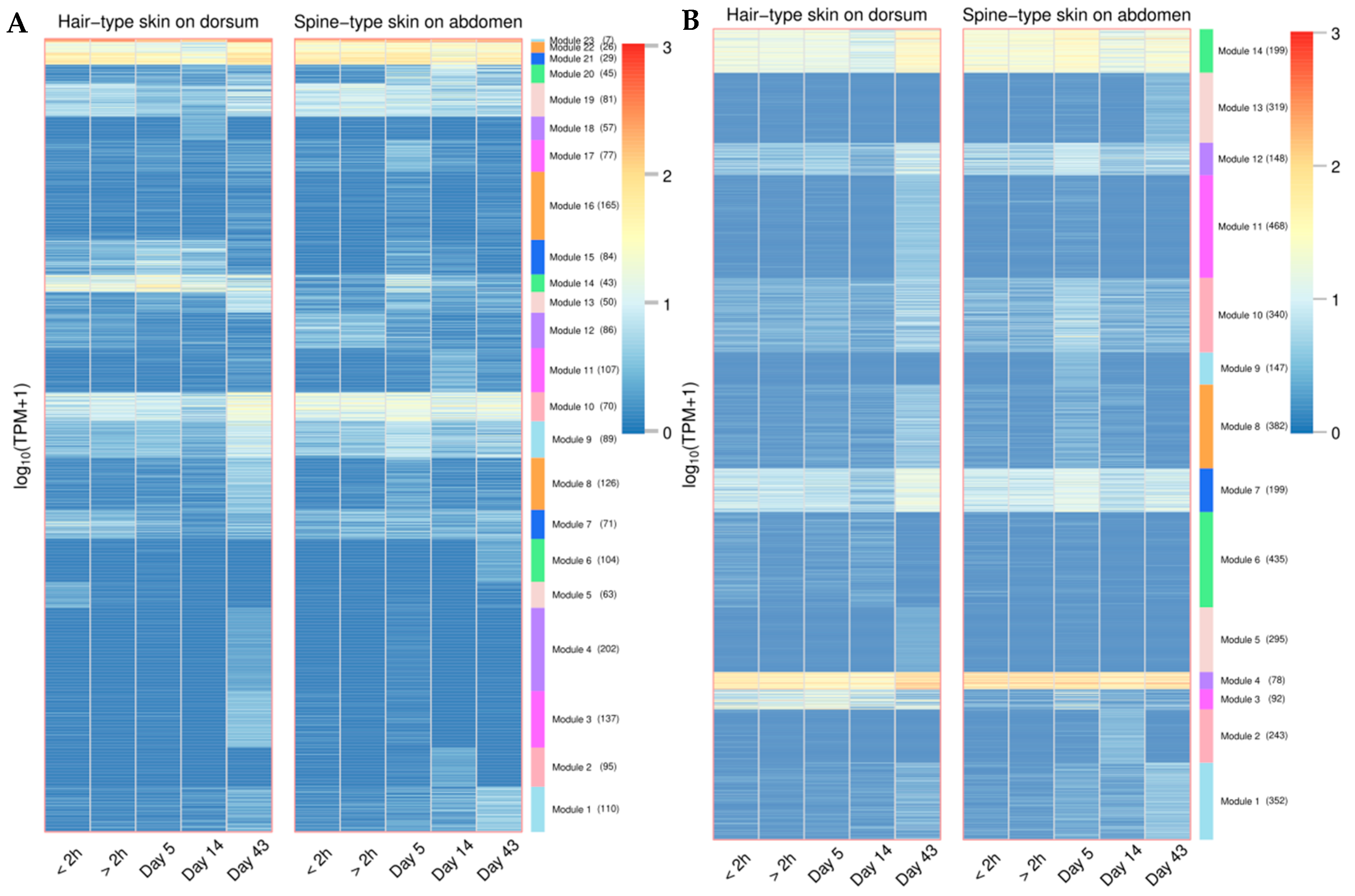
3.7. LINEs and SINEs Are Associated with Spine Formation in A. albiventris
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Unknown Subfamilies | TE Type | De Novo | Homologous | Length (bp) | Predicted Classification | Copy Number |
|---|---|---|---|---|---|---|
| UnL-9_aAlb | LINE | - | DR0188083 | 2610 | LINE_N1 | 147 |
| UnL-5_aAlb | - | DR0187941 | 6555 | 424 | ||
| UnL-3_aAlb | - | DR0187847 | 6294 | 492 | ||
| UnL-2_aAlb | - | DR0187849 | 5800 | 778 | ||
| UnL-1_aAlb | - | DR0188009 | 5966 | 778 | ||
| UnL-11_aAlb | - | DR0188066 | 3940 | 31 | ||
| UnL-7_aAlb | - | DR0187956 | 6171 | 230 | ||
| UnL-4_aAlb | rnd-6_family-1116 | - | 2894 | RTE-BovB | 483 | |
| UnL-8_aAlb | - | DR0187787 | 1092 | 151 | ||
| UnL-6_aAlb | rnd-6_family-2127 | - | 2609 | RTE-X | 338 | |
| UnL-10_aAlb | - | DR0187728 | 2877 | 40 | ||
| UnS-1_aAlb | SINE | - | DR0187830 | 279 | SINE_N1 | 151 |
| UnS-2_aAlb | - | DR0188050 | 196 | 123 | ||
| UnS-3_aAlb | rnd-1_family-337 | - | 208 | SINE_N2 | 17 | |
| UnS-4_aAlb | rnd-1_family-322 | - | 123 | MIR | 17 |







References
- Santana, E.M.; Jantz, H.E.; Best, T.L. Atelerix albiventris (Erinaceomorpha: Erinaceidae). Mamm. Species 2010, 42, 99–110. [Google Scholar] [CrossRef]
- Grzesiakowska, A.; Baran, P.; Kuchta-Gładysz, M.; Szeleszczuk, O. Cytogenetic karyotype analysis in selected species of the family. J. Vet. Res. 2019, 63, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Xu, J.; Zhu, M.; Lv, Z.; Ning, Z.; Yang, F. A haplotype-resolved genome reveals the genetic basis of spine formation in Atelerix albiventris. J. Genet. Genom. 2024, 51, 1529–1532. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Zhao, B.; Chen, Y.; D’Alessandro, E.; Chen, C.; Ji, T.; Song, C. Distinct retrotransposon evolution profile in the genome of rabbit (Oryctolagus cuniculus). Genome Biol. Evol. 2021, 13, evab168. [Google Scholar] [CrossRef]
- Beck, C.R.; Collier, P.; Macfarlane, C.; Malig, M.; Kidd, J.M.; Eichler, E.E.; Moran, J.V. LINE-1 retrotransposition activity in human genomes. Cell 2010, 141, 1159–1170. [Google Scholar] [CrossRef]
- Biémont, C. A brief history of the status of transposable elements: From junk DNA to major players in evolution. Genetics 2010, 186, 1085–1093. [Google Scholar] [CrossRef]
- Seibt, K.M.; Wenke, T.; Muders, K.; Truberg, B.; Schmidt, T. Short interspersed nuclear elements (SINEs) are abundant in Solanaceae and have a family-specific impact on gene structure and genome organization. Plant J. 2016, 86, 268–285. [Google Scholar] [CrossRef]
- Davidson, E.H.; Britten, R.J. Regulation of gene expression: Possible role of repetitive sequences. Science 1979, 204, 1052–1059. [Google Scholar] [CrossRef]
- Diehl, A.G.; Ouyang, N.; Boyle, A.P. Transposable elements contribute to cell and species-specific chromatin looping and gene regulation in mammalian genomes. Nat. Commun. 2020, 11, 1796. [Google Scholar] [CrossRef]
- Senft, A.D.; Macfarlan, T.S. Transposable elements shape the evolution of mammalian development. Nat. Rev. Genet. 2021, 22, 691–711. [Google Scholar] [CrossRef]
- Dolinoy, D.C.; Huang, D.; Jirtle, R.L. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc. Natl. Acad. Sci. USA 2007, 104, 13056–13061. [Google Scholar] [PubMed]
- Sharif, J.; Koseki, H.; Parrish, N.F. Bridging multiple dimensions: Roles of transposable elements in higher-order genome regulation. Curr. Opin. Genet. Dev. 2023, 80, 102035. [Google Scholar] [CrossRef] [PubMed]
- Wicker, T.; Gundlach, H.; Spannagl, M.; Uauy, C.; Borrill, P.; Ramírez-González, R.H.; Choulet, F. Impact of transposable elements on genome structure and evolution in bread wheat. Genome Biol. 2018, 19, 103. [Google Scholar] [CrossRef] [PubMed]
- Kroutter, E.N.; Belancio, V.P.; Wagstaff, B.J.; Roy-Engel, A.M. The RNA polymerase dictates ORF1 requirement and timing of LINE and SINE retrotransposition. PLoS Genet. 2009, 5, e1000458. [Google Scholar] [CrossRef]
- Kramerov, D.A.; Vassetzky, N.S. Origin and evolution of SINEs in eukaryotic genomes. Heredity 2011, 107, 487–495. [Google Scholar]
- Osmanski, A.B.; Paulat, N.S.; Korstian, J.; Grimshaw, J.R.; Halsey, M.; Sullivan, K.A.; Ray, D.A. Insights into mammalian TE diversity via the curation of 248 mammalian genome assemblies. Science 2023, 380, eabn1430. [Google Scholar]
- Taylor, M.S.; LaCava, J.; Mita, P.; Molloy, K.R.; Huang, C.R.L.; Li, D.; Dai, L. Affinity proteomics reveals human host factors implicated in discrete stages of LINE-1 retrotransposition. Cell 2013, 155, 1034–1048. [Google Scholar]
- Veniaminova, N.A.; Vassetzky, N.S.; Kramerov, D.A. B1 SINEs in different rodent families. Genomics 2007, 89, 678–686. [Google Scholar] [CrossRef][Green Version]
- Zhang, X.O.; Pratt, H.; Weng, Z. Investigating the potential roles of SINEs in the human genome. Annu. Rev. Genom. Hum. Genet. 2021, 22, 199–218. [Google Scholar]
- Ostertag, E.M.; Kazazian, H.H., Jr. Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 2001, 35, 501–538. [Google Scholar]
- Elbarbary, R.A.; Lucas, B.A.; Maquat, L.E. Retrotransposons as regulators of gene expression. Science 2016, 351, aac7247. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Scott, L.; Wichman, H.A. Tracing the history of LINE and SINE extinction in sigmodontine rodents. Mobile DNA 2019, 10, 22. [Google Scholar] [PubMed]
- Richardson, S.R.; Doucet, A.J.; Kopera, H.C.; Moldovan, J.B.; Garcia-Perez, J.L.; Moran, J.V. The influence of LINE-1 and SINE retrotransposons on mammalian genomes. Mobile DNA III 2015, 1165–1208. [Google Scholar] [CrossRef]
- Vassetzky, N.S.; Kramerov, D.A. SINEBase: A database and tool for SINE analysis. Nucleic Acids Res. 2013, 41, D83–D89. [Google Scholar]
- Konkel, M.K.; Walker, J.A.; Batzer, M.A. LINEs and SINEs of primate evolution. Evol. Anthropol. 2010, 19, 236–249. [Google Scholar]
- Kido, H.; Komarneni, S.; Roy, R. Preparation of La₂Zr₂O₇ by Sol-Gel Route. J. Am. Ceram. Soc. 1991, 74, 422–424. [Google Scholar]
- Kazazian, H.H., Jr. Mobile elements: Drivers of genome evolution. Science 2004, 303, 1626–1632. [Google Scholar]
- Manthey, J.D.; Moyle, R.G.; Boissinot, S. Multiple and independent phases of transposable element amplification in the genomes of piciformes (woodpeckers and allies). Genome Biol. Evol. 2018, 10, 1445–1456. [Google Scholar] [CrossRef]
- Zhao, P.; Gu, L.; Gao, Y.; Pan, Z.; Liu, L.; Li, X.; Wang, Z. Young SINEs in pig genomes impact gene regulation, genetic diversity, and complex traits. Commun. Biol. 2023, 6, 894. [Google Scholar]
- Lu, J.Y.; Chang, L.; Li, T.; Wang, T.; Yin, Y.; Zhan, G.; Shen, X. Homotypic clustering of L1 and B1/Alu repeats compartmentalizes the 3D genome. Cell Res. 2021, 31, 613–630. [Google Scholar]
- Haws, S.A.; Simandi, Z.; Barnett, R.J.; Phillips-Cremins, J.E. 3D genome, on repeat: Higher-order folding principles of the heterochromatinized repetitive genome. Cell 2022, 185, 2690–2707. [Google Scholar] [CrossRef] [PubMed]
- Román, A.C.; González-Rico, F.J.; Moltó, E.; Hernando, H.; Neto, A.; Vicente-Garcia, C.; Fernández-Salguero, P.M. Dioxin receptor and SLUG transcription factors regulate the insulator activity of B1 SINE retrotransposons via an RNA polymerase switch. Genome Res. 2011, 21, 422–432. [Google Scholar] [CrossRef] [PubMed]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Ren, B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef]
- Nora, E.P.; Goloborodko, A.; Valton, A.L.; Gibcus, J.H.; Uebersohn, A.; Abdennur, N.; Bruneau, B.G. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 2017, 169, 930–944. [Google Scholar] [CrossRef]
- Choudhary, M.N.; Friedman, R.Z.; Wang, J.T.; Jang, H.S.; Zhuo, X.; Wang, T. Co-opted transposons help perpetuate conserved higher-order chromosomal structures. Genome Biol. 2020, 21, 16. [Google Scholar] [CrossRef]
- Kentepozidou, E.; Aitken, S.J.; Feig, C.; Stefflova, K.; Ibarra-Soria, X.; Odom, D.T.; Flicek, P. Clustered CTCF binding is an evolutionary mechanism to maintain topologically associating domains. Genome Biol. 2020, 21, 5. [Google Scholar] [CrossRef]
- Chow, J.C.; Ciaudo, C.; Fazzari, M.J.; Mise, N.; Servant, N.; Glass, J.L.; Heard, E. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell 2010, 141, 956–969. [Google Scholar] [CrossRef]
- Mao, H.; Wang, H.; Liu, S.; Li, Z.; Yang, X.; Yan, J.; Qin, F. A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat. Commun. 2015, 6, 8326. [Google Scholar] [CrossRef]
- Attig, J.; Ruiz de los Mozos, I.; Haberman, N.; Wang, Z.; Emmett, W.; Zarnack, K.; Ule, J. Splicing repression allows the gradual emergence of new Alu-exons in primate evolution. eLife 2016, 5, e19545. [Google Scholar] [CrossRef]
- Sundaram, V.; Cheng, Y.; Ma, Z.; Li, D.; Xing, X.; Edge, P.; Wang, T. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 2014, 24, 1963–1976. [Google Scholar] [CrossRef]
- Trizzino, M.; Park, Y.; Holsbach-Beltrame, M.; Aracena, K.; Mika, K.; Caliskan, M.; Brown, C.D. Transposable elements are the primary source of novelty in primate gene regulation. Genome Res. 2017, 27, 1623–1633. [Google Scholar] [PubMed]
- Lin, L.; Shen, S.; Tye, A.; Cai, J.J.; Jiang, P.; Davidson, B.L.; Xing, Y. Diverse splicing patterns of exonized Alu elements in human tissues. PLoS Genet. 2008, 4, e1000225. [Google Scholar] [CrossRef] [PubMed]
- Yakovchuk, P.; Goodrich, J.A.; Kugel, J.F. B2 RNA and Alu RNA repress transcription by disrupting contacts between RNA polymerase II and promoter DNA within assembled complexes. Proc. Natl. Acad. Sci. USA 2009, 106, 5569–5574. [Google Scholar] [PubMed]
- Hadjiargyrou, M.; Delihas, N. The intertwining of transposable elements and non-coding RNAs. Int. J. Mol. Sci. 2013, 14, 13307–13328. [Google Scholar] [CrossRef]
- Roberts, J.T.; Cooper, E.A.; Favreau, C.J.; Howell, J.S.; Lane, L.G.; Mills, J.E.; Borchert, G.M. Continuing analysis of microRNA origins: Formation from transposable element insertions and noncoding RNA mutations. Mob. Genet. Elem. 2013, 3, e27755. [Google Scholar]
- Flynn, J.M.; Hubley, R.; Goubert, C.; Rosen, J.; Clark, A.G.; Feschotte, C.; Smit, A.F. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. USA 2020, 117, 9451–9457. [Google Scholar]
- Abrusán, G.; Grundmann, N.; DeMester, L.; Makalowski, W. TEclass-a tool for automated classification of unknown eukaryotic transposable elements. Bioinformatics 2009, 25, 1329–1330. [Google Scholar]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Goubert, C.; Craig, R.J.; Bilat, A.F.; Peona, V.; Vogan, A.A.; Protasio, A.V. A beginner’s guide to manual curation of transposable elements. Mob. DNA 2022, 13, 7. [Google Scholar]
- McGinnis, S.; Madden, T.L. BLAST: At the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004, 32, W20–W25. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.I.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Magis, C.; Taly, J.F.; Bussotti, G.; Chang, J.M.; Di Tommaso, P.; Erb, I.; Notredame, C. T-Coffee: Tree-based consistency objective function for alignment evaluation. Methods Mol. Biol. 2014, 117, 129–143. [Google Scholar]
- Dainat, J. AGAT: Another Gff Analysis Toolkit to Handle Annotations in Any GTF/GFF Format (Version v0.5.1). Zenodo 2021. Available online: https://github.com/NBISweden/AGAT (accessed on 6 December 2024).
- Wang, Q.; Li, M.; Wu, T.; Zhan, L.; Li, L.; Chen, M.; Yu, G. Exploring epigenomic datasets by ChIPseeker. Curr. Protoc. 2022, 2, e585. [Google Scholar] [CrossRef] [PubMed]
- Bolstad, B.M.; Irizarry, R.A.; Åstrand, M.; Speed, T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003, 19, 185–193. [Google Scholar] [CrossRef]
- Kumar, S.; Subramanian, S. Mutation rates in mammalian genomes. Proc. Natl. Acad. Sci. USA 2002, 99, 803–808. [Google Scholar]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.Y. ggtree: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Ramírez, F.; Bhardwaj, V.; Arrigoni, L.; Lam, K.C.; Grüning, B.A.; Villaveces, J.; Manke, T. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat. Commun. 2018, 9, 189. [Google Scholar] [CrossRef]
- Ni, P.; Nie, F.; Zhong, Z.; Xu, J.; Huang, N.; Zhang, J.; Wang, J. DNA 5-methylcytosine detection and methylation phasing using PacBio circular consensus sequencing. Nat. Commun. 2023, 14, 4054. [Google Scholar] [CrossRef]
- Kong, Y.; Rose, C.M.; Cass, A.A.; Williams, A.G.; Darwish, M.; Lianoglou, S.; Chen-Harris, H. Transposable element expression in tumors is associated with immune infiltration and increased antigenicity. Nat. Commun. 2019, 10, 5228. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [PubMed]
- Conesa, A.; Nueda, M.J.; Ferrer, A.; Talón, M. maSigPro: A method to identify significantly differential expression profiles in time-course microarray experiments. Bioinformatics 2006, 22, 1096–1102. [Google Scholar] [PubMed]
- Gibbons, J.D.; Chakraborti, S. Nonparametric Statistical Inference. Revised and Expanded, 4th ed.CRC Press: Boca Raton, FL, USA, 2003. [Google Scholar]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Yu, G. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. The Innovation 2021, 2, 3. [Google Scholar]
- Gilbert, N.; Labuda, D. CORE-SINEs: Eukaryotic short interspersed retroposing elements with common sequence motifs. Proc. Natl. Acad. Sci. USA 1999, 96, 2869–2874. [Google Scholar]
- Nishihara, H.; Smit, A.F.; Okada, N. Functional noncoding sequences derived from SINEs in the mammalian genome. Genome Res. 2006, 16, 864–874. [Google Scholar]
- Canapa, A.; Barucca, M.; Biscotti, M.A.; Forconi, M.; Olmo, E. Transposons, genome size, and evolutionary insights in animals. Cytogenet. Genome Res. 2016, 147, 217–239. [Google Scholar]
- Hayward, A.; Gilbert, C. Transposable elements. Curr. Biol. 2022, 32, R904–R909. [Google Scholar]
- Waterston, R.H. Initial sequencing and comparative analysis of the mouse genome. Nature 2002, 420, 520–562. [Google Scholar]
- Kawase, M.; Ichiyanagi, K. Mouse retrotransposons: Sequence structure, evolutionary age, genomic distribution and function. Genes Genet. Syst. 2024, 98, 337–351. [Google Scholar]
- Kumar, S.; Stecher, G.; Suleski, M.; Hedges, S.B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 2017, 34, 1812–1819. [Google Scholar] [CrossRef] [PubMed]
- Smit, A.F. The origin of interspersed repeats in the human genome. Curr. Opin. Genet. Dev. 1996, 6, 743–748. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Joyce, B.T.; Liu, L.; Zhang, Z.; Kibbe, W.A.; Zhang, W.; Hou, L. Prediction of genome-wide DNA methylation in repetitive elements. Nucleic Acids Res. 2017, 45, 8697–8711. [Google Scholar] [CrossRef]
- Zhou, W.; Liang, G.; Molloy, P.L.; Jones, P.A. DNA methylation enables transposable element-driven genome expansion. Proc. Natl. Acad. Sci. USA 2020, 117, 19359–19366. [Google Scholar] [CrossRef]
- Lucas, B.A.; Lavi, E.; Shiue, L.; Cho, H.; Katzman, S.; Miyoshi, K.; Maquat, L.E. Evidence for convergent evolution of SINE-directed Staufen-mediated mRNA decay. Proc. Natl. Acad. Sci. USA 2018, 115, 968–973. [Google Scholar] [CrossRef]
- Sironen, A.; Uimari, P.; Iso-Touru, T.; Vilkki, J. L1 insertion within SPEF2 gene is associated with increased litter size in the Finnish Yorkshire population. J. Anim. Breed. Genet. 2012, 129, 92–97. [Google Scholar] [CrossRef]
- Hancks, D.C.; Kazazian, H.H. Roles for retrotransposon insertions in human disease. Mob. DNA 2016, 7, 9. [Google Scholar] [CrossRef]
- Liu, C.; Ran, X.; Niu, X.; Li, S.; Wang, J.; Zhang, Q. Insertion of 275-bp SINE into first intron of PDIA4 gene is associated with litter size in Xiang pigs. Anim. Reprod. Sci. 2018, 195, 16–23. [Google Scholar] [CrossRef]
- Carrieri, C.; Cimatti, L.; Biagioli, M.; Beugnet, A.; Zucchelli, S.; Fedele, S.; Gustincich, S. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 2012, 491, 454–457. [Google Scholar] [CrossRef]
- Estécio, M.R.; Gallegos, J.; Dekmezian, M.; Lu, Y.; Liang, S.; Issa, J.P.J. SINE retrotransposons cause epigenetic reprogramming of adjacent gene promoters. Mol. Cancer Res. 2012, 10, 1332–1342. [Google Scholar] [CrossRef]
- Coufal, N.G.; Garcia-Perez, J.L.; Peng, G.E.; Yeo, G.W.; Mu, Y.; Lovci, M.T.; Gage, F.H. L1 retrotransposition in human neural progenitor cells. Nature 2009, 460, 1127–1131. [Google Scholar] [PubMed]
- Gebrie, A. Transposable elements as essential elements in the control of gene expression. Mob. DNA 2023, 14, 9. [Google Scholar]
- Garza, R.; Atacho, D.A.; Adami, A.; Gerdes, P.; Vinod, M.; Hsieh, P.; Jakobsson, J. LINE-1 retrotransposons drive human neuronal transcriptome complexity and functional diversification. Sci. Adv. 2023, 9, eadh9543. [Google Scholar]
- Gasparotto, E.; Burattin, F.V.; Di Gioia, V.; Panepuccia, M.; Ranzani, V.; Marasca, F.; Bodega, B. Transposable elements co-option in genome evolution and gene regulation. Int. J. Mol. Sci. 2023, 24, 2610. [Google Scholar] [CrossRef]
- Lanciano, S.; Cristofari, G. Measuring and interpreting transposable element expression. Nat. Rev. Genet. 2020, 21, 721–736. [Google Scholar]
- Zhang, X.O.; Gingeras, T.R.; Weng, Z. Genome-wide analysis of polymerase III-transcribed Alu elements suggests cell-type-specific enhancer function. Genome Res. 2019, 29, 1402–1414. [Google Scholar]
- Mori, Y.; Ichiyanagi, K. melRNA-seq for Expression Analysis of SINE RNAs and Other Medium-Length Non-Coding RNAs. Mob. DNA 2021, 12, 15. [Google Scholar] [CrossRef]


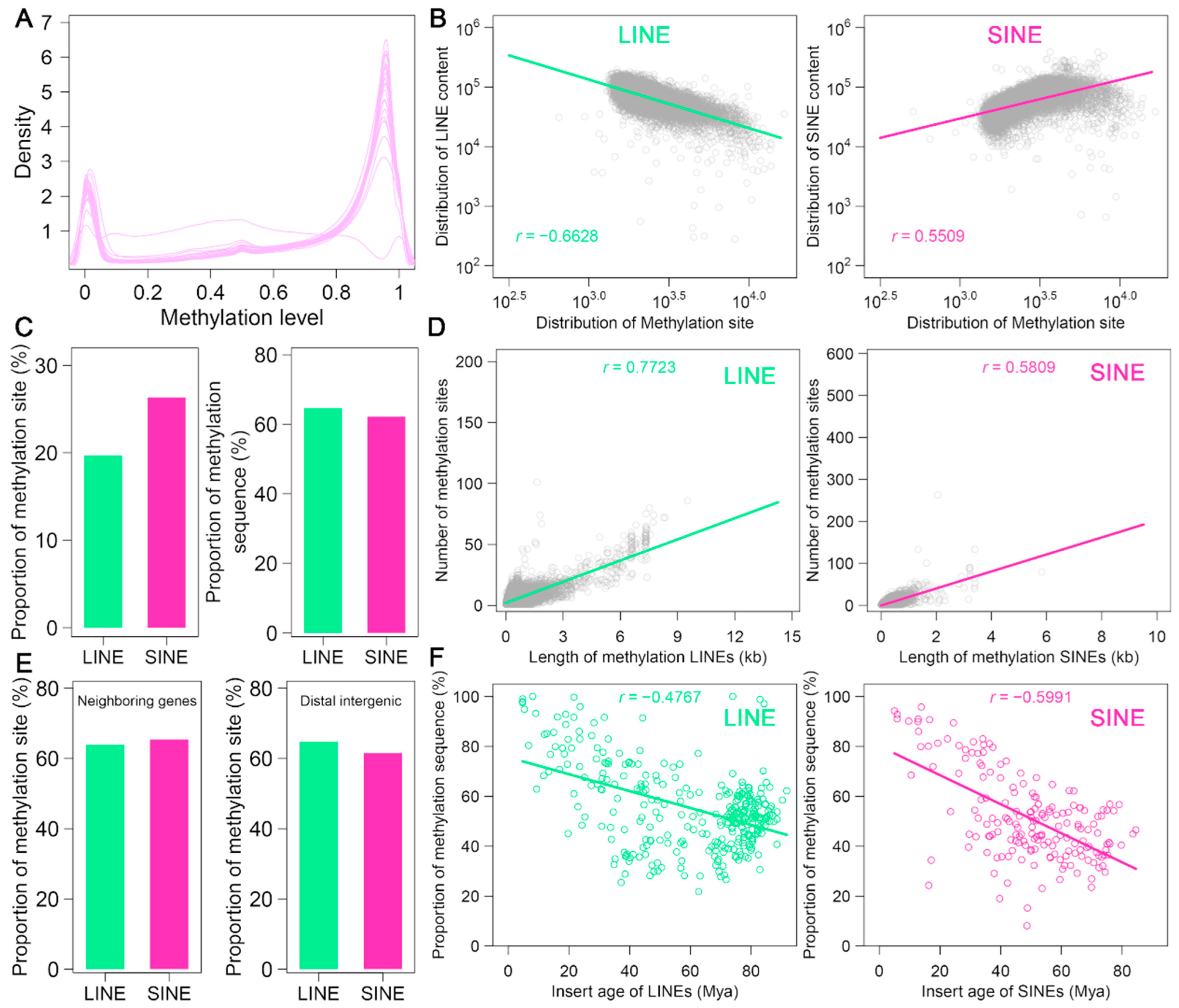
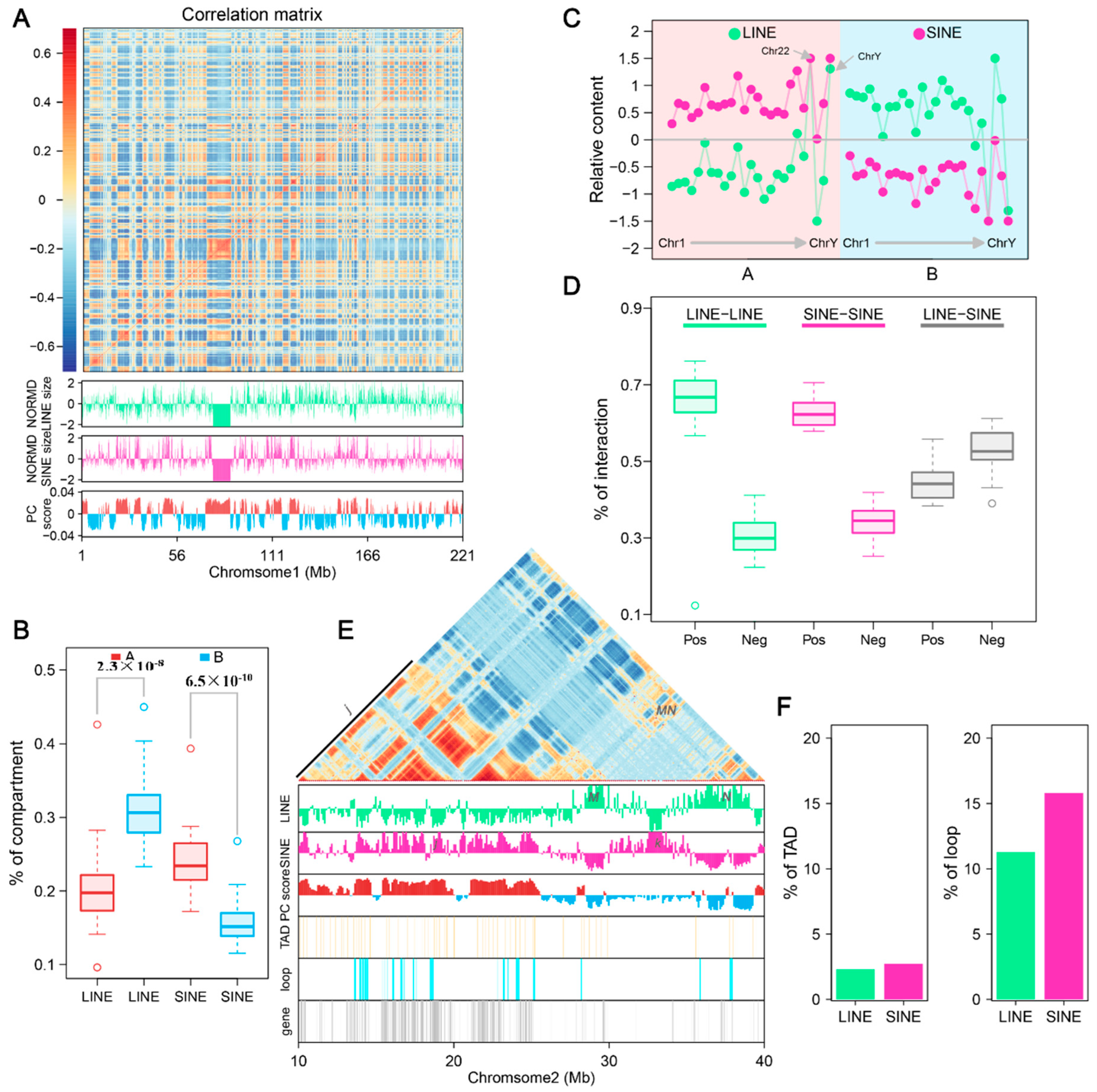

| TE Types | Family | Total Length (Mb) | Percent of the Genome (%) | Copy Number | Number of Cons | De Novo Predicted Cons | Homologous Predicted Cons |
|---|---|---|---|---|---|---|---|
| LINE | LINE1 | 709.342 | 26.768 | 157,890 | 320 | 162 | 158 |
| LINE2 | 3.251 | 0.123 | 20,612 | 38 | 2 | 36 | |
| I-Jockey | 2.232 | 0.084 | 4476 | 4 | 3 | 1 | |
| RTE-BovB | 0.934 | 0.035 | 1333 | 4 | 1 | 3 | |
| CR1 | 0.457 | 0.017 | 2569 | 23 | 0 | 23 | |
| RTE-X | 0.165 | 0.006 | 888 | 3 | 0 | 3 | |
| Dong-R4 | 0.025 | 0.001 | 110 | 1 | 0 | 1 | |
| L1-Tx1 | 0.005 | 0.0002 | 24 | 1 | 0 | 1 | |
| Unkown | 8.376 | 0.316 | 47,737 | 51 | 31 | 20 | |
| SINE | tRNA | 522.783 | 19.727 | 231,903 | 169 | 167 | 2 |
| B2 | 10.527 | 0.397 | 76,040 | 3 | 3 | 0 | |
| ID | 4.176 | 0.158 | 36,318 | 11 | 10 | 1 | |
| MIR | 4.018 | 0.152 | 33,881 | 19 | 4 | 15 | |
| 5S | 0.062 | 0.002 | 1388 | 2 | 2 | 0 | |
| Alu | 0.0438 | 0.002 | 233 | 2 | 1 | 1 | |
| 5S-Deu-L2 | 0.029 | 0.001 | 225 | 1 | 0 | 1 | |
| tRNA-RTE | 0.019 | 0.000749 | 174 | 1 | 0 | 1 | |
| tRNA-Deu | 0.007 | 0.000252 | 46 | 1 | 0 | 1 | |
| B4 | 0.0002 | 0.0001 | 3 | 1 | 0 | 1 | |
| Unkown | 0.332 | 0.013 | 5179 | 10 | 4 | 6 |
| ID | Chr | Start | End | Family | Adjacent DEG | Annotation | r2 |
|---|---|---|---|---|---|---|---|
| LINE | |||||||
| LINE_1120057 | chr4 | 143488317 | 143488819 | L1 | AA_009405.1 | FAM26E | 0.70 |
| LINE_247084 | chr11 | 56863933 | 56865234 | L1 | AA_020233.1 | - | 0.61 |
| LINE_247085 | chr11 | 56867207 | 56867409 | L1 | AA_020233.1 | - | 0.58 |
| LINE_247086 | chr11 | 56867597 | 56868351 | L1 | AA_020233.1 | - | 0.64 |
| LINE_247472 | chr11 | 57402587 | 57403490 | L1 | AA_020278.1 | - | 0.78 |
| LINE_421289 | chr14 | 29764936 | 29765085 | L2 | AA_024301.1 | - | −0.04 |
| LINE_486246 | chr15 | 48668411 | 48668813 | L1 | AA_026619.1 | DSG4 | 0.81 |
| LINE_535022 | chr16 | 35077514 | 35078004 | L2 | AA_027161.1 | - | −0.20 |
| LINE_552051 | chr16 | 66158943 | 66159959 | L1 | AA_028322.1 | NLRP10 | 0.77 |
| SINE | |||||||
| SINE_1151806 | chr2 | 62824431 | 62824620 | tRNA | AA_003621.1 | CDH3 | 0.20 |
| SINE_1727795 | chr4 | 143485902 | 143486092 | tRNA | AA_009405.1 | FAM26E | 0.47 |
| SINE_1727797 | chr4 | 143487233 | 143487444 | tRNA | AA_009405.1 | FAM26E | 0.63 |
| SINE_1866950 | chr5 | 144747966 | 144748260 | tRNA | AA_010439.1 | - | −0.51 |
| SINE_1916990 | chr6 | 53175805 | 53176119 | tRNA | AA_010921.1 | - | 0.91 |
| SINE_1933630 | chr6 | 71455240 | 71455642 | tRNA | AA_011248.1 | - | 0.96 |
| SINE_1997950 | chr7 | 3354262 | 3354512 | tRNA | AA_012513.1 | - | 0.29 |
| SINE_1997951 | chr7 | 3354611 | 3354803 | tRNA | AA_012513.1 | - | 0.62 |
| SINE_2047969 | chr7 | 54084191 | 54084453 | tRNA | AA_013182.1 | GPRC5D | 0.91 |
| SINE_2047970 | chr7 | 54085862 | 54086112 | tRNA | AA_013182.1 | GPRC5D | 0.91 |
| SINE_289188 | chr10 | 126357358 | 126357515 | tRNA | AA_018661.1 | ENC1 | 0.92 |
| SINE_355210 | chr11 | 56868368 | 56868560 | tRNA | AA_020233.1 | - | 0.44 |
| SINE_726222 | chr15 | 26599048 | 26599240 | tRNA | AA_026313.1 | AZGP1 | 0.91 |
| SINE_793552 | chr16 | 9695099 | 9695276 | MIR | AA_027002.1 | THRSP | 0.99 |
| SINE_812613 | chr16 | 35078151 | 35078359 | tRNA | AA_027161.1 | - | −0.15 |
| SINE_812614 | chr16 | 35078597 | 35079172 | tRNA | AA_027161.1 | - | −0.02 |
| SINE_812672 | chr16 | 35133514 | 35133879 | tRNA | AA_027161.1 | - | −0.29 |
| SINE_833011 | chr16 | 54486666 | 54486802 | tRNA | AA_028166.1 | LYVE1 | −0.54 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, M.; Zhou, J.; Chen, N.; Xu, J.; Wang, H.; Jiang, L.; Yang, F. Identification and Characterization of LINE and SINE Retrotransposons in the African Hedgehog (Atelerix albiventris, Erinaceidae) and Their Association with 3D Genome Organization and Gene Expression. Genes 2025, 16, 397. https://doi.org/10.3390/genes16040397
Zhu M, Zhou J, Chen N, Xu J, Wang H, Jiang L, Yang F. Identification and Characterization of LINE and SINE Retrotransposons in the African Hedgehog (Atelerix albiventris, Erinaceidae) and Their Association with 3D Genome Organization and Gene Expression. Genes. 2025; 16(4):397. https://doi.org/10.3390/genes16040397
Chicago/Turabian StyleZhu, Mengyuan, Jianxuan Zhou, Nannan Chen, Jianing Xu, Haipeng Wang, Libo Jiang, and Fengtang Yang. 2025. "Identification and Characterization of LINE and SINE Retrotransposons in the African Hedgehog (Atelerix albiventris, Erinaceidae) and Their Association with 3D Genome Organization and Gene Expression" Genes 16, no. 4: 397. https://doi.org/10.3390/genes16040397
APA StyleZhu, M., Zhou, J., Chen, N., Xu, J., Wang, H., Jiang, L., & Yang, F. (2025). Identification and Characterization of LINE and SINE Retrotransposons in the African Hedgehog (Atelerix albiventris, Erinaceidae) and Their Association with 3D Genome Organization and Gene Expression. Genes, 16(4), 397. https://doi.org/10.3390/genes16040397






