Transcriptome Analysis Reveals Key Genes Involved in Fatty Acid and Triacylglycerol Accumulation in Developing Sunflower Seeds
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Lipid Extraction and Targeted Lipidomic Analysis
2.3. RNA Extraction, cDNA Library Construction, and Transcriptome Sequencing
2.4. Sequence Alignment and Functional Annotation
2.5. Differentially Expressed Genes (DEGs) Analysis
2.6. Co-Expression Network Construction
2.7. Quantitative Reverse Transcriptase Polymerase Chain Reaction (RT-qPCR) Analysis
2.8. Statistical Analysis
3. Results
3.1. Fatty Acid Composition in Mature Sunflower Seeds
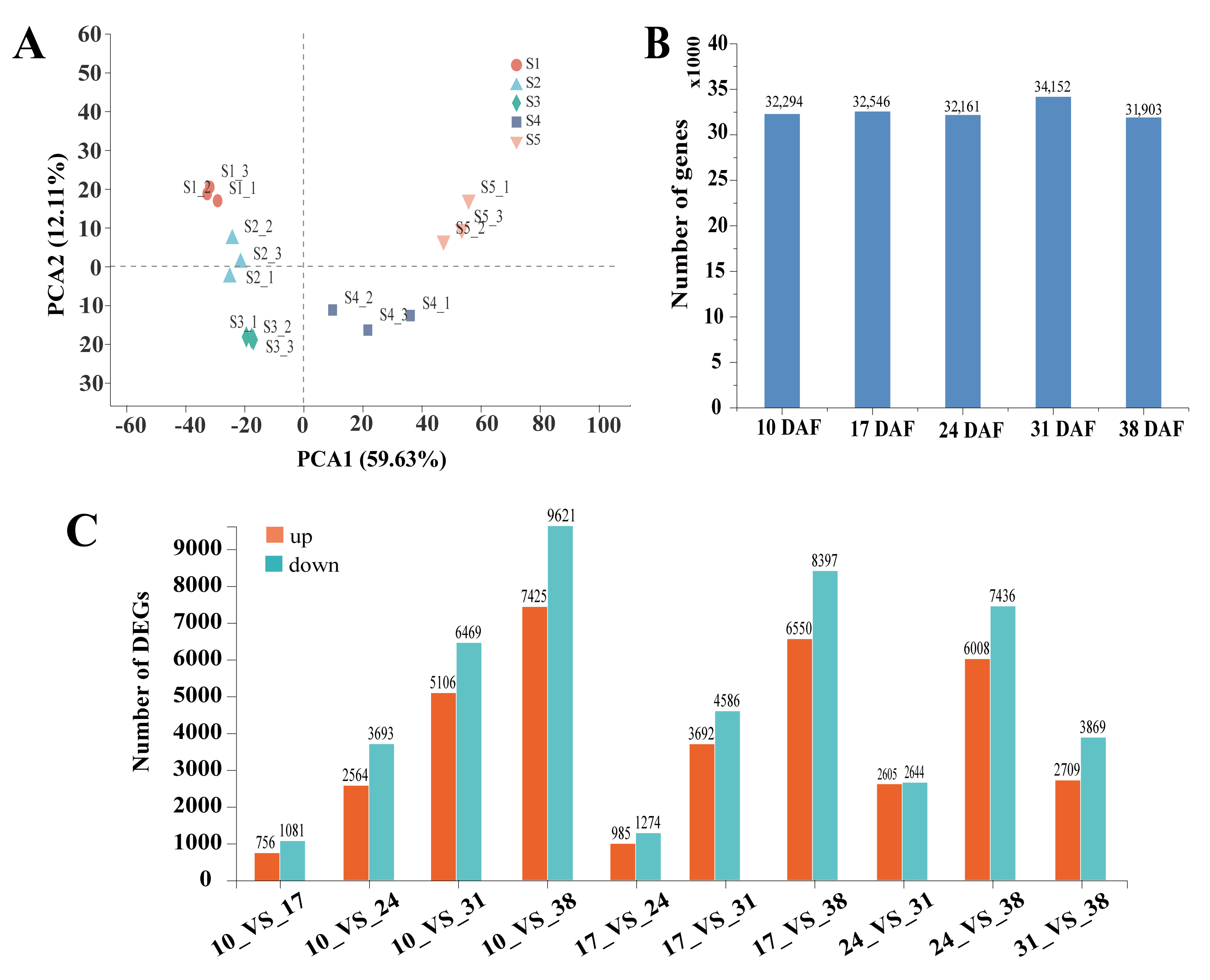
3.2. Illumina Sequencing and De Novo Assembly
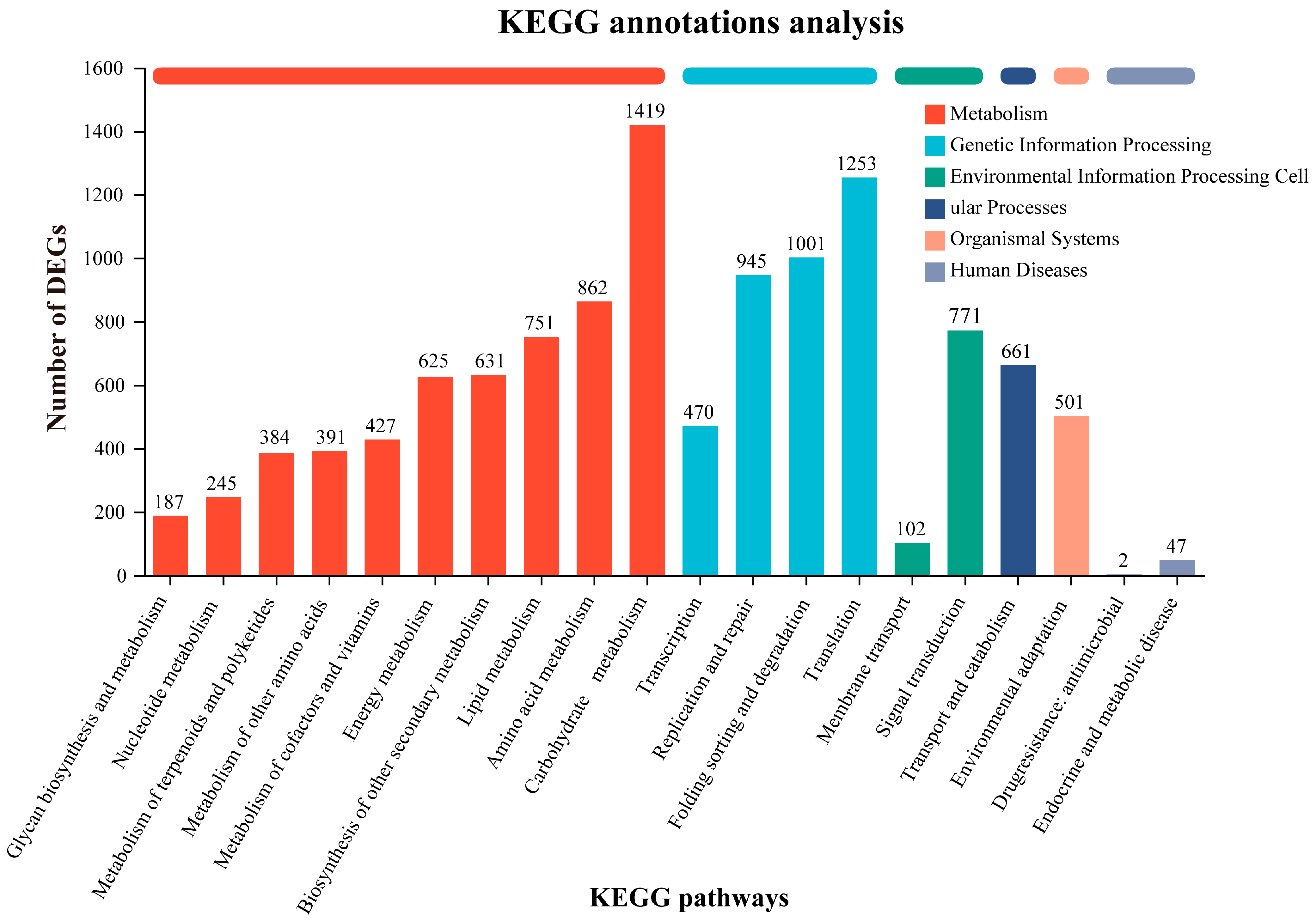
3.3. Gene Functional Annotation
3.4. Analysis of Differentially Expressed Genes in Developing Sunflower Seeds
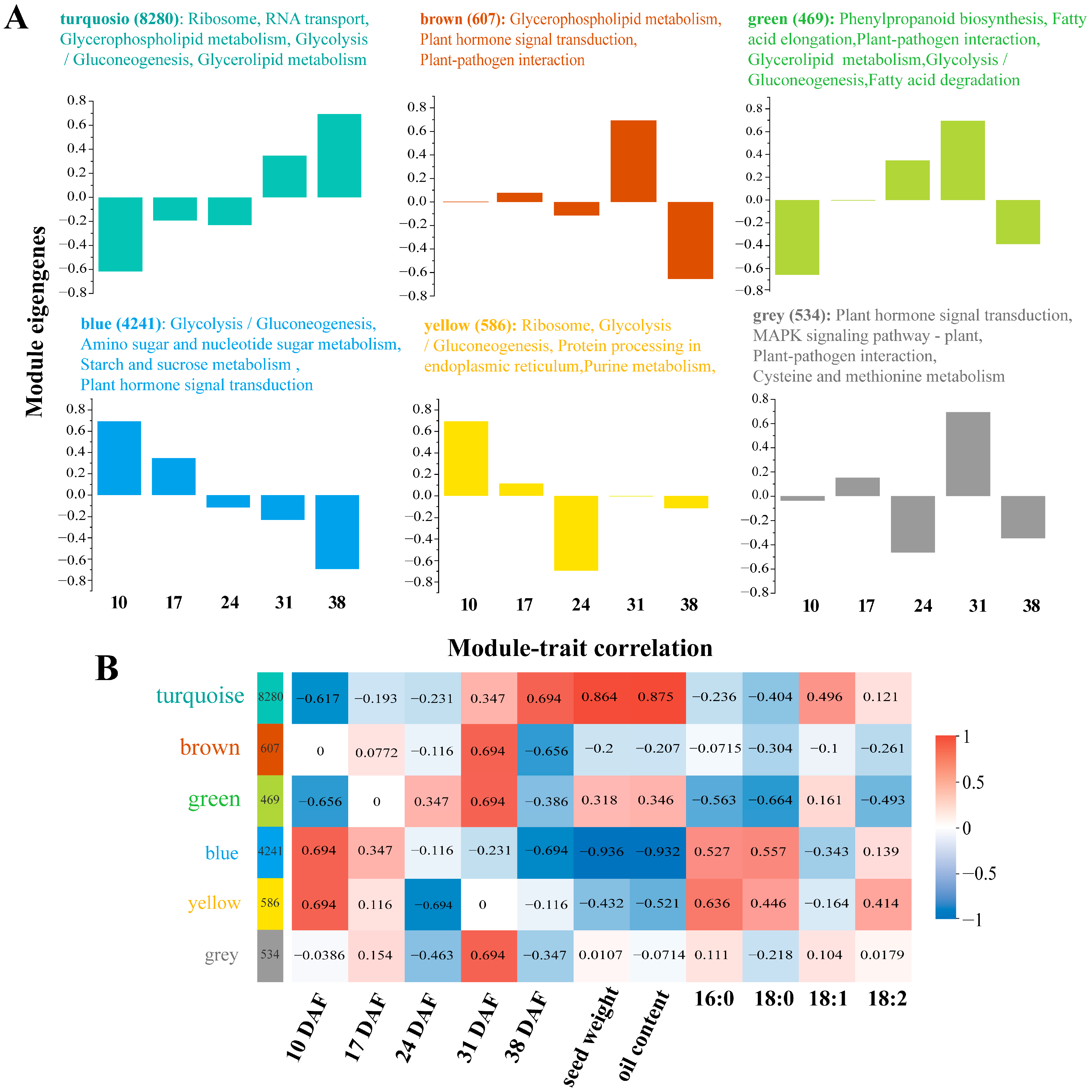
3.5. Weighted Gene Co-Expression Network Analysis
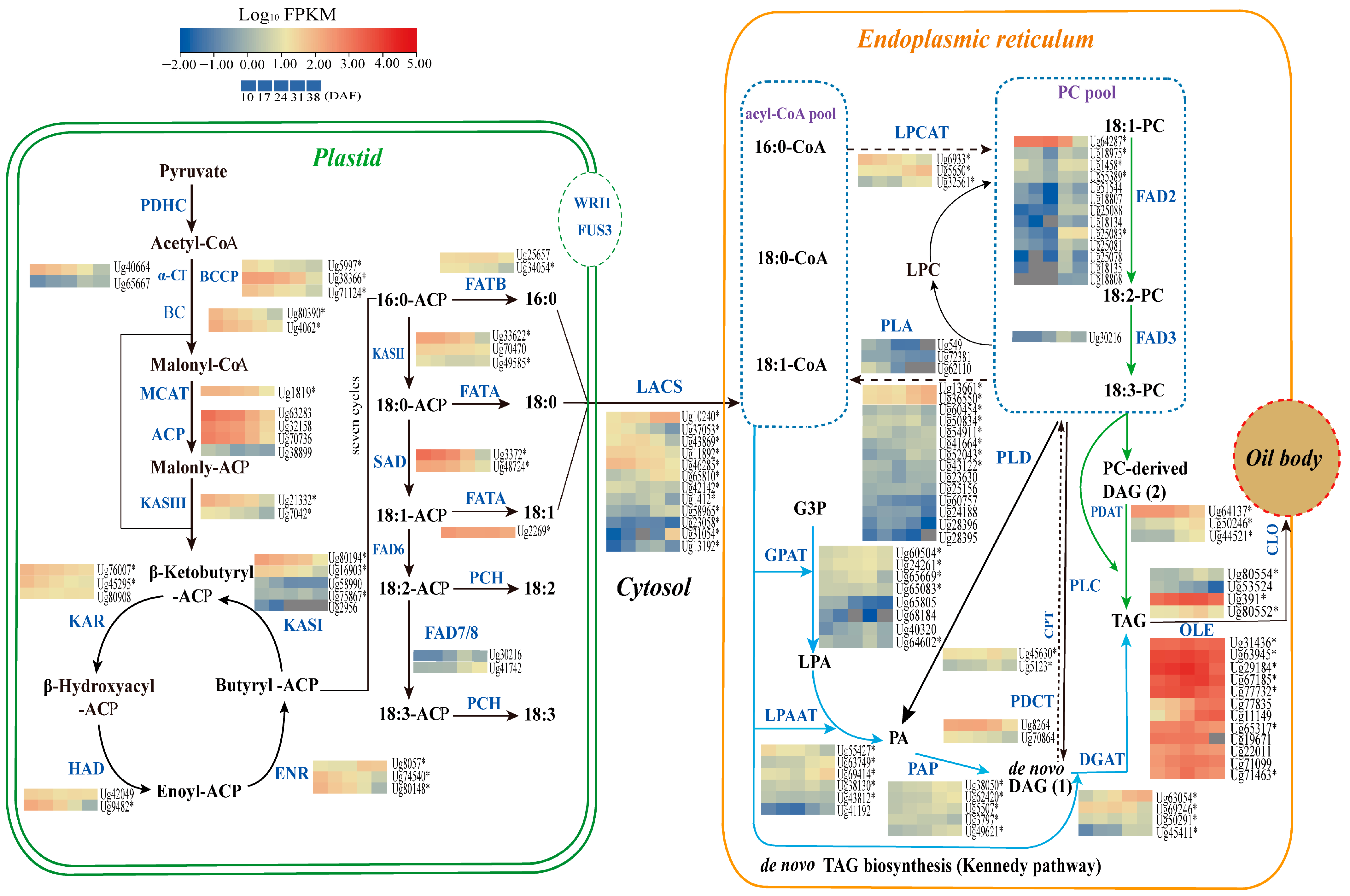
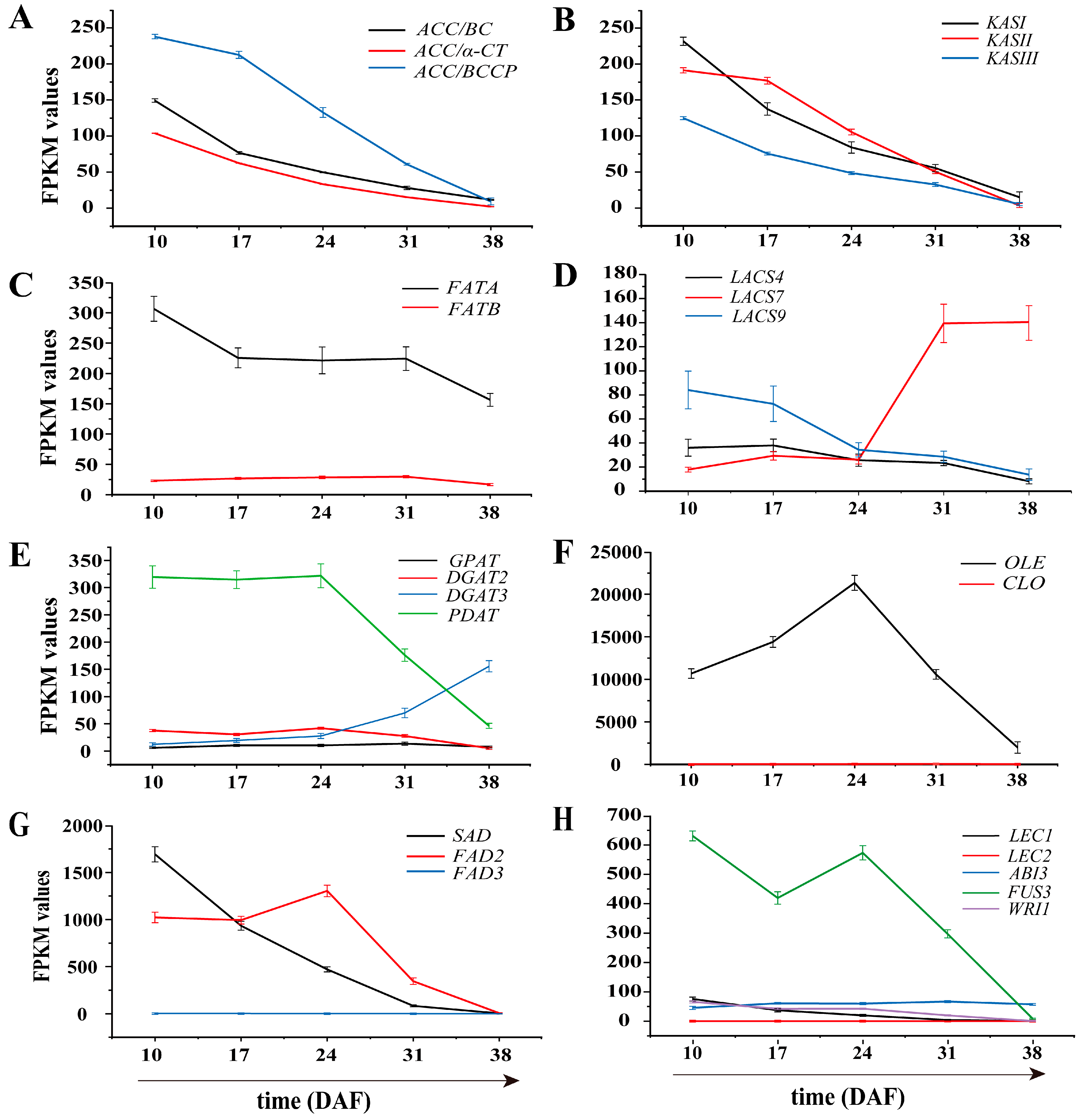
3.6. Identification of Genes Involved in Lipid Biosynthesis
3.6.1. Genes Involved in Fatty Acid Biosynthesis in Plastids
3.6.2. Genes Related to TAG and Oil Bodied in the Endoplasmic Reticulum
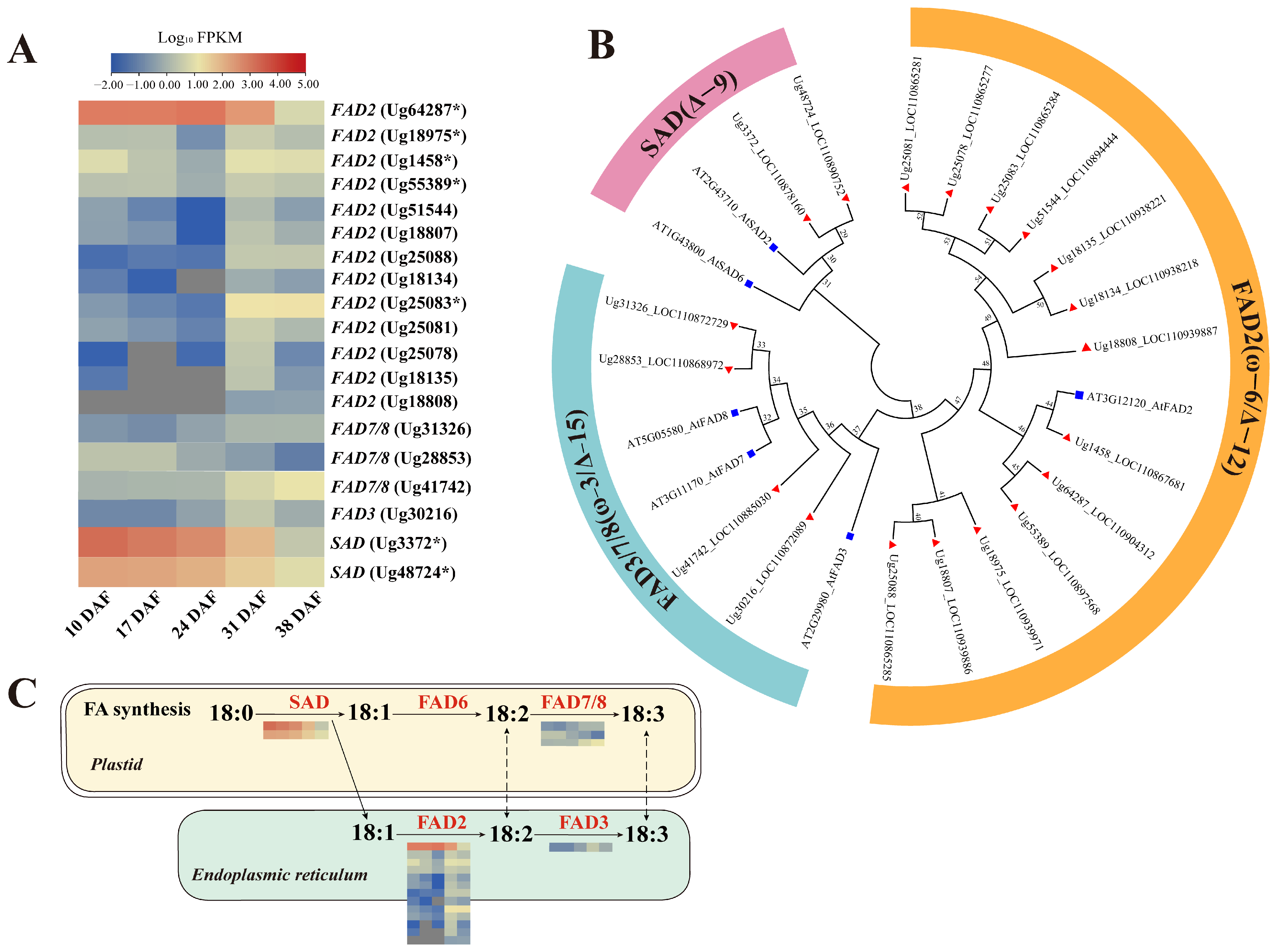
3.6.3. Genes Involved in Unsaturated Fatty Acid Synthesis
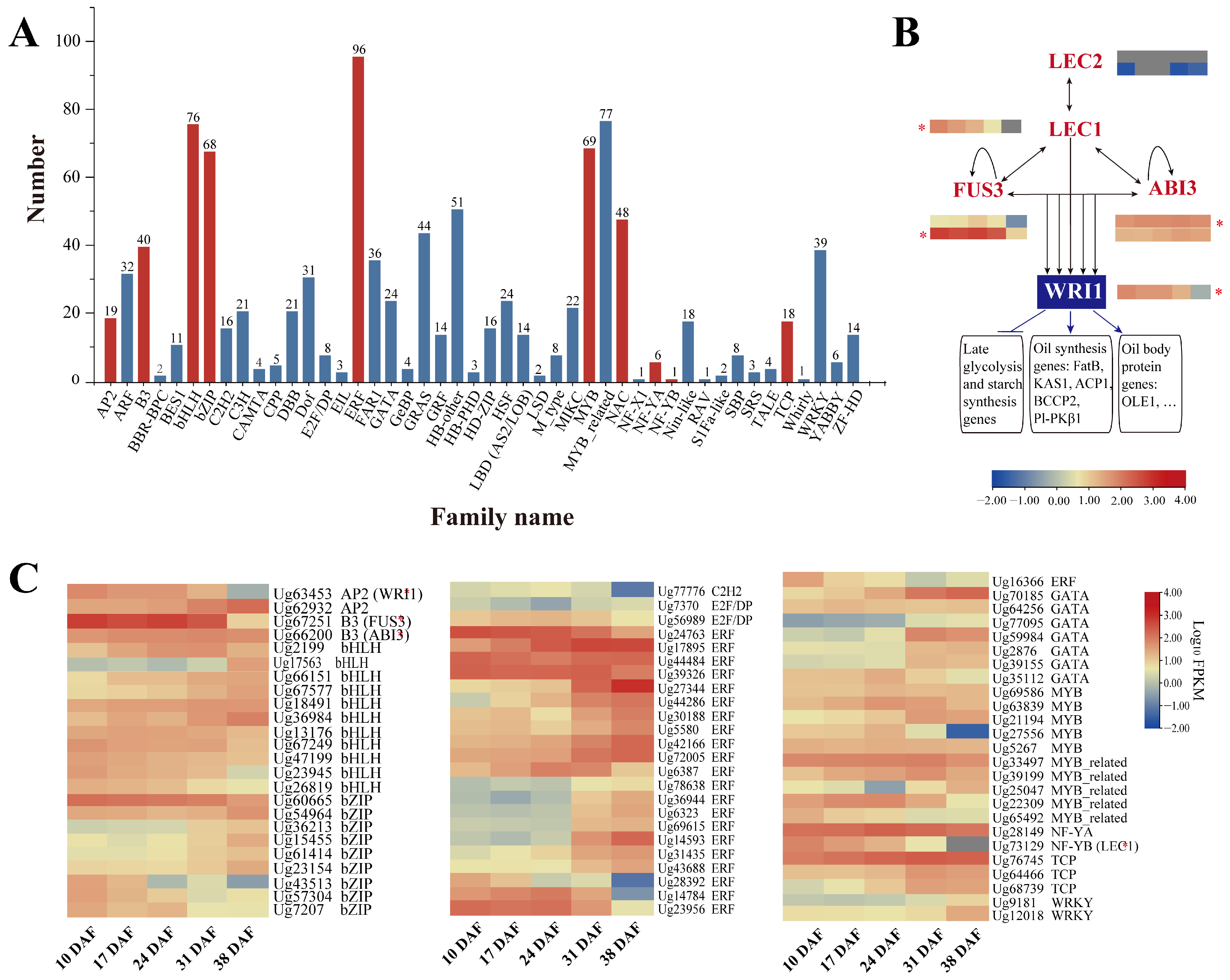
3.7. Transcription Factors Related to Lipid Synthesis
3.8. qPCR Analysis of DEGs Involved in Lipid Metabolism
4. Discussion
4.1. Role of Key De Novo Biosynthetic Enzymes in the Regulation of Fatty Acid Composition in Sunflower Seeds
4.2. Role of FAD and SAD Enzymes in Regulating Sunflower Seed Fatty Acid Desaturation and Composition
4.3. Key Enzymes and Transcription Factors Regulating Lipid Accumulation in Sunflower Seed
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Khan, S.; Choudhary, S.; Pandey, A.; Khan, M.K.; Thomas, G. Sunflower oil: Efficient oil source for human consumption. Emerg. Life Sci. Res. 2015, 1, 1–3. [Google Scholar]
- Seiler, G.J.; Qi, L.L.; Marek, L.F. Utilization of sunflower crop wild relatives for cultivated sunflower improvement. Crop Sci. 2017, 57, 1083–1101. [Google Scholar] [CrossRef]
- Akkaya, M.R. Prediction of fatty acid composition of sunflower seeds by near-infrared reflectance spectroscopy. J. Food Sci. Technol. 2018, 55, 2318–2325. [Google Scholar] [CrossRef]
- Flagella, Z.; Rotunno, T.; Tarantino, E.; Di Caterina, R.; De Caro, A. Changes in seed yield and oil fatty acid composition of high oleic sunflower (Helianthus annuus L.) hybrids in relation to the sowing date and the water regime. Eur. J. Agron. 2002, 17, 221–230. [Google Scholar] [CrossRef]
- Czumaj, A.; Sledzinski, T. Biological role of unsaturated fatty acid desaturases in health and disease. Nutrients 2020, 12, 356. [Google Scholar] [CrossRef]
- Petermann, A.B.; Reyna-Jeldes, M.; Ortega, L.; Coddou, C.; Yevenes, G.E. Roles of the unsaturated fatty acid docosahexaenoic acid in the central nervous system: Molecular and cellular insights. Int. J. Mol. Sci. 2022, 23, 5390. [Google Scholar] [CrossRef]
- Bates, P.D.; Stymne, S.; Ohlrogge, J. Biochemical pathways in seed oil synthesis. Curr. Opin. Plant Biol. 2013, 16, 358–364. [Google Scholar] [CrossRef]
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; Debono, A.; Durrett, T.P.; et al. Acyl-lipid metabolism. Arab. Book 2013, 11, e0161. [Google Scholar] [CrossRef]
- Xu, C.; Shanklin, J. Triacylglycerol metabolism, function, and accumulation in plant vegetative tissues. Annu. Rev. Plant Biol. 2016, 67, 179–206. [Google Scholar] [CrossRef]
- Dehesh, K.; Tai, H.; Edwards, P.; Byrne, J.; Jaworski, J.G. Overexpression of 3-ketoacyl-acyl-carrier protein synthase IIIs in plants reduces the rate of lipid synthesis. Plant Physiol. 2001, 125, 1103–1114. [Google Scholar] [CrossRef]
- Aznar-Moreno, J.A.; Venegas-Calerón, M.; Martínez-Force, E.; Garcés, R.; Salas, J.J. Acyl carrier proteins from sunflower (Helianthus annuus L.) seeds and their influence on FatA and FatB acyl-ACP thioesterase activities. Planta 2016, 244, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Xu, C.; Li-Beisson, Y.; Philippar, K. Fatty acid and lipid transport in plant cells. Trends Plant Sci. 2016, 21, 145–158. [Google Scholar] [PubMed]
- Kroon, J.T.; Wei, W.; Simon, W.J.; Slabas, A.R. Identification and functional expression of a type 2 acyl-CoA:diacylglycerol acyltransferase (DGAT2) in developing castor bean seeds which has high homology to the major triglyceride biosynthetic enzyme of fungi and animals. Phytochem. 2006, 67, 2541–2549. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, E.P. Biosynthesis of complex lipids. Fed. Proc. 1961, 20, 934. [Google Scholar]
- Ohlrogge, J.; Browse, J. Lipid Biosynthesis. Plant Cell 1995, 7, 957–970. [Google Scholar]
- Dahlqvist, A.; Ståhl, U.; Lenman, M.; Banas, A.; Lee, M.; Sandager, L.; Ronne, H.; Stymne, S. Phospholipid:diacylglycerol acyltransferase: An enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. USA 2000, 97, 6487–6492. [Google Scholar]
- Umate, P. Comparative genomics of the lipid-body-membrane proteins oleosin, caleosin and steroleosin in magnoliophyte, lycophyte and bryophyte. Genom. Proteom. Bioinform. 2012, 10, 345–353. [Google Scholar] [CrossRef]
- Dar, A.A.; Choudhury, A.R.; Kancharla, P.K.; Arumugam, N. The FAD2 gene in plants: Occurrence, regulation, and role. Front. Plant Sci. 2017, 8, 1789. [Google Scholar]
- Shah, F.H.; Cha, T.S. A mesocarp-and species-specific cDNA clone from oil palm encodes for sesquiterpene synthase. Plant Sci. 2000, 154, 153–160. [Google Scholar]
- Wang, H.; Guo, J.; Lambert, K.N.; Lin, Y. Developmental control of Arabidopsis seed oil biosynthesis. Planta 2007, 226, 773–783. [Google Scholar] [CrossRef]
- Focks, N.; Benning, C. wrinkled1: A novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 1998, 118, 91–101. [Google Scholar] [PubMed]
- Baud, S.; Mendoza, M.S.; To, A.; Harscot, E.; Lepiniec, L.; Dubreucq, B. WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 2007, 50, 825–838. [Google Scholar] [PubMed]
- Marchive, C.; Nikovics, K.; To, A.; Lepiniec, L.; Baud, S. Transcriptional regulation of fatty acid production in higher plants: Molecular bases and biotechnological outcomes. Eur. J. Lipid Sci. Technol. 2014, 116, 1332–1343. [Google Scholar] [CrossRef]
- Ridenour, J.B.; Smith, J.E.; Bluhm, B.H. The HAP complex governs fumonisin biosynthesis and maize kernel pathogenesis in fusarium verticillioides. J. Food Protect. 2016, 79, 1498–1507. [Google Scholar]
- To, A.; Valon, C.; Savino, G.; Guilleminot, J.; Devic, M.; Giraudat, J.; Parcy, F. A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 2006, 18, 1642–1651. [Google Scholar]
- Mu, J.; Tan, H.; Zheng, Q.; Fu, F.; Liang, Y.; Zhang, J.; Yang, X.; Wang, T.; Chong, K.; Wang, X.J.; et al. LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 2008, 148, 1042–1054. [Google Scholar]
- Zhang, M.; Cao, X.; Jia, Q.; Ohlrogge, J. FUSCA3 activates triacylglycerol accumulation in Arabidopsis seedlings and tobacco BY2 cells. Plant J. 2016, 88, 95–107. [Google Scholar]
- Lim, A.R.Q.; Kong, Q.; Singh, S.K.; Guo, L.; Yuan, L.; Ma, W. Sunflower WRINKLED1 plays a key role in transcriptional regulation of oil biosynthesis. Int. J. Mol. Sci. 2022, 23, 3054. [Google Scholar] [CrossRef]
- Badouin, H.; Gouzy, J.; Grassa, C.J.; Murat, F.; Staton, S.E.; Cottret, L.; Lelandais-Brière, C.; Owens, G.L.; Carrère, S.; Mayjonade, B.; et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 2017, 546, 148–152. [Google Scholar]
- Premnath, A.; Narayana, M.; Ramakrishnan, C.; Kuppusamy, S.; Chockalingam, V. Mapping quantitative trait loci controlling oil content, oleic acid and linoleic acid content in sunflower (Helianthus annuus L.). Mol. Breed. 2016, 36, 106. [Google Scholar] [CrossRef]
- Chernova, A.I.; Gubaev, R.F.; Singh, A.; Sherbina, K.; Goryunova, S.V.; Martynova, E.U.; Goryunov, D.V.; Boldyrev, S.V.; Vanyushkina, A.A.; Anikanov, N.A.; et al. Genotyping and lipid profiling of 601 cultivated sunflower lines reveals novel genetic determinants of oil fatty acid content. BMC Genom. 2021, 22, 505. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Xiang, L.; Zhang, L.; Maimaiti, Y.; Luo, W.; Lei, Z. Transcriptome sequencing reveals key genes for sunflower unsaturated fatty acid synthesis. Agronomy 2023, 13, 3. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Guo, S.; Zuo, Y.; Zhang, Y.; Wu, C.; Su, W.; Jin, W.; Yu, H.; An, Y.; Li, Q. Large-scale transcriptome comparison of sunflower genes responsive to Verticillium dahliae. BMC Genom. 2017, 18, 42. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Liu, Q.; Wu, M.; Zhang, B.; Shrestha, P.; Petrie, J.R.; Green, A.G.; Singh, S.P. Genetic enhancement of palmitic acid accumulation in cotton seed oil through RNAi down-regulation of ghKAS2 encoding β-ketoacyl-ACP synthase II (KASII). Plant Biotechnol. 2017, 15, 132–143. [Google Scholar] [CrossRef]
- Baud, S.; Lepiniec, L. Regulation of de novo fatty acid synthesis in maturing oilseeds of Arabidopsis. Plant Physiol. Biochem. 2009, 47, 448–455. [Google Scholar] [CrossRef]
- Xu, H.; Li, D.; Hao, Y.; Guo, X.; Lu, J.; Zhang, T. Genome-wide analysis of DGAT gene family in Perilla frutescens and functional characterization of PfDGAT2-2 and PfDGAT3-1 in Arabidopsis. Plant Sci. 2022, 324, 111426. [Google Scholar] [CrossRef] [PubMed]
- Turchetto-Zolet, A.C.; Christoff, A.P.; Kulcheski, F.R.; Loss-Morais, G.; Margis, R.; Margis-Pinheiro, M. Diversity and evolution of plant diacylglycerol acyltransferase (DGATs) unveiled by phylogenetic, gene structure and expression analyses. Genet. Mol. Biol. 2016, 39, 524–538. [Google Scholar] [PubMed]
- Hernández, M.L.; Whitehead, L.; He, Z.; Gazda, V.; Gilday, A.; Kozhevnikova, E.; Vaistij, F.E.; Larson, T.R.; Graham, I.A. A cytosolic acyltransferase contributes to triacylglycerol synthesis in sucrose-rescued Arabidopsis seed oil catabolism mutants. Plant Physiol. 2012, 160, 215–225. [Google Scholar] [PubMed]
- Beaudoin, F.; Lacey, D.J.; Napier, J.A. The biogenesis of the plant seed oil body: Oleosin protein is synthesised by ER-bound ribosomes. Plant Physiol. Biochem. 1999, 37, 481–490. [Google Scholar] [CrossRef]
- Frandsen, G.I.; Mundy, J.; Tzen, J.T.C. Oil bodies and their associated proteins, oleosin and caleosin. Physiol. Plant 2001, 112, 301–307. [Google Scholar]
- Naested, H.; Frandsen, G.I.; Jauh, G.Y.; Hernandez-Pinzon, I.; Nielsen, H.B.; Murphy, D.J.; Rogers, J.C.; Mundy, D.J. Caleosins: Ca2+-binding proteins associated with lipid bodies. Plant Mol. Biol. 2000, 44, 463–476. [Google Scholar]
- Fofana, B.; Cloutier, S.; Duguid, S.; Ching, J.; Rampitsch, C. Gene expression of stearoyl-ACP desaturase and delta12 fatty acid desaturase 2 is modulated during seed development of flax (Linum usitatissimum). Lipids 2006, 41, 705–712. [Google Scholar]
- Pérez-vich, B.; Fernández-martínez, J.M.; Grondona, M.; Knapp, S.J.; Berry, S.T. Stearoyl-ACP and oleoyl-PC desaturase genes cosegregate with quantitative trait loci underlying high stearic and high oleic acid mutant phenotypes in sunflower. Theor. Appl. Genet. 2002, 104, 338–349. [Google Scholar]
- Stone, S.L.; Kwong, L.W.; Yee, K.M.; Pelletier, J.; Lepiniec, L.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA 2001, 98, 11806–11811. [Google Scholar] [CrossRef]
- Nikolau, B.J.; Ohlrogge, J.B.; Wurtele, E.S. Plant biotin-containing carboxylases. Arch. Biochem. Biophys. 2003, 414, 211–222. [Google Scholar]
- Wei, Q.; Li, J.; Zhang, L.; Wu, P.; Chen, Y.; Li, M.; Jiang, H.; Wu, G. Cloning and characterization of a β-ketoacyl-acyl carrier protein synthase II from Jatropha curcas. J. Plant Physiol. 2012, 169, 816–824. [Google Scholar] [PubMed]
- Yu, N.; Xiao, W.F.; Zhu, J.; Chen, X.Y.; Peng, C.C. The Jatropha Curcas KASIII gene alters fatty acid composition of seeds in Arabidopsis thaliana. Biol. Plant 2015, 59, 773–782. [Google Scholar]
- Wang, Q.; Huang, W.; Jiang, Q.; Lian, J.; Liu, Z. Lower levels of expression of FATA2 gene promote longer siliques with modified seed oil content in Arabidopsis thaliana. Plant Mol. Biol. Rep. 2013, 31, 1368–1375. [Google Scholar]
- Moreno-Pérez, A.J.; Venegas-Calerón, M.; Vaistij, F.E.; Salas, J.J.; Larson, T.R.; Garcés, R.; Graham, I.A.; Martínez-Force, E. Reduced expression of FatA thioesterases in Arabidopsis affects the oil content and fatty acid composition of the seeds. Planta 2012, 235, 629–639. [Google Scholar]
- Iba, K.; Gibson, S.; Nishiuchi, T.; Fuse, T.; Nishimura, M.; Arondel, V.; Hugly, S.; Somerville, C. A gene encoding a chloroplast omega-3 fatty acid desaturase complements alterations in fatty acid desaturation and chloroplast copy number of the fad7 mutant of Arabidopsis thaliana. J. Biol. Chem. 1993, 268, 24099–24105. [Google Scholar]
- Shah, S.; Xin, Z.G.; Browse, J. Overexpression of the FAD3 desaturase gene in a mutant of Arabidopsis. Plant Physiol. 1997, 114, 1533–1539. [Google Scholar]
- Abdullah, H.M.; Akbari, P.; Paulose, B.; Schnell, D.; Qi, W.; Park, Y.; Pareek, A.; Dhankher, O.P. Transcriptome profiling of Camelina sativa to identify genes involved in triacylglycerol biosynthesis and accumulation in the developing seeds. Biotechnol. Biofuels 2016, 9, 136. [Google Scholar]
- Yang, S.; Miao, L.; He, J.; Zhang, K.; Li, Y.; Gai, J. Dynamic transcriptome changes related to oil accumulation in developing soybean seeds. Int. J. Mol. Sci. 2019, 20, 2202. [Google Scholar] [CrossRef]
- Li, S.S.; Wang, L.S.; Shu, Q.Y.; Wu, J.; Chen, L.G.; Shao, S.; Yin, D.D. Fatty acid composition of developing tree peony (Paeonia section Moutan DC.) seeds and transcriptome analysis during seed development. BMC Genom. 2015, 16, 208. [Google Scholar]
- Fei, X.; Ma, Y.; Hu, H.; Wei, A. Transcriptome analysis and GC-MS profiling of key genes in fatty acid synthesis of Zanthoxylum bungeanum seeds. Ind. Crops Prod. 2020, 156, 112870. [Google Scholar]
- Xie, D.; Dai, Z.; Yang, Z.; Tang, Q.; Deng, C.; Xu, Y.; Wang, J.; Chen, J.; Zhao, D.; Zhang, S.; et al. Combined genome-wide association analysis and transcriptome sequencing to identify candidate genes for flax seed fatty acid metabolism. Plant Sci. 2019, 286, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.-Z.; Wang, L.; Fu, J.-M.; Wuyun, T.-N.; Du, H.-Y.; Tan, X.-F.; Zou, F.; Li, F.-D. Transcriptome sequencing discovers genes related to fatty acid biosynthesis in the seeds of Eucommia ulmoides. Genes Genom. 2016, 38, 275–283. [Google Scholar] [CrossRef]
- Kazaz, S.; Barthole, G.; Domergue, F.; Ettaki, H.; To, A.; Vasselon, D.; De Vos, D.; Belcram, K.; Lepiniec, L.; Baud, S. Differential activation of partially redundant Δ9 stearoyl-ACP desaturase genes is critical for omega-9 monounsaturated fatty acid biosynthesis during seed development in Arabidopsis. Plant Cell 2020, 32, 3613–3637. [Google Scholar] [CrossRef]
- Salas, J.J.; Martínez-Force, E.; Garcés, R. Biochemical characterization of a high-palmitoleic acid Helianthus annuus mutant. Plant Physiol. Biochem. 2004, 42, 373–381. [Google Scholar] [CrossRef]
- Pleite, R.; Martínez-Force, E.; Garcés, R. Increase of the stearic acid content in high-oleic sunflower (Helianthus annuus) seeds. J. Agric. Food Chem. 2006, 54, 9383–9388. [Google Scholar]
- Perry, H.J.; Harwood, J.L. Changes in the lipid content of developing seeds of Brassica napus. Phytochemistry 1993, 32, 1411–1415. [Google Scholar]
- Liu, D.; Ji, H.; Yang, Z. Functional characterization of three novel genes encoding diacylglycerol acyltransferase (DGAT) from oil-rich tubers of Cyperus esculentus. Plant Cell Physiol. 2020, 61, 118–129. [Google Scholar] [CrossRef]
- Lu, C.L.; de Noyer, S.B.; Hobbs, D.H.; Kang, J.; Wen, Y.; Krachtus, D.; Hills, M.J. Expression pattern of diacylglycerol acyltransferase-1, an enzyme involved in triacylglycerol biosynthesis, in Arabidopsis thaliana. Plant Mol. Biol. 2003, 52, 31–41. [Google Scholar]
- Shockey, J.M.; Gidda, S.K.; Chapital, D.C.; Kuan, J.C.; Dhanoa, P.K.; Bland, J.M.; Rothstein, S.J.; Mullen, R.T.; Dyer, J.M. Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 2006, 18, 2294–2313. [Google Scholar] [CrossRef]
- Zhou, L.; Wu, Q.; Yang, Y.; Li, Q.; Li, R.; Ye, J. Regulation of oil biosynthesis and genetic improvement in plants: Advances and prospects. Genes 2024, 15, 1125. [Google Scholar] [CrossRef]
- Wurie, H.R.; Bucketf, L.; Zammit, V.A. Diacylglycerol acyhransferase 2 acts upstream of diacylglycerol acyhransferase 1 and utilizes nascent diglycerides and de novo synthesized fatty acids in HepG2 cells. FEBS J. 2012, 279, 3033–3047. [Google Scholar] [CrossRef] [PubMed]
- Lotan, T.; Ohto, M.; Yee, K.M.; West, M.A.; Lo, R.; Kwong, R.W.; Yamagishi, K.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 1998, 93, 1195–1205. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Caldo, K.M.P.; Pal-Nath, D.; Ozga, J.; Lemieux, M.J.; Weselake, R.J.; Chen, G. Properties and biotechnological applications of acyl-coa:diacylglycerol acyltransferase and phospholipid:diacylglycerol acyltransferase from terrestrial plants and microalgae. Lipids 2018, 53, 663–688. [Google Scholar] [CrossRef]
- Mueller, S.P.; Unger, M.; Guender, L.; Fekete, A.; Mueller, M.J. Phospholipid:diacylglycerol acyltransferase-mediated triacylglyerol synthesis augments basal thermotolerance. Plant Physiol. 2017, 175, 486–497. [Google Scholar] [CrossRef]
- Klińska-Bąchor, S.; Kędzierska, S.; Demski, K.; Banaś, A. Phospholipid:diacylglycerol acyltransferase1-overexpression stimulates lipid turnover, oil production and fitness in cold-grown plants. BMC Plant Biol. 2023, 23, 370. [Google Scholar] [CrossRef]
- Zhang, M.; Fan, J.; Taylor, D.C.; Ohlrogge, J.B. DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 2009, 21, 3885–3901. [Google Scholar] [CrossRef]
- Banaś, W.; Sanchez Garcia, A.; Banaś, A.; Stymne, S. Activities of acyl-CoA:diacylglycerol acyltransferase (DGAT) and phospholipid:diacylglycerol acyltransferase (PDAT) in microsomal preparations of developing sunflower and safflower seeds. Planta 2013, 237, 1627–1636. [Google Scholar] [CrossRef]
- Pan, X.; Siloto, R.M.; Wickramarathna, A.D.; Mietkiewska, E.; Weselake, R.J. Identification of a pair of phospholipid:diacylglycerol acyltransferases from developing flax (Linum usitatissimum L.) seed catalyzing the selective production of trilinolenin. J. Biol. Chem. 2013, 288, 24173–24188. [Google Scholar] [CrossRef]
- Zang, X.; Geng, X.; Ma, L.; Wang, N.; Pei, W.; Wu, M.; Zhang, J.; Yu, J. A genome-wide analysis of the phospholipid: Diacylglycerol acyltransferase gene family in Gossypium. BMC Genom. 2019, 20, 402. [Google Scholar] [CrossRef]
- Tan, H.; Yang, X.; Zhang, F.; Zheng, X.; Qu, C.; Mu, J.; Fu, F.; Li, J.; Guan, R.; Zhang, H.; et al. Enhanced seed oil production in canola by conditional expression of Brassica napus LEAFY COTYLEDON1 and LEC1-LIKE in developing seeds. Plant Physiol. 2011, 156, 1577–1588. [Google Scholar] [CrossRef]
- Tian, R.; Wang, F.; Zheng, Q.; Niza, V.M.A.G.E.; Downie, A.B.; Perry, S.E. Direct and indirect targets of the Arabidopsis seed transcription factor ABSCISIC ACID INSENSITIVE3. Plant J. 2020, 103, 1679–1694. [Google Scholar]
- Yang, Z.; Liu, X.; Wang, K.; Li, Z.; Jia, Q.; Zhao, C.; Zhang, M. ABI3 with or without FUS3 highly up-regulates lipid droplet proteins and activates oil accumulation. J. Exp. Bot. 2021, 73, 2077–2092. [Google Scholar]

| Fatty Acids | Formula | Percentage (%) |
|---|---|---|
| lauric acid | C12:0 | 0.01 |
| myristic acid | C14:0 | 0.07 |
| pentadecanoic acid | C15:0 | 0.02 |
| palmitic acid | C16:0 | 7.46 |
| heptadecanoic acid | C17:0 | 0.06 |
| stearic acid | C18:0 | 5.04 |
| arachidonic acid | C20:0 | 0.36 |
| eicosanoic acid | C21:0 | 0.01 |
| behenic acid | C22:0 | 0.03 |
| pantotrizoic acid | C23:0 | 0.04 |
| lignoceric acid | C24:0 | 0.29 |
| myristic acrylic acid | C14:1N5 | 0.07 |
| pentadecenoic acid | C15:1N5 | 0.06 |
| palmitoleic acid | C16:1N7 | 0.13 |
| heptadecenoic acid | C17:1N7 | 0.04 |
| trans-oleic acid | C18:1TN9 | 0.03 |
| oleic acid | C18:1N9 | 37.25 |
| linoleic acid | C18:2N6 | 48.47 |
| linolenic acid | C18:3N3 | 0.06 |
| eicosenoic acid | C20:1N9 | 0.15 |
| eicosapentaenoic acid | C20:5N3 | 0.34 |
| docosapentaenoic acid | C22:5N6 | 0.01 |
| tetracosenoic acid | C24:1N9 | 0.01 |
| docosahexaenoic acid (DHA) | C22:6N3 | 0.01 |
| Total_ SFAs | 13.39 | |
| Total_ MUFAs | 37.73 | |
| Total_ PUFAs | 48.88 |
| Enzyme | KEGG Annotation | Ug ID | Gene Name | 10 DAF | 17 DAF | 24 DAF | 31 DAF | 38 DAF | Model |
|---|---|---|---|---|---|---|---|---|---|
| ACC/α-CT | acetyl-coenzyme A carboxylase carboxyl transferase subunit α | Ug40664 | LOC110884458 | 52.17 | 38.65 | 22.77 | 19.89 | 13.02 | blue |
| Ug65667 | LOC110909894 | 103.79 | 62.47 | 33.27 | 15.24 | 2.2 | blue | ||
| ACC/BC | biotin carboxylase 2, chloroplastic-like | Ug80390 | LOC110920936 | 96.35 | 44.51 | 25.83 | 11.01 | 2.84 | blue |
| Ug4062 | LOC110882513 | 149.27 | 76.77 | 49.89 | 28.14 | 11.67 | blue | ||
| ACC/BCCP | biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic-like | Ug5997 | LOC110917264 | 25.373 | 15.25 | 7.36 | 6.19 | 3.49 | blue |
| Ug38366 | LOC110879644 | 237.86 | 212.6 | 132.74 | 60.93 | 9.06 | blue | ||
| Ug71124 | LOC110915869 | 62.46 | 38.75 | 20.41 | 11.74 | 4.26 | blue | ||
| MCAT | malonyl CoA-acyl carrier protein transacylase-like | Ug1819 | LOC110869205 | 110.83 | 106.79 | 72.17 | 53.85 | 14.11 | blue |
| HAD | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase | Ug9482 | LOC110928978 | 162.82 | 96.32 | 37.41 | 8.29 | 0.94 | blue |
| ENR/FABI | enoyl-[acyl-carrier-protein] reductase [NADH] | Ug8057 | LOC110927155 | 11.85 | 9.85 | 10.04 | 52.21 | 43.95 | turquosio |
| Ug80148 | LOC110920857 | 103.98 | 48.75 | 33.04 | 10.11 | 1.25 | blue | ||
| Ug74540 | LOC110918310 | 77.22 | 39.44 | 21.94 | 11.02 | 3.84 | blue | ||
| KAR | 3-oxoacyl-ACP reductase | Ug76007 | LOC110926295 | 94.51 | 57.81 | 44.01 | 29.67 | 22.61 | blue |
| Ug45295 | LOC110886898 | 74.61 | 41.17 | 23.86 | 15.71 | 10.65 | blue | ||
| KASI | 3-oxoacyl-[acyl-carrier-protein] synthase I | Ug80194 | LOC110925958 | 231.82 | 137.51 | 84.1 | 55.46 | 14.97 | blue |
| Ug16903 | LOC110937383 | 39.87 | 35.72 | 20.45 | 12.02 | 3.97 | blue | ||
| Ug75867 | LOC110923177 | 1.1 | 1.39 | 0.99 | 1.84 | 0.73 | brown | ||
| KASII | 3-oxoacyl-[acyl-carrier-protein] synthase II | Ug33622 | LOC110877021 | 191.49 | 177 | 105.5 | 50.4 | 3.36 | blue |
| Ug49585 | LOC110891189 | 10.87 | 7.85 | 4.83 | 4.87 | 6.25 | yellow | ||
| KASIII | 3-oxoacyl-[acyl-carrier-protein] synthase III | Ug21332 | LOC110941199 | 124.88 | 75.5 | 48.56 | 32.63 | 5.19 | blue |
| Ug7042 | LOC110913024 | 15.96 | 11.36 | 5.54 | 3.92 | 0.96 | blue | ||
| FAD2 | delta (12) fatty acid desaturase | Ug64287 | LOC110904312 | 1022.51 | 994.4 | 1304.58 | 346.14 | 5.73 | blue |
| Ug25083 | LOC110865284 | 0.26 | 0.1 | 0.05 | 18.88 | 20.01 | turquosio | ||
| Ug1458 | LOC110867681 | 7.07 | 2.2 | 0.65 | 8.93 | 7.59 | turquosio | ||
| Ug55389 | LOC110897568 | 1.96 | 2.17 | 0.75 | 3.08 | 2.2 | turquosio | ||
| Ug18975 | LOC110939971 | 1.72 | 1.85 | 0.16 | 3.43 | 1.66 | grey | ||
| SAD | stearoyl-[acyl-carrier-protein] 9-desaturase | Ug3372 | LOC110878160 | 1693.36 | 934.07 | 470.82 | 84.21 | 3.08 | blue |
| Ug48724 | LOC110890752 | 192.41 | 174.93 | 113.85 | 40.04 | 9.64 | blue | ||
| FATA | oleoyl-acyl carrier protein thioesterase | Ug2269 | LOC110871748 | 306.52 | 225.67 | 221.38 | 224.42 | 156.45 | turquosio |
| FATB | palmitoyl-acyl carrier protein thioesterase | Ug34054 | LOC110877217 | 16.91 | 17.45 | 8.78 | 7.46 | 2.19 | blue |
| LACS | long chain acyl-CoA synthetase 1 | Ug65810 | LOC110910053 | 2.13 | 3.87 | 6.94 | 24.48 | 18.81 | turquosio |
| long chain acyl-CoA synthetase 2 | Ug43869 | LOC110886089 | 16.23 | 27.99 | 30.52 | 10.7 | 1.57 | blue | |
| long chain acyl-CoA synthetase 4 | Ug11892 | LOC110930246 | 36.09 | 38.02 | 25.77 | 23.43 | 8.24 | blue | |
| Ug58965 | LOC110899500 | 0 | 0.51 | 0.39 | 3.69 | 5.33 | turquosio | ||
| long chain acyl-CoA synthetase 6, peroxisomal | Ug31054 | LOC110872567 | 0.02 | 0.07 | 0 | 0.12 | 40.11 | turquosio | |
| Ug42142 | LOC110885243 | 7.33 | 5.63 | 2.58 | 3.86 | 2.65 | yellow | ||
| long chain acyl-CoA synthetase 7, peroxisomal | Ug10240 | LOC110929342 | 17.95 | 29.45 | 26.21 | 139.35 | 140.43 | turquosio | |
| Ug1412 | LOC110895747 | 2.15 | 1.38 | 2.51 | 4.92 | 0.47 | brown | ||
| long chain acyl-CoA synthetase 9, chloroplastic- | Ug37053 | LOC110878808 | 18.53 | 10.99 | 4.26 | 1.53 | 0.28 | blue | |
| Ug46285 | LOC110889498 | 84.03 | 72.58 | 34.41 | 28.61 | 13.77 | blue | ||
| GPAT | glycerol-3-phosphate acyltransferase | Ug65669 | LOC110909897 | 3.95 | 5.02 | 8.88 | 9.99 | 1.14 | brown |
| Ug64602 | LOC110904530 | 1.67 | 0.84 | 0.84 | 2.89 | 0.69 | brown | ||
| Ug65083 | LOC110904838 | 3.92 | 3.76 | 3.24 | 4.68 | 2.62 | brown | ||
| Ug60504 | LOC110900391 | 4.92 | 6.71 | 9.99 | 12.04 | 7.39 | brown | ||
| Ug24261 | LOC110864776 | 5.86 | 10.34 | 10.34 | 13.55 | 7.49 | green | ||
| ATS1 | glycerol-3-phosphate acyltransferase, chloroplastic | Ug11854 | LOC110930224 | 1.29 | 0.96 | 0.83 | 4.61 | 4.85 | turquosio |
| Ug59423 | LOC110899775 | 6.57 | 5.32 | 4.42 | 5 | 8.2 | grey | ||
| Ug70832 | LOC110916084 | 1.38 | 2.01 | 0.63 | 1.57 | 7.94 | turquosio | ||
| LPAAT | lysophosphatidic acid acyltransferase | Ug55427 | LOC110897582 | 26.12 | 13.03 | 12.27 | 7.04 | 1.9 | blue |
| Ug38130 | LOC110879498 | 2.97 | 4.31 | 3.61 | 6.47 | 2.69 | green | ||
| Ug43812 | LOC110886059 | 2.41 | 1.74 | 1.55 | 1.92 | 1.55 | blue | ||
| Ug69414 | LOC110912534 | 10.01 | 8.46 | 4.57 | 16.5 | 33.87 | turquosio | ||
| Ug63749 | LOC110903942 | 2.68 | 3.53 | 3.67 | 10.19 | 16.89 | turquosio | ||
| PAP | phosphatidate phosphatase PAH1-like | Ug3797 | LOC110880605 | 4.51 | 4.41 | 3.01 | 3.11 | 1.43 | blue |
| Ug49621 | LOC110891213 | 1.38 | 1.87 | 2.28 | 5.22 | 7.4 | turquosio | ||
| Ug38050 | LOC110879450 | 2.91 | 4.93 | 5.27 | 12.33 | 9.89 | turquosio | ||
| Ug5507 | LOC110914598 | 2.62 | 3.99 | 5.01 | 7.52 | 4.91 | turquosio | ||
| Ug62420 | LOC110908856 | 2.38 | 3.21 | 3.5 | 9.06 | 15.91 | turquosio | ||
| PDAT | phospholipid: diacylglycerol acyltransferase 1 | Ug44521 | LOC110886458 | 1.22 | 1.45 | 1.31 | 8.07 | 15.62 | turquosio |
| Ug50246 | LOC110893689 | 3.61 | 7.15 | 6.85 | 14.4 | 24.3 | turquosio | ||
| phospholipid: diacylglycerol acyltransferase 2 | Ug64137 | LOC110904203 | 319.61 | 314.75 | 321.83 | 176.02 | 46.18 | blue | |
| LPCAT | lysophospholipid acyltransferase | Ug32561 | LOC110873391 | 1.35 | 3.28 | 1.93 | 10.18 | 12 | turquosio |
| Ug6933 | LOC110922258 | 70.16 | 43.99 | 25.82 | 14.16 | 7.09 | blue | ||
| Ug5650 | LOC110915451 | 14.74 | 16.7 | 18.71 | 40.04 | 72.81 | turquosio | ||
| DGAT | diacylglycerol O-acyltransferase 1 | Ug45411 | LOC110886978 | 0.13 | 0.46 | 0.49 | 4.05 | 3.91 | turquosio |
| Ug50291 | LOC110893727 | 5.5 | 7.51 | 4.45 | 8.21 | 4.79 | yellow | ||
| diacylglycerol O-acyltransferase 2 | Ug69246 | LOC110912448 | 37.46 | 30.43 | 41.78 | 27.58 | 5.05 | blue | |
| Ug63054 | LOC110909196 | 3.39 | 9.05 | 23.09 | 84.76 | 125.24 | turquosio | ||
| diacylglycerol O-acyltransferase 3, cytosolic | Ug21368 | LOC110941225 | 12.08 | 19.43 | 27.61 | 69.91 | 155.47 | turquosio | |
| CPI | cycloeucalenol cycloisomerase | Ug38774 | LOC110879898 | 8.71 | 7.21 | 6.3 | 6.9 | 3.62 | blue |
| CPT | choline/ethanolaminephosphotransferase | Ug45630 | LOC110889136 | 18.58 | 16.5 | 14.76 | 27.11 | 9.44 | brown |
| Ug5123 | LOC110912689 | 1.75 | 2.54 | 3.01 | 7.18 | 2.73 | turquosio | ||
| PLD | phospholipase D | Ug13661 | LOC110935648 | 22.04 | 28.03 | 19.28 | 69.34 | 119.35 | turquosio |
| Ug43122 | LOC110885693 | 1.6 | 1.4 | 0.37 | 1.3 | 1.167 | yellow | ||
| Ug36550 | LOC110878503 | 23.17 | 28.45 | 22.51 | 56.19 | 113.88 | yellow | ||
| Ug54911 | LOC110897355 | 4.35 | 2.84 | 1.22 | 2.12 | 3.1 | yellow | ||
| Ug50834 | LOC110894046 | 2.89 | 4.22 | 3.61 | 6.29 | 1.32 | brown | ||
| Ug41664 | LOC110884979 | 0.7 | 1.19 | 1.73 | 2.55 | 2.77 | turquosio | ||
| Ug52043 | LOC110894725 | 0.37 | 0.76 | 0.75 | 3.65 | 2.12 | turquosio | ||
| Ug60454 | LOC110900357 | 1.18 | 2.25 | 1.69 | 2.66 | 0.88 | brown | ||
| Caleosin (CLO) | peroxygenase | Ug391 | LOC110943549 | 1628.64 | 2634.68 | 4621.25 | 4012.81 | 1703.27 | turquosio |
| Ug80554 | LOC110922725 | 1.45 | 1.3 | 2.37 | 7.53 | 2.97 | turquosio | ||
| Ug80552 | LOC110922319 | 12.52 | 11.84 | 27.84 | 44.47 | 12.47 | green | ||
| oleosin (OLE) | oleosin | Ug67185 | LOC110911264 | 2918.2 | 4725.85 | 10,221.61 | 11,148.61 | 3956.6 | green |
| Ug65317 | LOC110906283 | 681.18 | 686.94 | 1068.37 | 480.97 | 340.3 | green | ||
| Ug63945 | LOC110904071 | 7917.12 | 7774.56 | 9851.5 | 7118.1 | 3225.92 | green | ||
| Ug29184 | LOC110869166 | 10,678.39 | 14,402.46 | 21,354.37 | 10,587.85 | 1985.85 | green | ||
| Ug31436 | LOC110871077 | 1858.28 | 1716.85 | 2878.29 | 1815.63 | 2101.36 | green | ||
| Ug71463 | LOC110919066 | 241.72 | 503.79 | 1575.6 | 610.37 | 429.39 | green | ||
| Ug77732 | LOC110921783 | 4105.23 | 5143.99 | 7891.14 | 3065.98 | 1777.43 | green |
| Enzyme | KEGG Annotation | Ug ID | Gene Name | 10 DAF | 17 DAF | 24 DAF | 31 DAF | 38 DAF |
|---|---|---|---|---|---|---|---|---|
| LEC1 | nuclear transcription factor Y subunit B-6-like (NF-YB) | Ug73129 | LOC110918598 | 78.70 | 39.24 | 20.43 | 4.24 | 0 |
| LEC2 | B3 domain-containing transcription factor LEC2-like | Ug68124 | LOC110909750 | 0.02 | 0 | 0 | 0.02 | 0.04 |
| Ug68126 | LOC110913834 | 0 | 0 | 0 | 0 | 0 | ||
| FUS3 | B3 domain-containing transcription factor FUS3-like | Ug64049 | LOC110906026 | 4.23 | 5.43 | 10.28 | 5.05 | 0.16 |
| Ug67251 | LOC110911305 | 630.63 | 419.49 | 573.11 | 297.36 | 8.39 | ||
| ABI3 | B3 domain-containing transcription factor ABI3-like | Ug66200 | LOC110910470 | 45.38 | 60.76 | 59.76 | 66.66 | 57.29 |
| Ug39718 | LOC110883938 | 19.18 | 17.90 | 24.46 | 35.48 | 30.41 | ||
| WRI1 | ethylene-responsive transcription factor WRI1 | Ug63453 | LOC110903740 | 65.87 | 41.94 | 42.60 | 19.73 | 0.58 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, W.; Zeng, L.; Yang, X.; Chen, D.; Sun, L. Transcriptome Analysis Reveals Key Genes Involved in Fatty Acid and Triacylglycerol Accumulation in Developing Sunflower Seeds. Genes 2025, 16, 393. https://doi.org/10.3390/genes16040393
Meng W, Zeng L, Yang X, Chen D, Sun L. Transcriptome Analysis Reveals Key Genes Involved in Fatty Acid and Triacylglycerol Accumulation in Developing Sunflower Seeds. Genes. 2025; 16(4):393. https://doi.org/10.3390/genes16040393
Chicago/Turabian StyleMeng, Wanqiu, Linglu Zeng, Xiuli Yang, Dawei Chen, and Li Sun. 2025. "Transcriptome Analysis Reveals Key Genes Involved in Fatty Acid and Triacylglycerol Accumulation in Developing Sunflower Seeds" Genes 16, no. 4: 393. https://doi.org/10.3390/genes16040393
APA StyleMeng, W., Zeng, L., Yang, X., Chen, D., & Sun, L. (2025). Transcriptome Analysis Reveals Key Genes Involved in Fatty Acid and Triacylglycerol Accumulation in Developing Sunflower Seeds. Genes, 16(4), 393. https://doi.org/10.3390/genes16040393




