A Super Enhancer-Derived Enhancer RNA Acts Together with CTCF/Cohesin in Trans to Regulate Erythropoiesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Mouse E14.5 Fetal Liver Cell Ex Vivo Differentiation
2.3. 5′- and 3′-Rapid Amplification of the cDNA Ends (5′-RACE and 3′-RACE)
2.4. RNA Fluorescent in Situ Hybridization (FISH)
2.5. shRNA Knock Down
2.6. Flow Cytometry Assays
2.7. CRISPR-Cas9 Knock out
2.8. Subcellular Fractionation
2.9. RT-qPCR
2.10. ChIP-qPCR
2.11. fCLiP-qPCR
2.12. RNA Pull-Down Experiment
2.13. ChIRP Sequencing
2.14. 3C-qPCR
2.15. Data Analysis
2.15.1. Analysis of Public Datasets
2.15.2. Gene Ontology Enrichment Analysis
2.15.3. ChIRP Sequencing Data Analysis
3. Results
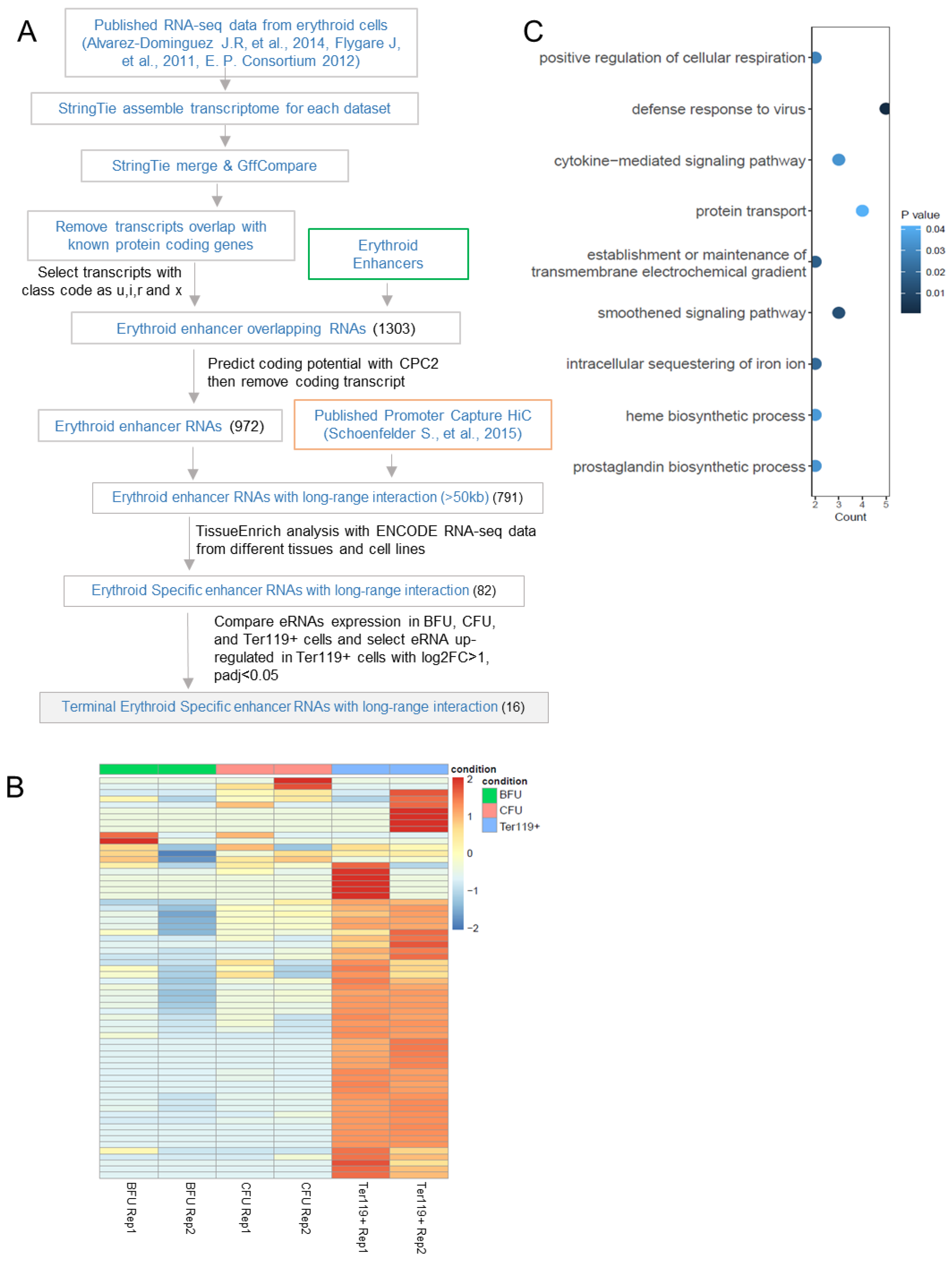
3.1. Identification of Enhancer RNAs Expressed in Erythroblasts Involved in Long-Range Interactions
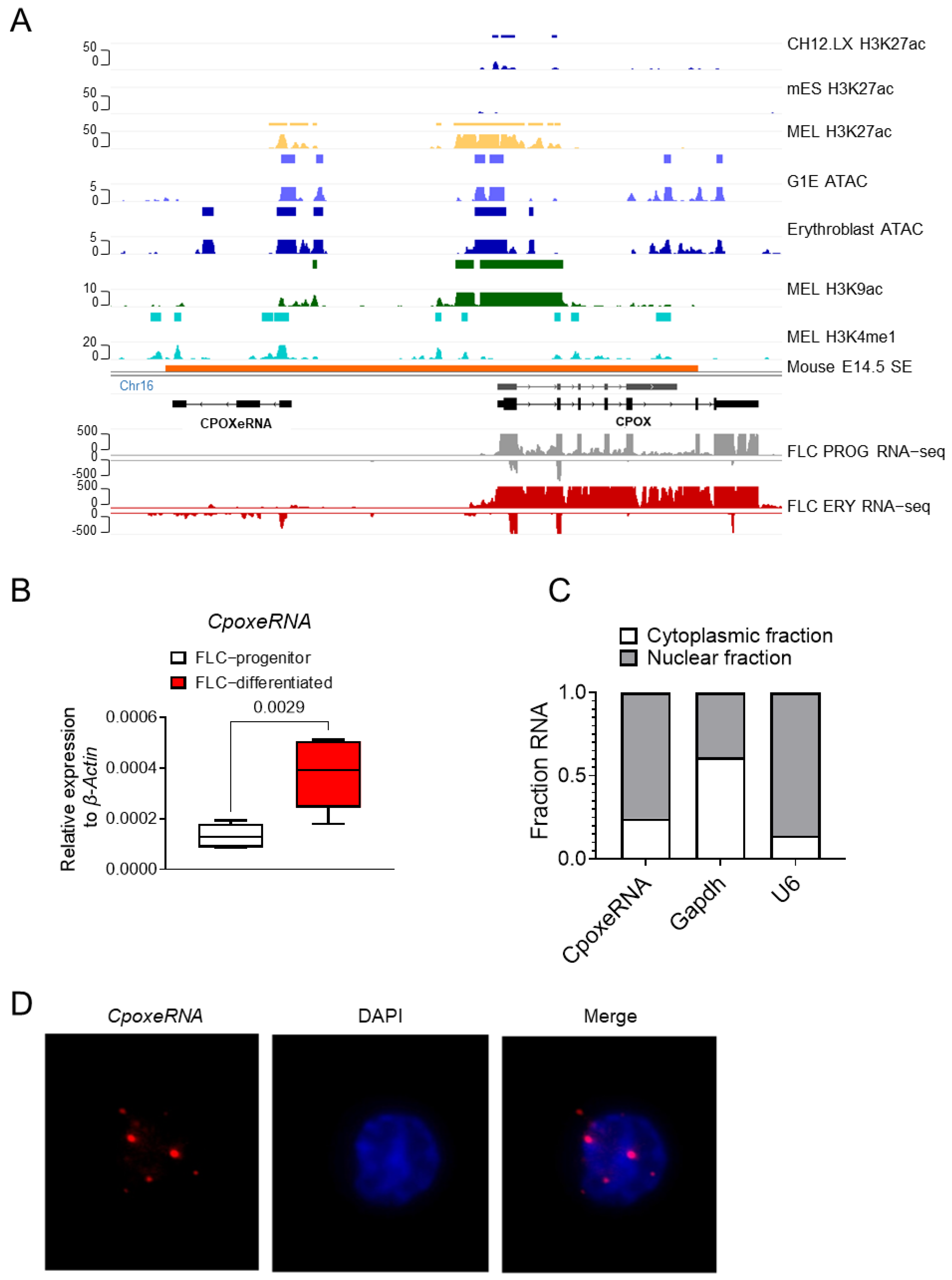
3.2. CpoxeRNA Is an Erythroid-Specific Enhancer RNA
3.3. CpoxeRNA Mediates Chromatin Interactions Between Its Genomic Locus and TAD Boundaries
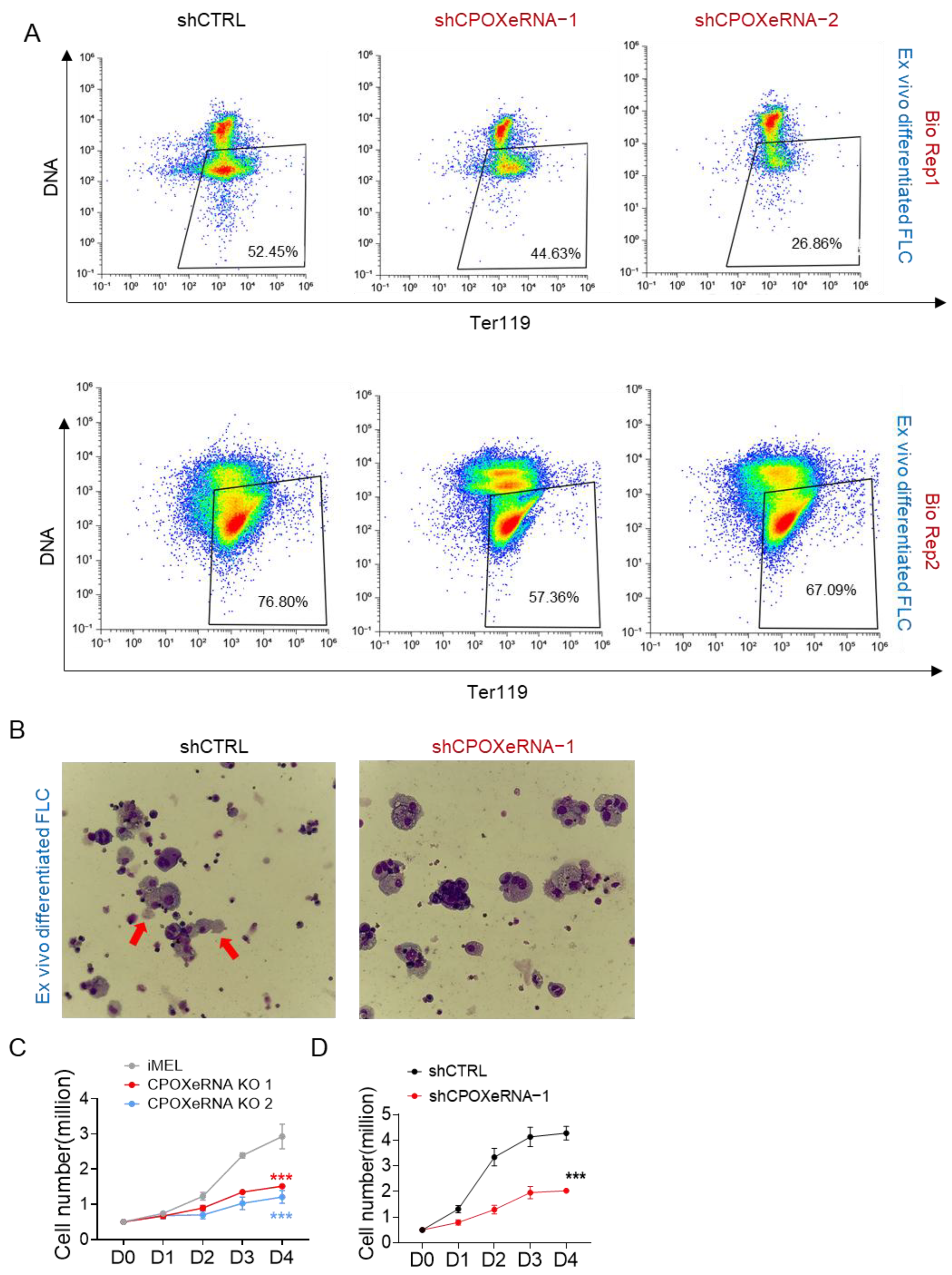
3.4. CpoxeRNA Is Critical for Survival and Proliferation During Terminal Erythropoiesis
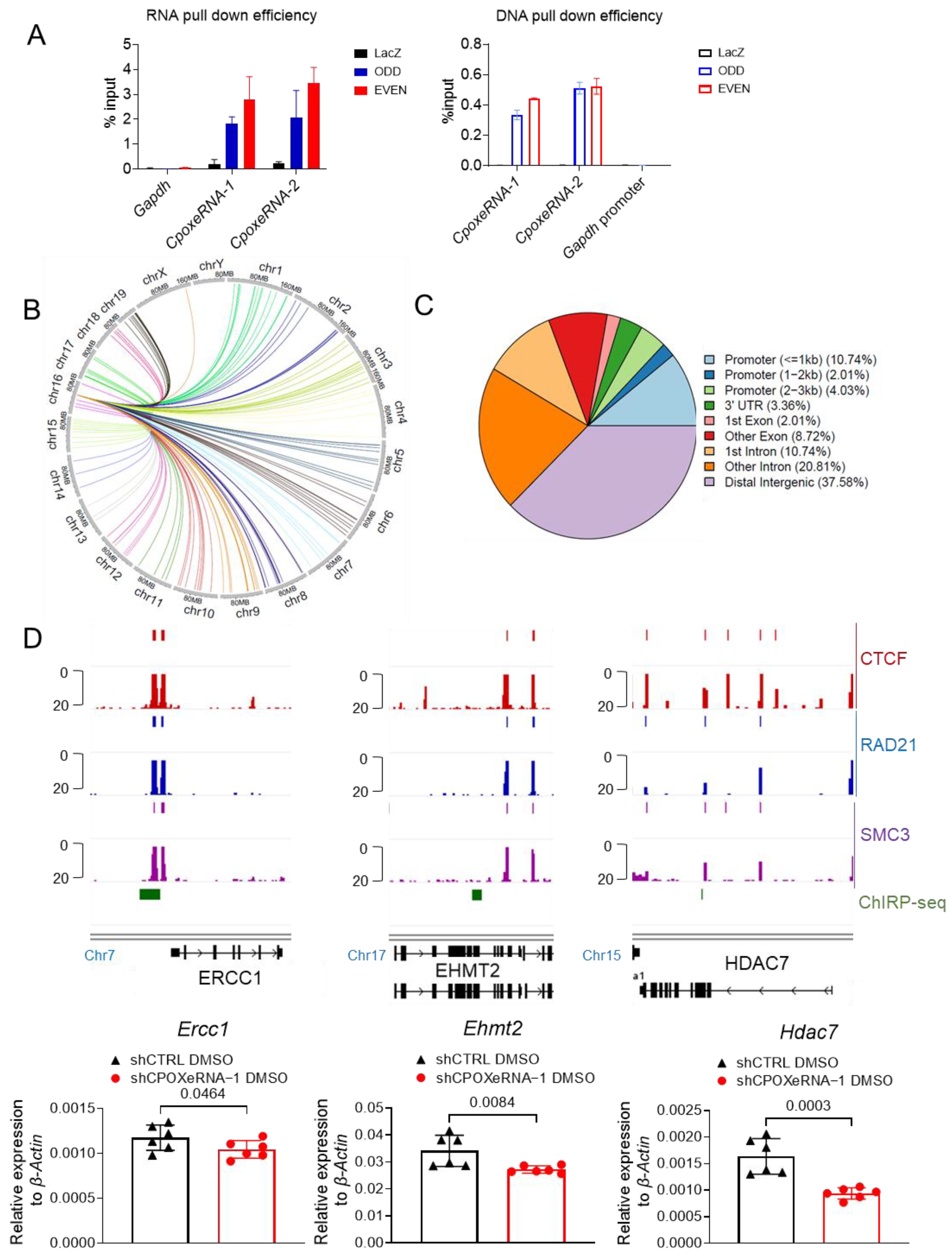
3.5. CpoxeRNA Interacts with the CTCF/Cohesin Complex to Act in Trans to Regulate Erythropoiesis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andersson, R.; Gebhard, C.; Miguel-Escalada, I.; Hoof, I.; Bornholdt, J.; Boyd, M.; Chen, Y.; Zhao, X.; Schmidl, C.; Suzuki, T.; et al. An atlas of active enhancers across human cell types and tissues. Nature 2014, 507, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Core, L.J.; Martins, A.L.; Danko, C.G.; Waters, C.T.; Siepel, A.; Lis, J.T. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat. Genet. 2014, 46, 1311–1320. [Google Scholar] [CrossRef] [PubMed]
- De Santa, F.; Barozzi, I.; Mietton, F.; Ghisletti, S.; Polletti, S.; Tusi, B.K.; Muller, H.; Ragoussis, J.; Wei, C.L.; Natoli, G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010, 8, e1000384. [Google Scholar] [CrossRef]
- Kim, T.K.; Hemberg, M.; Gray, J.M.; Costa, A.M.; Bear, D.M.; Wu, J.; Harmin, D.A.; Laptewicz, M.; Barbara-Haley, K.; Kuersten, S.; et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 2010, 465, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Pefanis, E.; Wang, J.; Rothschild, G.; Lim, J.; Kazadi, D.; Sun, J.; Federation, A.; Chao, J.; Elliott, O.; Liu, Z.P.; et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 2015, 161, 774–789. [Google Scholar] [CrossRef]
- Ling, J.; Ainol, L.; Zhang, L.; Yu, X.; Pi, W.; Tuan, D. HS2 enhancer function is blocked by a transcriptional terminator inserted between the enhancer and the promoter. J. Biol. Chem. 2004, 279, 51704–51713. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.; Elefant, F.; Liebhaber, S.A.; Cooke, N.E. Locus control region transcription plays an active role in long-range gene activation. Mol. Cell 2006, 23, 365–375. [Google Scholar] [CrossRef]
- Lam, M.T.; Cho, H.; Lesch, H.P.; Gosselin, D.; Heinz, S.; Tanaka-Oishi, Y.; Benner, C.; Kaikkonen, M.U.; Kim, A.S.; Kosaka, M.; et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature 2013, 498, 511–515. [Google Scholar] [CrossRef]
- Li, W.; Notani, D.; Ma, Q.; Tanasa, B.; Nunez, E.; Chen, A.Y.; Merkurjev, D.; Zhang, J.; Ohgi, K.; Song, X.; et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 2013, 498, 516–520. [Google Scholar] [CrossRef]
- Schaukowitch, K.; Joo, J.Y.; Liu, X.; Watts, J.K.; Martinez, C.; Kim, T.K. Enhancer RNA facilitates NELF release from immediate early genes. Mol. Cell 2014, 56, 29–42. [Google Scholar] [CrossRef]
- Mousavi, K.; Zare, H.; Dell’orso, S.; Grontved, L.; Gutierrez-Cruz, G.; Derfoul, A.; Hager, G.L.; Sartorelli, V. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol. Cell 2013, 51, 606–617. [Google Scholar] [CrossRef] [PubMed]
- Pnueli, L.; Rudnizky, S.; Yosefzon, Y.; Melamed, P. RNA transcribed from a distal enhancer is required for activating the chromatin at the promoter of the gonadotropin alpha-subunit gene. Proc. Natl. Acad. Sci. USA 2015, 112, 4369–4374. [Google Scholar] [CrossRef] [PubMed]
- Hah, N.; Murakami, S.; Nagari, A.; Danko, C.G.; Kraus, W.L. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013, 23, 1210–1223. [Google Scholar] [CrossRef]
- Chepelev, I.; Wei, G.; Wangsa, D.; Tang, Q.; Zhao, K. Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res. 2012, 22, 490–503. [Google Scholar] [CrossRef]
- Heintzman, N.D.; Hon, G.C.; Hawkins, R.D.; Kheradpour, P.; Stark, A.; Harp, L.F.; Ye, Z.; Lee, L.K.; Stuart, R.K.; Ching, C.W.; et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 2009, 459, 108–112. [Google Scholar] [CrossRef]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936. [Google Scholar] [CrossRef]
- Rada-Iglesias, A.; Bajpai, R.; Swigut, T.; Brugmann, S.A.; Flynn, R.A.; Wysocka, J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 2011, 470, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Garcia-Bassets, I.; Benner, C.; Li, W.; Su, X.; Zhou, Y.; Qiu, J.; Liu, W.; Kaikkonen, M.U.; Ohgi, K.A.; et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 2011, 474, 390–394. [Google Scholar] [CrossRef]
- Pekowska, A.; Benoukraf, T.; Zacarias-Cabeza, J.; Belhocine, M.; Koch, F.; Holota, H.; Imbert, J.; Andrau, J.C.; Ferrier, P.; Spicuglia, S. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 2011, 30, 4198–4210. [Google Scholar] [CrossRef]
- Koch, F.; Fenouil, R.; Gut, M.; Cauchy, P.; Albert, T.K.; Zacarias-Cabeza, J.; Spicuglia, S.; de la Chapelle, A.L.; Heidemann, M.; Hintermair, C.; et al. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat. Struct. Mol. Biol. 2011, 18, 956–963. [Google Scholar] [CrossRef]
- Adelman, K.; Lis, J.T. Promoter-proximal pausing of RNA polymerase II: Emerging roles in metazoans. Nat. Rev. Genet. 2012, 13, 720–731. [Google Scholar] [CrossRef] [PubMed]
- Deforzh, E.; Kharel, P.; Zhang, Y.; Karelin, A.; El Khayari, A.; Ivanov, P.; Krichevsky, A.M. HOXDeRNA activates a cancerous transcription program and super enhancers via genome-wide binding. Mol. Cell 2024, 84, 3950–3966.e3956. [Google Scholar] [CrossRef]
- Tsai, P.F.; Dell’Orso, S.; Rodriguez, J.; Vivanco, K.O.; Ko, K.D.; Jiang, K.; Juan, A.H.; Sarshad, A.A.; Vian, L.; Tran, M.; et al. A Muscle-Specific Enhancer RNA Mediates Cohesin Recruitment and Regulates Transcription In trans. Mol. Cell 2018, 71, 129–141.e128. [Google Scholar] [CrossRef]
- Alvarez-Dominguez, J.R.; Hu, W.; Yuan, B.; Shi, J.; Park, S.S.; Gromatzky, A.A.; Oudenaarden, A.v.; Lodish, H.F. Global discovery of erythroid long noncoding RNAs reveals novel regulators of red cell maturation. Blood 2014, 123, 570–581. [Google Scholar] [CrossRef]
- Yang, S.; Sun, G.; Wu, P.; Chen, C.; Kuang, Y.; Liu, L.; Zheng, Z.; He, Y.; Gu, Q.; Lu, T.; et al. WDR82-binding long noncoding RNA lncEry controls mouse erythroid differentiation and maturation. J. Exp. Med. 2022, 219, e20211688. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Diaz, L.F.; Miller, M.J.; Leadem, B.; Krivega, I.; Dean, A. An enhancer RNA recruits KMT2A to regulate transcription of Myb. Cell Rep. 2024, 43, 114378. [Google Scholar] [CrossRef]
- Ivaldi, M.; Diaz, L.; Chakalova, L.; Lee, J.; Krivega, I.; Dean, A. Fetal γ-globin genes are regulated by the BGLT3 long noncoding RNA locus. Blood 2018, 132, 1963–1973. [Google Scholar] [CrossRef]
- Liu, J.; Li, Y.; Tong, J.; Gao, J.; Guo, Q.; Zhang, L.; Wang, B.; Zhao, H.; Wang, H.; Jiang, E.; et al. Long non-coding RNA-dependent mechanism to regulate heme biosynthesis and erythrocyte development. Nat. Commun. 2018, 9, 4386. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Dominguez, J.R.; Knoll, M.; Gromatzky, A.A.; Lodish, H.F. The Super-Enhancer-Derived alncRNA-EC7/Bloodlinc Potentiates Red Blood Cell Development in trans. Cell Rep. 2017, 19, 2503–2514. [Google Scholar] [CrossRef]
- Liu, G.; Kim, J.; Nguyen, N.; Zhou, L.; Dean, A. Long noncoding RNA GATA2AS influences human erythropoiesis by transcription factor and chromatin landscape modulation. Blood 2024, 143, 2300–2313. [Google Scholar] [CrossRef]
- Pan, H.; Jin, M.; Ghadiyaram, A.; Kaur, P.; Miller, H.E.; Ta, H.M.; Liu, M.; Fan, Y.; Mahn, C.; Gorthi, A.; et al. Cohesin SA1 and SA2 are RNA binding proteins that localize to RNA containing regions on DNA. Nucleic Acids Res. 2020, 48, 5639–5655. [Google Scholar] [CrossRef] [PubMed]
- Porter, H.; Li, Y.; Neguembor, M.V.; Beltran, M.; Varsally, W.; Martin, L.; Cornejo, M.T.; Pezic, D.; Bhamra, A.; Surinova, S.; et al. Cohesin-independent STAG proteins interact with RNA and R-loops and promote complex loading. eLife 2023, 12, e79386. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Yang, T.; Shao, C.; Cao, Z.; Zhang, H. LncRNA MIAT activates vascular endothelial growth factor A through RAD21 to promote nerve injury repair in acute spinal cord injury. Mol. Cell Endocrinol. 2021, 528, 111244. [Google Scholar] [CrossRef] [PubMed]
- Arnold, P.R.; Wells, A.D.; Li, X.C. Diversity and Emerging Roles of Enhancer RNA in Regulation of Gene Expression and Cell Fate. Front. Cell Dev. Biol. 2019, 7, 377. [Google Scholar] [CrossRef]
- Saldana-Meyer, R.; Rodriguez-Hernaez, J.; Escobar, T.; Nishana, M.; Jacome-Lopez, K.; Nora, E.P.; Bruneau, B.G.; Tsirigos, A.; Furlan-Magaril, M.; Skok, J.; et al. RNA Interactions Are Essential for CTCF-Mediated Genome Organization. Mol. Cell 2019, 76, 412–422.e415. [Google Scholar] [CrossRef] [PubMed]
- Hansen, A.S.; Hsieh, T.S.; Cattoglio, C.; Pustova, I.; Saldana-Meyer, R.; Reinberg, D.; Darzacq, X.; Tjian, R. Distinct Classes of Chromatin Loops Revealed by Deletion of an RNA-Binding Region in CTCF. Mol. Cell 2019, 76, 395–411.e313. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.K.; Blanco, M.R.; Walkup, W.G.; Bonesteele, G.; Urbinati, C.R.; Banerjee, A.K.; Chow, A.; Ettlin, O.; Strehle, M.; Peyda, P.; et al. Denaturing purifications demonstrate that PRC2 and other widely reported chromatin proteins do not appear to bind directly to RNA in vivo. Mol. Cell 2024, 84, 1271–1289.e1212. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Blum, R.; Rosenberg, M.; Lee, J.T. Re-analysis of CLAP data affirms PRC2 as an RNA binding protein. bioRxiv 2024. [Google Scholar] [CrossRef]
- Scotto-Lavino, E.; Du, G.; Frohman, M.A. 5′ end cDNA amplification using classic RACE. Nat. Protoc. 2006, 1, 2555–2562. [Google Scholar] [CrossRef]
- Scotto-Lavino, E.; Du, G.; Frohman, M.A. 3′ end cDNA amplification using classic RACE. Nat. Protoc. 2006, 1, 2742–2745. [Google Scholar] [CrossRef]
- Ji, P.; Jayapal, S.R.; Lodish, H.F. Enucleation of cultured mouse fetal erythroblasts requires Rac GTPases and mDia2. Nat. Cell Biol. 2008, 10, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Hattangadi, S.M.; Burke, K.A.; Lodish, H.F. Homeodomain-interacting protein kinase 2 plays an important role in normal terminal erythroid differentiation. Blood 2010, 115, 4853–4861. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [PubMed]
- Krivega, I.; Dean, A. Chromatin Immunoprecipitation (ChIP) with Erythroid Samples. Methods Mol. Biol. 2018, 1698, 229–236. [Google Scholar] [CrossRef]
- Kim, B.; Kim, V.N. fCLIP-seq for transcriptomic footprinting of dsRNA-binding proteins: Lessons from DROSHA. Methods 2019, 152, 3–11. [Google Scholar] [CrossRef]
- Chu, C.; Quinn, J.; Chang, H.Y. Chromatin isolation by RNA purification (ChIRP). J. Vis. Exp. 2012, 61, 3912. [Google Scholar] [CrossRef]
- Hagege, H.; Klous, P.; Braem, C.; Splinter, E.; Dekker, J.; Cathala, G.; de Laat, W.; Forne, T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc. 2007, 2, 1722–1733. [Google Scholar] [CrossRef]
- Krivega, I.; Dean, A. Chromosome Conformation Capture (3C and Higher) with Erythroid Samples. Methods Mol. Biol. 2018, 1698, 237–243. [Google Scholar] [CrossRef]
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Flygare, J.; Rayon Estrada, V.; Shin, C.; Gupta, S.; Lodish, H.F. HIF1alpha synergizes with glucocorticoids to promote BFU-E progenitor self-renewal. Blood 2011, 117, 3435–3444. [Google Scholar] [CrossRef]
- Kang, Y.J.; Yang, D.C.; Kong, L.; Hou, M.; Meng, Y.Q.; Wei, L.; Gao, G. CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef] [PubMed]
- Schoenfelder, S.; Furlan-Magaril, M.; Mifsud, B.; Tavares-Cadete, F.; Sugar, R.; Javierre, B.M.; Nagano, T.; Katsman, Y.; Sakthidevi, M.; Wingett, S.W.; et al. The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Res. 2015, 25, 582–597. [Google Scholar] [CrossRef]
- Harmston, N.; Ing-Simmons, E.; Perry, M.; Baresic, A.; Lenhard, B. GenomicInteractions: An R/Bioconductor package for manipulating and investigating chromatin interaction data. BMC Genom. 2015, 16, 963. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Tuteja, G. TissueEnrich: Tissue-specific gene enrichment analysis. Bioinformatics 2019, 35, 1966–1967. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Gorkin, D.U.; Barozzi, I.; Zhao, Y.; Zhang, Y.; Huang, H.; Lee, A.Y.; Li, B.; Chiou, J.; Wildberg, A.; Ding, B.; et al. An atlas of dynamic chromatin landscapes in mouse fetal development. Nature 2020, 583, 744–751. [Google Scholar] [CrossRef]
- Gao, T.; Qian, J. EnhancerAtlas 2.0: An updated resource with enhancer annotation in 586 tissue/cell types across nine species. Nucleic Acids Res. 2020, 48, D58–D64. [Google Scholar] [CrossRef]
- Durand, N.C.; Shamim, M.S.; Machol, I.; Rao, S.S.; Huntley, M.H.; Lander, E.S.; Aiden, E.L. Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Syst. 2016, 3, 95–98. [Google Scholar] [CrossRef]
- Durand, N.C.; Robinson, J.T.; Shamim, M.S.; Machol, I.; Mesirov, J.P.; Lander, E.S.; Aiden, E.L. Juicebox Provides a Visualization System for Hi-C Contact Maps with Unlimited Zoom. Cell Syst. 2016, 3, 99–101. [Google Scholar] [CrossRef]
- Doynova, M.D.; Markworth, J.F.; Cameron-Smith, D.; Vickers, M.H.; O’Sullivan, J.M. Linkages between changes in the 3D organization of the genome and transcription during myotube differentiation in vitro. Skelet. Muscle 2017, 7, 5. [Google Scholar] [CrossRef] [PubMed]
- Dennis, G., Jr.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, R60. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Dale, R.K.; Pedersen, B.S.; Quinlan, A.R. Pybedtools: A flexible Python library for manipulating genomic datasets and annotations. Bioinformatics 2011, 27, 3423–3424. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; He, Q.Y. ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015, 31, 2382–2383. [Google Scholar] [CrossRef]
- Wang, Y.; Jia, L.; Tian, G.; Dong, Y.; Zhang, X.; Zhou, Z.; Luo, X.; Li, Y.; Yao, W. shinyCircos-V2.0: Leveraging the creation of Circos plot with enhanced usability and advanced features. Imeta 2023, 2, e109. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Ye, R.; Cao, C.; Xue, Y. Enhancer RNA: Biogenesis, function, and regulation. Essays Biochem. 2020, 64, 883–894. [Google Scholar] [CrossRef]
- Xie, B.; Dean, A. Noncoding function of super enhancer derived Cpox pre-mRNA in modulating neighbouring gene expression and chromatin interactions. RNA Biol. 2025, 22, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Conway, A.J.; Brown, F.C.; Fullinfaw, R.O.; Kile, B.T.; Jane, S.M.; Curtis, D.J. A mouse model of hereditary coproporphyria identified in an ENU mutagenesis screen. Dis. Model. Mech. 2017, 10, 1005–1013. [Google Scholar] [CrossRef] [PubMed]
- Prasher, J.M.; Lalai, A.S.; Heijmans-Antonissen, C.; Ploemacher, R.E.; Hoeijmakers, J.H.; Touw, I.P.; Niedernhofer, L.J. Reduced hematopoietic reserves in DNA interstrand crosslink repair-deficient Ercc1−/− mice. EMBO J. 2005, 24, 861–871. [Google Scholar] [CrossRef]
- Zhang, H.; Emerson, D.J.; Gilgenast, T.G.; Titus, K.R.; Lan, Y.; Huang, P.; Zhang, D.; Wang, H.; Keller, C.A.; Giardine, B.; et al. Chromatin structure dynamics during the mitosis-to-G1 phase transition. Nature 2019, 576, 158–162. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, B.; Dean, A. A Super Enhancer-Derived Enhancer RNA Acts Together with CTCF/Cohesin in Trans to Regulate Erythropoiesis. Genes 2025, 16, 389. https://doi.org/10.3390/genes16040389
Xie B, Dean A. A Super Enhancer-Derived Enhancer RNA Acts Together with CTCF/Cohesin in Trans to Regulate Erythropoiesis. Genes. 2025; 16(4):389. https://doi.org/10.3390/genes16040389
Chicago/Turabian StyleXie, Bingning, and Ann Dean. 2025. "A Super Enhancer-Derived Enhancer RNA Acts Together with CTCF/Cohesin in Trans to Regulate Erythropoiesis" Genes 16, no. 4: 389. https://doi.org/10.3390/genes16040389
APA StyleXie, B., & Dean, A. (2025). A Super Enhancer-Derived Enhancer RNA Acts Together with CTCF/Cohesin in Trans to Regulate Erythropoiesis. Genes, 16(4), 389. https://doi.org/10.3390/genes16040389





