Analysis of WRKY Gene Family in Acer fabri and Their Expression Patterns Under Cold Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. qRT-PCR Gene Expression Validation
2.3. De Novo Assembly of RNA-Seq Reads and Quantitative Analysis of Gene Expression
2.4. RNA Extraction, cDNA Library Construction, and Sequencing
2.5. Measurement of Physiological Indicators
2.6. Identification of Members of the A. fabri WRKY Gene Family
2.7. Bioinformatics Analysis of the WRKY Gene Family in A. fabri
2.8. Expression Analysis of A. fabri WRKY Genes
3. Results
3.1. General Characteristics of the A. fabri WRKY Gene Family
3.2. Analysis of the Conserved Domains of the WRKY Gene Family in A. fabri
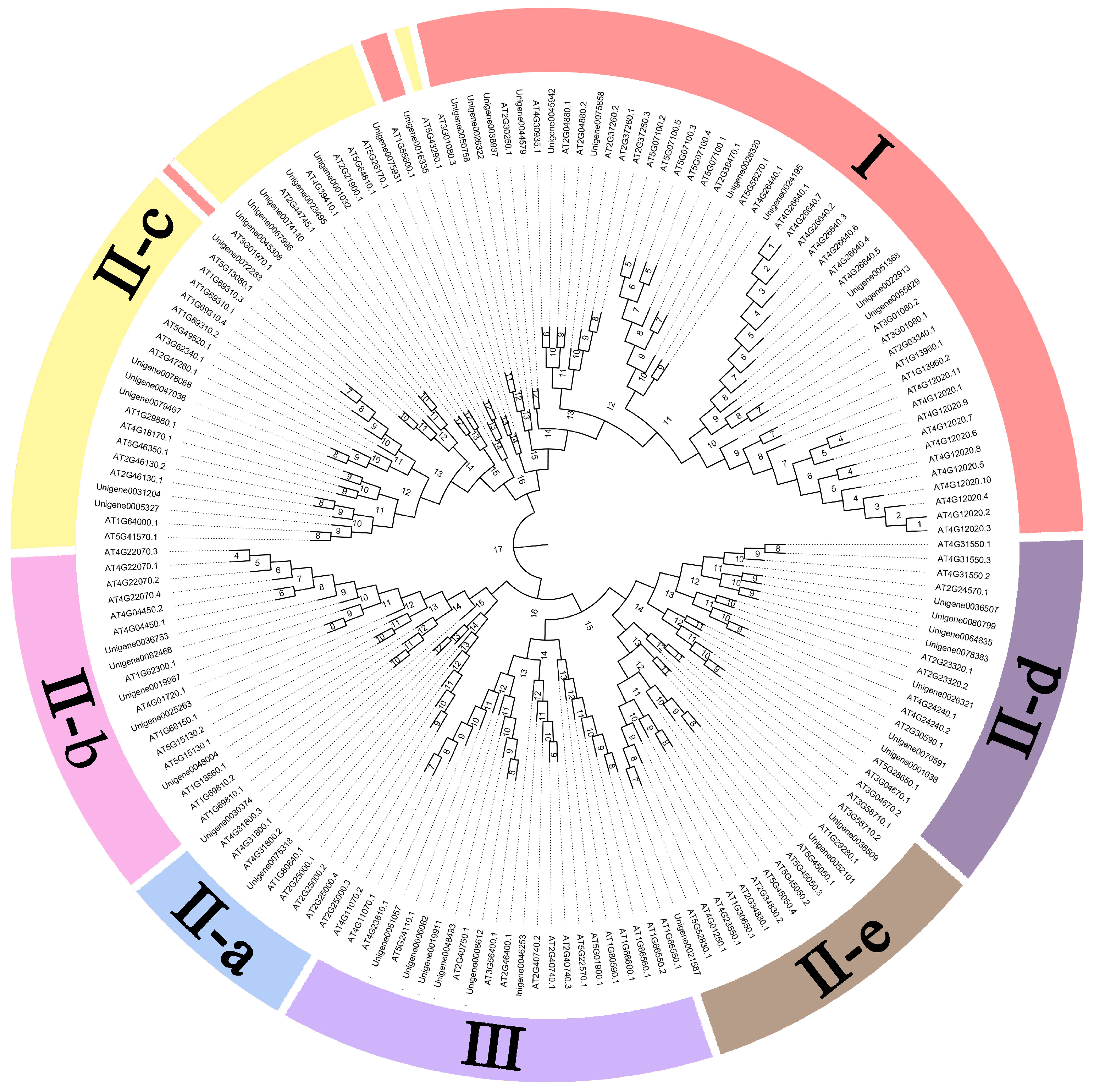
3.3. Evolutionary Analysis of the WRKY Gene Family
3.4. Expression Analysis of the WRKY Gene Family Members Under Cold Stress
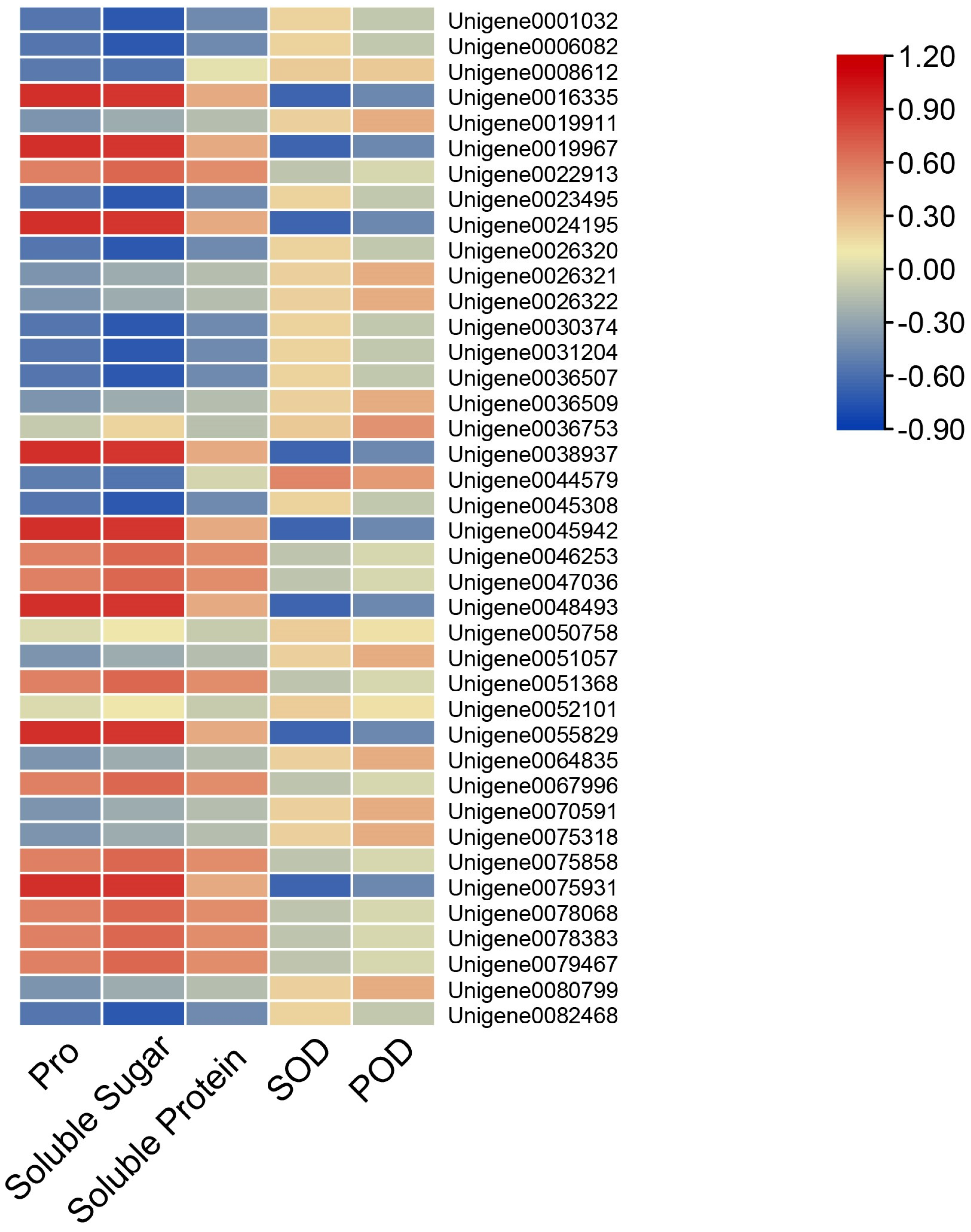
3.5. The Correlation Analysis Between AfWEKYs Gene Expression and Physiological Indicators
3.6. Real-Time Fluorescence Quantitative PCR (qRT-PCR) Verification of the WRKY Gene Family Members in A. fabri
4. Discussion
4.1. Functional Divergence of the WRKY Gene Family in A. fabri and Other Woody Plants
4.2. Functional Verification and Regulation Network of A. fabri
4.3. Evolutionary Drivers of Functional Diversification in the A. fabri WRKY Gene Family
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| SOD | superoxide dismutase |
| POD | peroxidase |
| Pro | proline |
| SS | soluble sugars |
| SP | soluble proteins |
References
- Liu, G.; Gu, H.; Cai, H.; Guo, C.; Chen, Y.; Wang, L.; Chen, G. Integrated Transcriptome and Biochemical Analysis Provides New Insights into the Leaf Color Change in Acer fabri. Forests 2023, 14, 1638. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Maiden, N.A.; Syd Ali, N.; Ahmad, K.; Atan, S.; Wong, M.Y. Growth and physiological responses of Hevea brasiliensis to Rigidoporus microporus infection. J. Rubber Res. 2022, 25, 213–221. [Google Scholar] [CrossRef]
- Wang, X.J.; Li, W.G.; Gao, X.S.; Wu, C.T.; Zhang, X.F. Physiological Characteristics of Hevea brasiliensis in Response to Low Temperature Stress and Its Regulation Mechanisms. Plant Physiol. 2012, 159, 123–145. [Google Scholar] [CrossRef]
- Tian, Y.H.; Yuan, H.F.; Xie, J.; Deng, J.W.; Dao, X.S.; Zheng, Y.L. Effect of diurnal irradiance on night-chilling tolerance of six rubber cultivars. Photosynthetica 2016, 54, 374–380. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, M.; Zhang, X.; Hao, B.; Kaushik, S.K.; Pan, Y. WRKY gene family evolution in Arabidopsis thaliana. Genetica 2011, 139, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, J.; Zhou, X.; Liu, S.; Zhuang, Y. Identification of WRKY gene family and characterization of cold stress-responsive WRKY genes in eggplant. PeerJ 2020, 8, e8777. [Google Scholar] [CrossRef]
- Yang, Y.L.; Cushman, S.A.; Wang, S.C.; Wang, F.; Li, Q.; Liu, H.L.; Li, Y. Genome-wide investigation of the WRKY transcription factor gene family in weeping forsythia: Expression profile and cold and drought stress responses. Genetica 2023, 151, 153–165. [Google Scholar] [CrossRef]
- Dai, Z.; Wei, M.; Zhang, B.; Yuan, Y.; Zhang, B. VuWRKY, a group I WRKY gene from Vaccinium uliginosum, confers tolerance to cold and salt stresses in plant. Plant Cell Tissue Organ. Cult. 2021, 147, 157–168. [Google Scholar] [CrossRef]
- Li, X.; Wang, Y.; Zhang, J.; Liu, H.; Chen, F.; Sun, W. Transcriptomic and metabolomic analysis reveals the role of MYB transcription factors in anthocyanin biosynthesis during leaf color transition of Acer palmatum. BMC Plant Biol. 2022, 22, 512–528. [Google Scholar] [CrossRef]
- Yue, Y.; Du, J.; Li, Y.; Thomas, H.R.; Frank, M.H.; Wang, L.; Hu, H. Insight into the petunia Dof transcription factor family reveals a new regulator of male-sterility. Ind. Crops Prod. 2021, 161, 113196. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, L.; Liu, Y.; Shang, X.; Fang, S. Identification and Expression Analysis of R2R3-MYB Family Genes Associated with Salt Tolerance in Cyclocarya paliurus. Int. J. Mol. Sci. 2022, 23, 3429. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Li, J.; Wu, K.; Li, L.; Ma, G.; Fang, L.; Zeng, S. Transcriptomic analysis reveals biosynthesis genes and transcription factors related to leaf anthocyanin biosynthesis in Aglaonema commutatum. BMC Genom. 2023, 24, 567. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, X.; Wang, Z.; Chen, J.; Zhang, H.; Li, Y. The WRKY Transcription Factor Family: From Evolution to Functional Diversification in Plant Stress Responses. Trends Plant Sci. 2023, 28, 123–135. [Google Scholar] [CrossRef]
- Wang, L.; Chen, G.; Liu, G.; Gu, H.; Cai, H.; Chen, Y. Evolutionary and Functional Insights into the WRKY Gene Family: A Key Player in Plant Adaptation to Environmental Stresses. Trends Genet. 2023, 39, 456–468. [Google Scholar] [CrossRef]
- Zhou, Y.; Qin, X.; Zhang, D.; Chen, F.; Yue, Y.; Wang, L. Comprehensive Analysis of the WRKY Gene Family in Plants: Roles in Development, Stress Responses, and Beyond. Int. J. Mol. Sci. 2024, 25, 5369. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, Y.; Shang, X.; Fang, S. Comparative Analysis of Protein Identification Tools in Proteomics Research. Int. J. Mol. Sci. 2023, 24, 7896. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Cell-PLoc 2.0: An Improved Package of Web-Servers for Predicting Subcellular Localization of Proteins in Various Organisms. Nat. Sci. 2010, 2, 1090–1103. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for Motif Discovery and Searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Chen, C.; Xia, R.; Chen, H.; He, Y. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Chen, Y.; Kong, X.; Yang, L.; Fu, M.; Zhang, S. Genome-Wide Identification of WRKY Family Genes and the Expression Profiles in Response to Nitrogen Deficiency in Poplar. Genes 2022, 13, 2324. [Google Scholar] [CrossRef] [PubMed]
- Aguayo, P.; Lagos, C.; Conejera, D.; Medina, D.; Fernández, M.; Valenzuela, S. Transcriptome-wide identification of WRKY family genes and their expression under cold acclimation in Eucalyptus globulus. Trees 2019, 33, 1313–1327. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, Y.; Li, J.; Wang, S.; Liu, Z.; Zhou, J.; Zhu, J.K. WRKY transcription factors in plant abiotic stress responses: Molecular mechanisms and crosstalk with phytohormone signaling. Int. J. Mol. Sci. 2022, 23, 8435. [Google Scholar] [CrossRef]
- Long, L.; Gu, L.; Wang, S.; Cai, H.; Wu, J.; Wang, J.; Yang, M. Progress in the understanding of WRKY transcription factors in woody plants. Int. J. Biol. Macromol. 2023, 242, 124379. [Google Scholar] [CrossRef]
- Wang, H.; Cheng, X.; Yin, D.; Chen, D.; Luo, C.; Liu, H.; Huang, C. Advances in the Research on Plant WRKY Transcription Factors Responsive to External Stresses. Curr. Issues Mol. Biol. 2023, 45, 2861–2880. [Google Scholar] [CrossRef]
- Zhang, R.; Liu, S.; Wang, X.; Chen, L.; Li, H.; Hu, Z.; Zhou, Y. Comparative transcriptome and metabolome analysis reveals the role of WRKY and bHLH transcription factors in autumn leaf coloration of Acer truncatum. Hortic. Res. 2023, 10, uhad045. [Google Scholar] [CrossRef]
- Li, Y.; Li, X.; Wei, J.; Cai, K.; Zhang, H.; Ge, L.; Ren, Z.; Zhao, C.; Zhao, X. Genome-Wide Identification and Analysis of the WRKY Gene Family and Cold Stress Response in Acer truncatum. Genes 2021, 12, 1867. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, T.; Liang, X.; Ye, Q.; Wang, Y.; Han, J.; Han, D. MbWRKY53, a M. baccata WRKY Transcription Factor, Contributes to Cold and Drought Stress Tolerance in Transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 2024, 25, 7626. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Zhao, Y.; Jiang, Q.; Yang, J.; Zhao, W.; Taylor, I.A.; Peng, Y.L.; Wang, D.; Liu, J. Structural basis of dimerization and dual W-box DNA recognition by rice WRKY domain. Nucleic Acids Res. 2019, 47, 4308–4318. [Google Scholar] [CrossRef]
- Guo, M.; Yang, F.; Liu, C.; Zou, J.; Qi, Z.; Fotopoulos, V.; Lu, G.; Yu, J.; Zhou, J. A single-nucleotide polymorphism in WRKY33 promoter is associated with the cold sensitivity in cultivated tomato. New Phytol. 2022, 236, 989–1005. [Google Scholar] [CrossRef]
- Wang, Y.; Dong, B.; Wang, N.; Zheng, Z.; Yang, L.; Zhong, S.; Fang, Q.; Xiao, Z.; Zhao, H. A WRKY Transcription Factor PmWRKY57 from Prunus mume Improves Cold Tolerance in Arabidopsis thaliana. Mol. Biotechnol. 2023, 65, 1359–1368. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.J.; Chen, J.X.; Zhang, J.Q.; Guo, G.Y.; Zhang, H.X.; Zhao, X.L.; Lu, S.F.; Xu, H.W.; Hou, D.Y. Gene Expression and Interaction Analysis of FsWRKY4 and FsMAPK3 in Forsythia suspensa. Plants 2023, 12, 3415. [Google Scholar] [CrossRef]
- Llorca, C.M.; Potschin, M.; Zentgraf, U. bZIPs and WRKYs: Two large transcription factor families executing two different functional strategies. Front. Plant Sci. 2014, 5, 169. [Google Scholar] [CrossRef]
- Song, Y.; Ai, C.R.; Jing, S.J.; Yu, D.Q. Research Progress on Functional Analysis of Rice WRKY Genes. Rice Sci. 2010, 17, 60–72. [Google Scholar] [CrossRef]
- Dong, X.; Yang, Y.; Zhang, Z.; Xiao, Z.; Bai, X.; Gao, J.; Hur, Y.; Hao, S.; He, F. Genome-Wide Identification of WRKY Genes and Their Response to Cold Stress in Coffea canephora. Forests 2019, 10, 335. [Google Scholar] [CrossRef]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Ayadi, M.; Hanana, M.; Kharrat, N.; Merchaoui, H.; Marzoug, R.B.; Lauvergeat, V.; Rebaï, A.; Mzid, R. The WRKY Transcription Factor Family in Citrus: Valuable and Useful Candidate Genes for Citrus Breeding. Appl. Biochem. Biotechnol. 2016, 180, 516–543. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.N.; Liu, Y.; Xin, Z.Z.; Zhang, D.Z.; Ge, B.M.; Yang, R.P.; Wang, Z.F.; Yang, L.; Tang, B.P.; Zhou, C.L. Genome-wide identification and characterization of the WRKY gene family in potato (Solanum tuberosum). Biochem. Syst. Ecol. 2017, 71, 212–218. [Google Scholar] [CrossRef]
- Filiz, E.; Kurt, F. Expression and Co-expression Analyses of WRKY, MYB, bHLH and bZIP Transcription Factor Genes in Potato (Solanum tuberosum) Under Abiotic Stress Conditions: RNA-seq Data Analysis. Potato Res. 2021, 64, 721–741. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, G.; Zhou, Y.; Zhang, D.; Chen, F.; Qin, X.; Cai, H.; Gu, H.; Yue, Y.; Wang, L.; Liu, G. Analysis of WRKY Gene Family in Acer fabri and Their Expression Patterns Under Cold Stress. Genes 2025, 16, 344. https://doi.org/10.3390/genes16030344
Chen G, Zhou Y, Zhang D, Chen F, Qin X, Cai H, Gu H, Yue Y, Wang L, Liu G. Analysis of WRKY Gene Family in Acer fabri and Their Expression Patterns Under Cold Stress. Genes. 2025; 16(3):344. https://doi.org/10.3390/genes16030344
Chicago/Turabian StyleChen, Gongwei, Yixiao Zhou, Dandan Zhang, Fengyuan Chen, Xuyang Qin, Hongyu Cai, Heng Gu, Yuanzheng Yue, Lianggui Wang, and Guohua Liu. 2025. "Analysis of WRKY Gene Family in Acer fabri and Their Expression Patterns Under Cold Stress" Genes 16, no. 3: 344. https://doi.org/10.3390/genes16030344
APA StyleChen, G., Zhou, Y., Zhang, D., Chen, F., Qin, X., Cai, H., Gu, H., Yue, Y., Wang, L., & Liu, G. (2025). Analysis of WRKY Gene Family in Acer fabri and Their Expression Patterns Under Cold Stress. Genes, 16(3), 344. https://doi.org/10.3390/genes16030344




