Phylogeny and Specific Determination of Gloydius halys-intermedius Complex Based on Complete Mitochondrial Genes
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and DNA Extraction
2.2. Genome Sequencing, Assembly, and Annotation
2.3. Phylogenetic Tree Construction and Pairwise Distance Estimation
2.4. Molecular Species Delimitation
3. Results
3.1. The Composition of the Mitochondrial Genome
3.2. Phylogenetic Relationships
3.3. Species Delimitation
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Abbreviations | Full Names |
|---|---|
| ND2 | NADH dehydrogenase subunit 2 |
| COX1 | cytochrome c oxidase subunit I |
| COX2 | cytochrome c oxidase subunit II |
| ATP8 | ATP synthase F0 subunit 8 |
| ATP6 | ATP synthase F0 subunit 6 |
| COX3 | cytochrome c oxidase subunit III |
| ND3 | NADH dehydrogenase subunit 3 |
| ND4L | NADH dehydrogenase subunit 4L |
| ND4 | NADH dehydrogenase subunit 4 |
| ND5 | NADH dehydrogenase subunit 5 |
| ND6 | NADH dehydrogenase subunit 6 |
| CYTB | cytochrome b |
| ND1 | NADH dehydrogenase subunit 1 |
Appendix B
Appendix B.1
| No. | Species | Voucher Number |
|---|---|---|
| 1 | G. caucasicus | B474 |
| 2 | G. cognatus | IA |
| 3 | G. halys | KM186844 |
| 4 | G. intermedius 2 | KM434236 |
| 5 | G. ussuriensis | KP262412 |
| 6 | G. intermedius 1 | MW143075 |
| 7 | G. shedaoensis | Q7 |
| 8 | G. stejnegeri | S6 |
| 9 | G. brevicaudus | B6 |
| 10 | G. himalayanus | MK559438 |
| 11 | G. changdaoensis | MT731652 |
| 12 | G. shedaoensis | KT726956 |
| 13 | O. okinavensis | AB175670-out |
Appendix B.2
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.005 | 0.005 | 0.004 | 0.009 | 0.005 | 0.005 | 0.004 | 0.008 | 0.008 | 0.005 | 0.005 | 0.009 | |
| 2 | 0.045 | 0.005 | 0.005 | 0.009 | 0.005 | 0.005 | 0.005 | 0.008 | 0.008 | 0.005 | 0.005 | 0.01 | |
| 3 | 0.037 | 0.037 | 0.004 | 0.009 | 0.004 | 0.004 | 0.004 | 0.009 | 0.009 | 0.005 | 0.005 | 0.009 | |
| 4 | 0.036 | 0.04 | 0.026 | 0.009 | 0.003 | 0.003 | 0.004 | 0.009 | 0.008 | 0.005 | 0.003 | 0.009 | |
| 5 | 0.108 | 0.113 | 0.112 | 0.112 | 0.009 | 0.009 | 0.009 | 0.008 | 0.008 | 0.009 | 0.009 | 0.009 | |
| 6 | 0.039 | 0.042 | 0.026 | 0.021 | 0.114 | 0.004 | 0.004 | 0.009 | 0.008 | 0.005 | 0.004 | 0.009 | |
| 7 | 0.035 | 0.044 | 0.03 | 0.015 | 0.114 | 0.026 | 0.005 | 0.009 | 0.008 | 0.005 | 0.003 | 0.009 | |
| 8 | 0.033 | 0.041 | 0.031 | 0.033 | 0.114 | 0.034 | 0.038 | 0.008 | 0.009 | 0.005 | 0.005 | 0.009 | |
| 9 | 0.116 | 0.123 | 0.125 | 0.127 | 0.106 | 0.119 | 0.129 | 0.119 | 0.009 | 0.009 | 0.009 | 0.009 | |
| 10 | 0.114 | 0.118 | 0.125 | 0.121 | 0.114 | 0.122 | 0.117 | 0.119 | 0.132 | 0.008 | 0.008 | 0.008 | |
| 11 | 0.042 | 0.054 | 0.051 | 0.047 | 0.109 | 0.049 | 0.046 | 0.048 | 0.118 | 0.115 | 0.006 | 0.009 | |
| 12 | 0.042 | 0.049 | 0.036 | 0.019 | 0.112 | 0.029 | 0.011 | 0.044 | 0.127 | 0.123 | 0.052 | 0.009 | |
| 13 | 0.137 | 0.137 | 0.139 | 0.14 | 0.128 | 0.137 | 0.137 | 0.139 | 0.147 | 0.12 | 0.129 | 0.14 |
Appendix C
| Species | Voucher Number |
|---|---|
| O. okinavensis * | AB175670-Ovophis okinavensis |
| G. shedaoensis | MK064736_G._shedaoensis_CHS329 |
| MK064611_G._shedaoensis_CHS090 | |
| MK064610_G._shedaoensis_CHS089 | |
| KY040587_G._s_shedaoensis_JS1408D2 | |
| KT726956_G._shedaoensis_shedaoensis | |
| G. intermedius | MK064735_G_intermedius_CHS327 |
| MK064733_G_intermedius_CHS324 | |
| MK064614_G_intermedius_CHS095 | |
| KY040589_G_s_qianshanensis_JS1306Q4 | |
| KY040588_G_s_qianshanensis_JS150722 | |
| KM434236_G_intermedius | |
| G. stejnegeri | MK064732_G_stejnegeri_CHS323 |
| MK064731_G_stejnegeri_CHS322 | |
| KY040601_G_stejnegeri_JS1508S4 | |
| KY040600_G_stejnegeri_JS1409S3 | |
| G_stejnegeri_S6 | |
| G. ussuriensis | MK064730_G_ussuriensis_CHS318 |
| MK064729_G_ussuriensis_CHS316 | |
| MK064612_G_ussuriensis_CHS092 | |
| KP262412_G_ussuriensis | |
| G. brevicaudus | MH220717_G_brevicaudus_ZJ121106 |
| MH220697_G_brevicaudus_ZJ100802 | |
| MH153664_G_brevicaudus_DWF4 | |
| MH153663_G_brevicaudus_DWF3 | |
| MH153662_G_brevicaudus_DWF2 | |
| MF099695_G_brevicaudus_DWF04 | |
| MF099694_G_brevicaudus_DWF03 | |
| MF099693_G_brevicaudus_DWF02 | |
| KY040616_G_brevicaudus_DL70 | |
| KR046009_G_brevicaudus_SR153 | |
| KR046007_G_brevicaudus_SR151 | |
| KR046006_G_brevicaudus_SR150 | |
| KR046005_G_brevicaudus_SR149 | |
| G. halys | KY040592_G_halys_JSSD1607H9 |
| KY040591_G_halys_JSSD1508X3 | |
| KY040590_G_halys_SYNU1301908 | |
| KM186844_G_halys | |
| G. intermedius | JQ798882_G_intermedius_NIBRRP0000100199 |
| JQ798881_G_intermedius_NIBRRP0000100251 | |
| MW143075_G_intermedius | |
| G. changdaoensi | MT731652_G_changdaoensis |
| G. caucasicus | G_caucasicus_B474 |
References
- Shi, J.; Wang, G.; Chen, X.; Fang, Y.; Ding, L.; Huang, S.; Hou, M.; Liu, J.; Li, P. A new moth-preying alpine pit viper species from Qinghai-Tibetan Plateau (Viperidae, Crotalinae). Amphib. Reptil. 2017, 38, 517–532. [Google Scholar] [CrossRef]
- Shi, J.S.; Liu, J.C.; Giri, R.; Owens, J.B.; Santra, V.; Kuttalam, S.; Selvan, M.; Guo, K.J.; Malhotra, A. Molecular phylogenetic analysis of the genus Gloydius (Squamata, Viperidae, Crotalinae), with description of two new alpine species from Qinghai-Tibet Plateau, China. Zookeys 2021, 1061, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Yang, D.; Zhang, W.; Peng, L.-F.; Orlov, N.L.; Jiang, F.; Ding, L.; Hou, M.; Huang, X.; Huang, S.; et al. A New Species of the Gloydius strauchi Complex (Crotalinae: Viperidae: Serpentes) from Qinghai, Sichuan, and Gansu, China. Russ. J. Herpetol. 2018, 25, 126–138. [Google Scholar] [CrossRef]
- Zhang, M.H.; Shi, S.C.; Li, C.; Yan, P.; Wang, P.; Ding, L.; Du, J.; Plenkovic-Moraj, A.; Jiang, J.P.; Shi, J.S. Exploring cryptic biodiversity in a world heritage site: A new pitviper (Squamata, Viperidae, Crotalinae) from Jiuzhaigou, Aba, Sichuan, China. Zookeys 2022, 1114, 59–76. [Google Scholar] [CrossRef] [PubMed]
- Nikolai, L.O.; Barabanov, A. Analysis of Nomenclature, Classification, and Distribution of the Agkistrodon halys—Agkistrodon intermedius Complexes: A Critical Review. Russ. J. Herpetol. 1999, 6, 167–192. [Google Scholar]
- Gloyd, H.K.; Conant, R. Snakes of the Agkistrodon complex: A monographic review. In Contribution to Herpetology; Society for the Study of Amphibians and Reptiles: Oxford, UK, 1990; Volume 6. [Google Scholar]
- Wu, Y.; Li, K.; Liu, Q.; Chen, S.; Cai, B. The complete mitochondrial genome of the Asian pitviper Gloydius changdaoensis (Squamata, Viperidae). Mitochondrial DNA B Resour. 2020, 5, 3276–3277. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, A.; Mukherjee, S.; Chowdhury, S.; Purkayastha, J. Geographic Distribution: Gloydius himalayanus (Himalayan Pitviper). Herpetol. Rev. 2018, 49, 505. [Google Scholar]
- Xu, C.; Xie, F.; Liu, Y.; Zhao, S.; Wang, Y.; Ma, T.; Zhao, T. Sequencing and analysis of the complete mitochondrial genome of Gloydius saxatilis (Squamata: Viperidae: Crotalinae). Mitochondrial DNA Part A 2015, 27, 2361–2362. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Zhao, S.; Dou, H. The complete mitochondrial genome of Gloydius intermedius (Squamata: Viperidae: Crotalinae) from China. Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016, 27, 2373–2374. [Google Scholar] [CrossRef]
- Xu, R.; Dujsebayeva, T.N.; Chen, D.; Mijidsuren, B.; Xu, F.; Guo, X. Phylogeography and Ecological Niche Modeling of the Alashan Pit Viper (Gloydius cognatus; Reptilia, Viperidae) in Northwest China and Adjacent Areas. Animals 2023, 13, 3726. [Google Scholar] [CrossRef]
- Guo, P.; Liu, Q.; Wu, Y.; Zhu, F.; Zhong, G. Pitvipers of China; Science Press: Beijing, China, 2021. [Google Scholar]
- Shi, J.; Yang, D.; Zhang, W.; Qi, S.; Li, P.; Ding, L. Distribution and Infraspecies Taxonomy of Gloydius halys-Gloydius intermedius Complex in China (Serpentes: Crotalinae). Chin. J. Zool. 2016, 51, 777–798. [Google Scholar]
- Xu, Y.; Liu, Q.; Myers, E.A.; Wang, L.; Huang, S.; He, Y.; Peng, P. Molecular Phylogeny of the Genus Gloydius (Serpentes: Crotalinae). Asian Herpetol. Res. 2012, 3, 127–132. [Google Scholar] [CrossRef]
- Wu, Y.-H.; Hou, S.-B.; Yuan, Z.-Y.; Jiang, K.; Huang, R.-Y.; Wang, K.; Liu, Q.; Yu, Z.-B.; Zhao, H.-P.; Zhang, B.-L.; et al. DNA barcoding of Chinese snakes reveals hidden diversity and conservation needs. Mol. Ecol. Resour. 2023, 23, 1124–1141. [Google Scholar] [CrossRef] [PubMed]
- Kuttalam, S.; Santra, V.; Owens, J.B.; Selvan, M.; Mukherjee, N.; Graham, S.; Togridou, A.; Bharti, O.K.; Shi, J.; Shanker, K.; et al. Phylogenetic and morphological analysis of Gloydius himalayanus (Serpentes, Viperidae, Crotalinae), with the description of a new species. Eur. J. Taxon. 2022, 852, 1–30. [Google Scholar] [CrossRef]
- Zeng, Y.; Wu, Y.; Liu, Q.; Yu, M.; Li, K.; Wang, J.; Guo, P.; Shu, G. A Report on Reproduction of Gloydius halys. Chin. J. Zool. 2024, 59, 639. [Google Scholar]
- Guo, P.; Che, J. Snakes in Qinghai-Xizang Plateau; Science Press: Beijing, China, 2024. [Google Scholar]
- Lee, Y.S.; Do, M.S.; Kim, W.; Jeon, H.S.; Lee, S.-C.; Jung, J.-H.; An, J. Phylogenetic relationships between three Korean pit viper Gloydius (Serpentes: Crotalinae) species using mitochondrial DNA genes. Genes Genom. 2022, 44, 517–526. [Google Scholar] [CrossRef]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, Automatic barcode gap discovery for primary species delimitation. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Jiang, K.; Guo, P.; Huang, S.; Rao, D.; Ding, L.; Takeuchi, H.; Che, J.; Zhang, Y.-p.; Myers, E.A.; et al. Assessing species boundaries and the phylogenetic position of the rare Szechwan ratsnake, Euprepiophis perlaceus (Serpentes: Colubridae), using coalescent-based methods. Mol. Phylogen. Evol. 2014, 70, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef] [PubMed]
- Meng, G.; Li, Y.; Yang, C.; Liu, S. MitoZ: A toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 2019, 47, e63. [Google Scholar] [CrossRef]
- Jin, J.-J.; Yu, W.-B.; Yang, J.-B.; Song, Y.; DePamphilis, C.W.; Yi, T.-S.; Li, D.-Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Puillandre, N.; Brouillet, S.; Achaz, G. ASAP: Assemble species by automatic partitioning. Mol. Ecol. Resour. 2021, 21, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, T.; Barraclough, T.G. Delimiting species using single-locus data and the generalized mixed Yule coalescent approach: A revised method and evaluation on simulated data sets. Syst. Biol. 2013, 62, 707–724. [Google Scholar] [CrossRef] [PubMed]
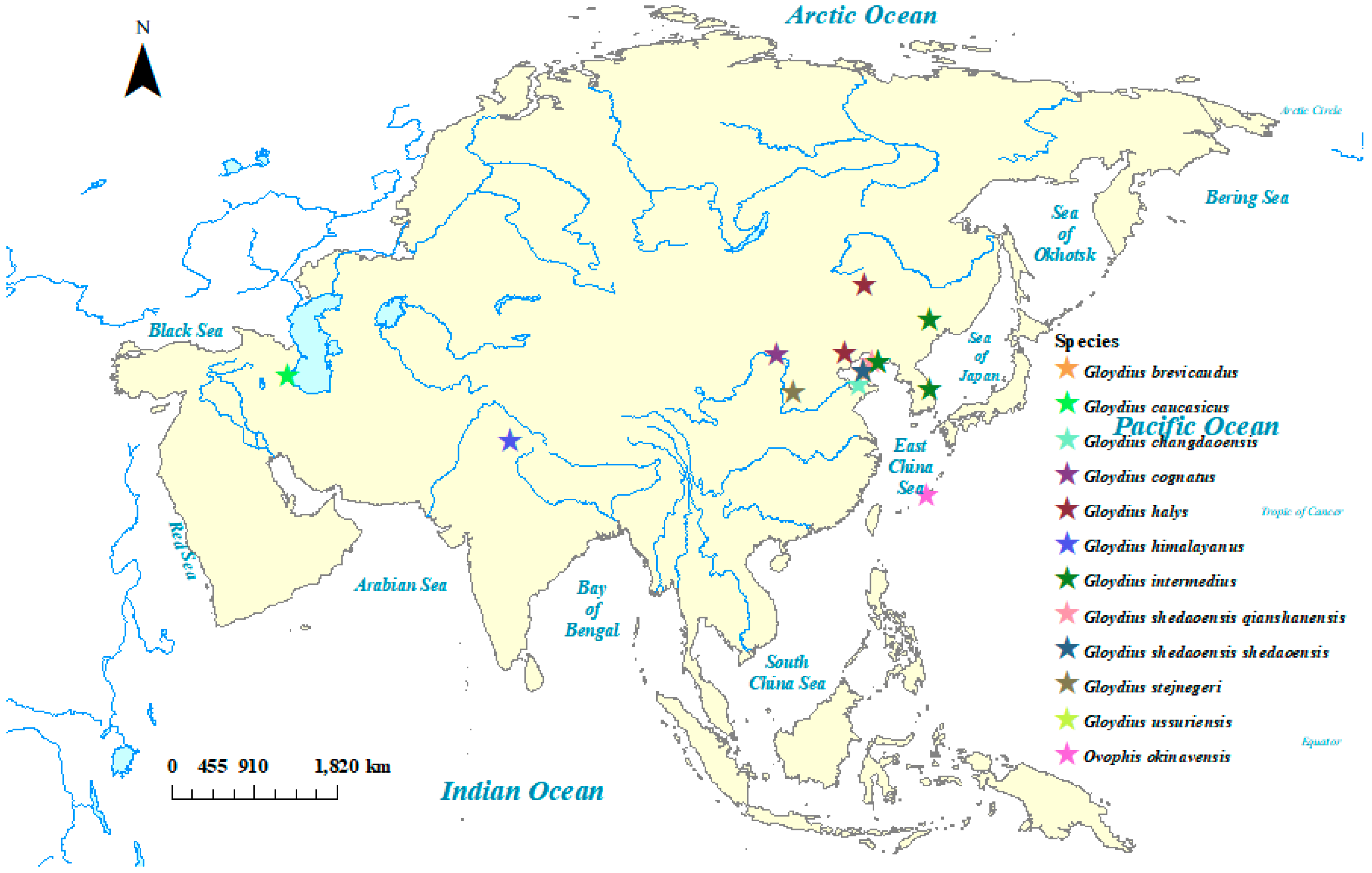
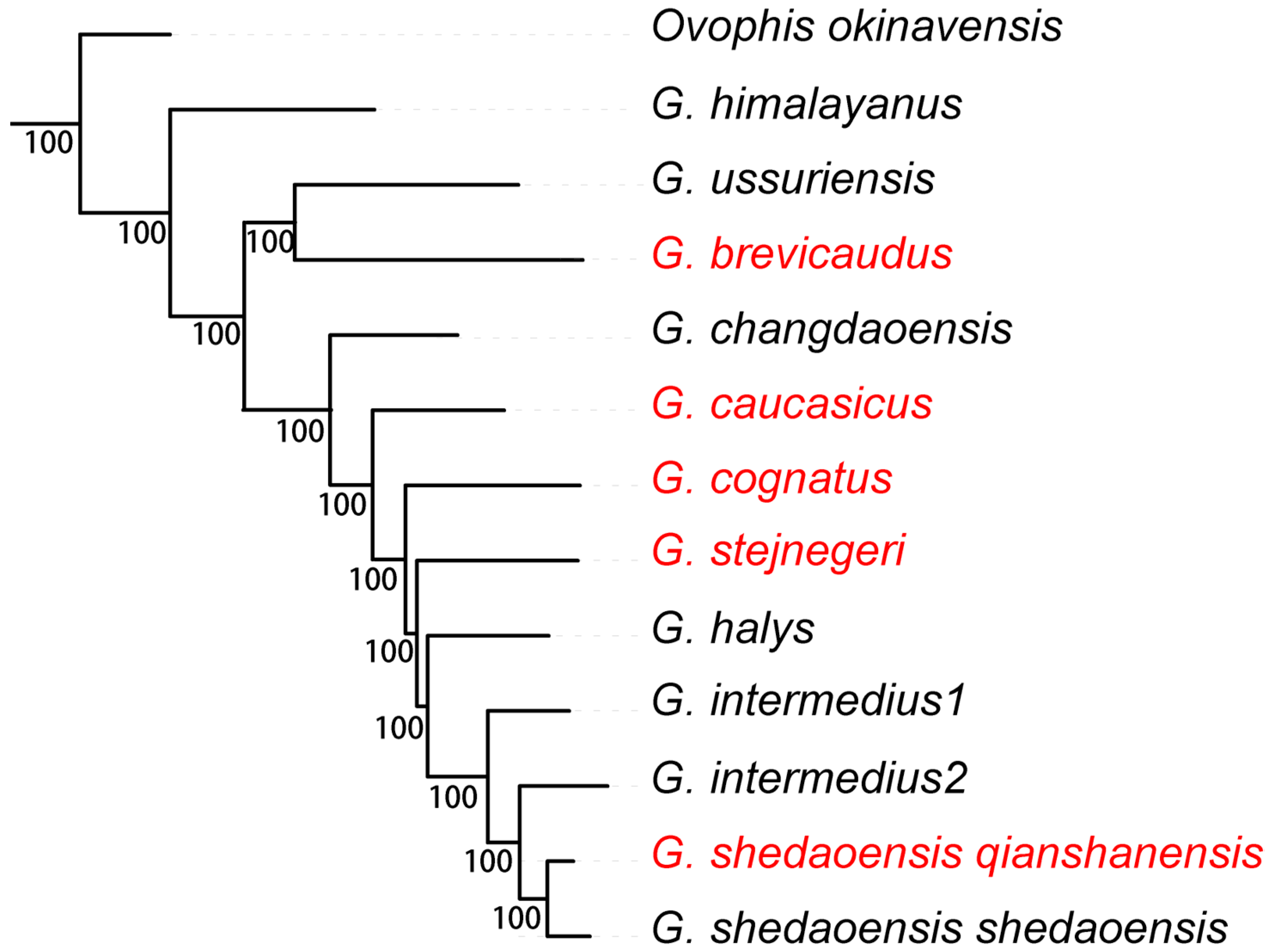
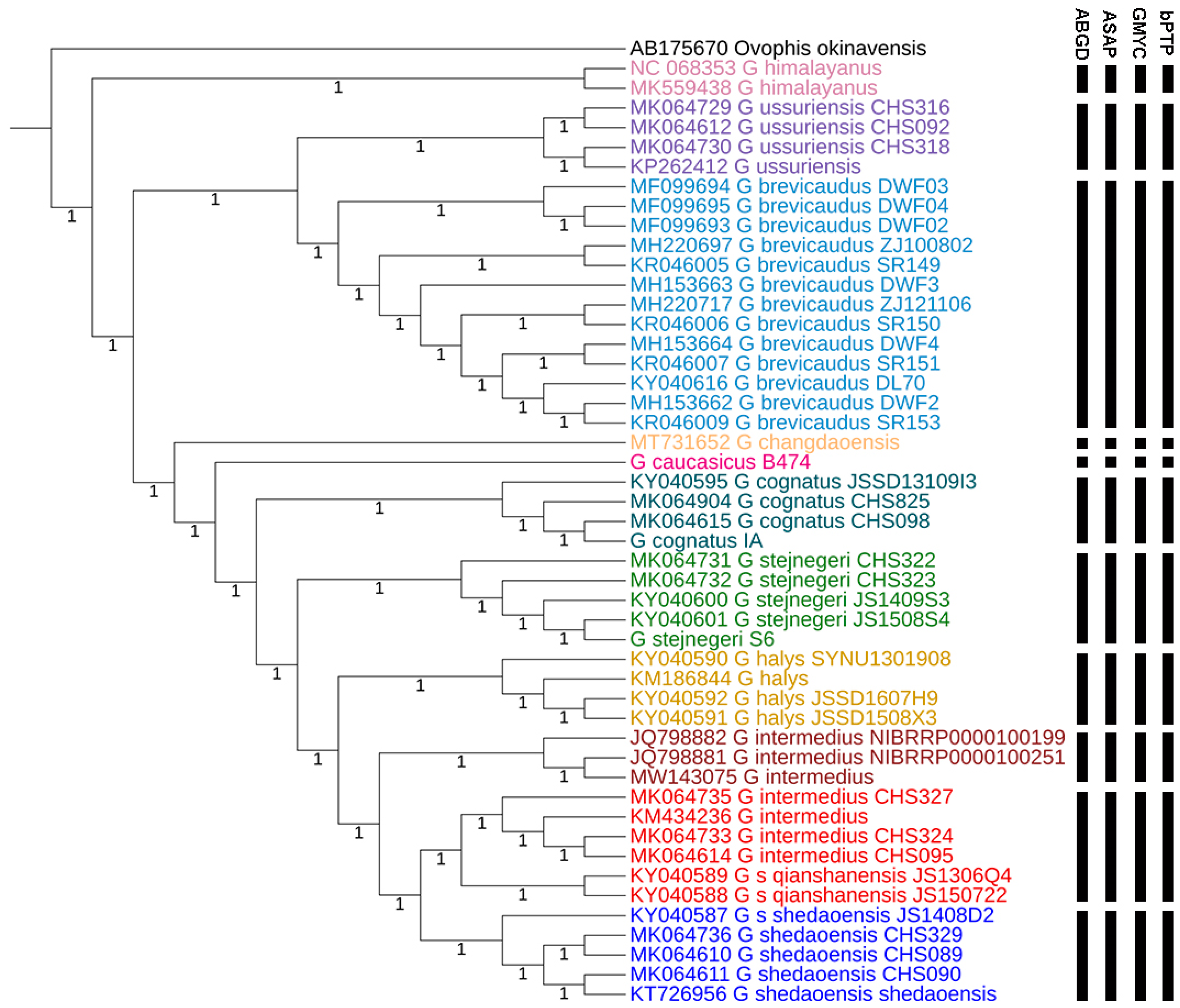
| Vouchers | Species | Locations |
|---|---|---|
| AB175670 | Ovophis okinavensis * | Japan, Okinawa Islands |
| B474 | G. caucasicus | Azerbaijan, Lankaran Region |
| B6 | Gloydius brevicaudus | Liaoning, China |
| IA | G. cognatus | Bayan Obo, Inner Mongolia, China |
| Q7 | Gloydius shedaoensis | Wafangdian, Liaoning, China |
| S6 | Gloydius stejnegeri | Lingshi, Shanxi, China |
| KM186844 | Gloydius halys | Inner Mongolia, China |
| KM434236 | Gloydius intermedius 2 | Heilongjiang, China |
| KP262412 | Gloydius ussuriensis | Heilongjiang, China |
| KT726956 | Gloydius shedaoensis | Lvshun, Liaoning, China |
| MK559438 | Gloydius himalayanus | Himachal Pradesh, India |
| MT731652 | Gloydius changdaoensis | Changdao, Shandong, China |
| MW143075 | Gloydius intermedius 1 | Samcheok-si, Gangwon-do, South Korea |
| Samples | Species | Length /bp | T% | C% | A% | G% | AT Skew | CG Skew |
|---|---|---|---|---|---|---|---|---|
| B474 | G. caucasicus | 17,224 | 26.0 | 28.8 | 32.2 | 13.1 | 0.11 | 0.37 |
| B6 | Gloydius brevicaudus | 16,655 | 26.1 | 28.5 | 32.2 | 13.2 | 0.10 | 0.37 |
| IA | G. cognatus | 17,228 | 25.9 | 28.8 | 32.4 | 12.9 | 0.11 | 0.38 |
| Q7 | Gloydius shedaoensis | 17,216 | 25.8 | 28.9 | 32.4 | 12.9 | 0.11 | 0.38 |
| S6 | Gloydius stejnegeri | 17,225 | 26.0 | 28.7 | 32.4 | 12.9 | 0.11 | 0.38 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, L.; Xia, Z.; Liu, N.; Hou, S.; Lv, C.; Tang, L.; Feng, S.; Shi, J.; Bai, M. Phylogeny and Specific Determination of Gloydius halys-intermedius Complex Based on Complete Mitochondrial Genes. Genes 2025, 16, 276. https://doi.org/10.3390/genes16030276
Jin L, Xia Z, Liu N, Hou S, Lv C, Tang L, Feng S, Shi J, Bai M. Phylogeny and Specific Determination of Gloydius halys-intermedius Complex Based on Complete Mitochondrial Genes. Genes. 2025; 16(3):276. https://doi.org/10.3390/genes16030276
Chicago/Turabian StyleJin, Lijie, Zuyao Xia, Ning Liu, Shengyue Hou, Chuandong Lv, Lianyou Tang, Shuguang Feng, Jingsong Shi, and Ming Bai. 2025. "Phylogeny and Specific Determination of Gloydius halys-intermedius Complex Based on Complete Mitochondrial Genes" Genes 16, no. 3: 276. https://doi.org/10.3390/genes16030276
APA StyleJin, L., Xia, Z., Liu, N., Hou, S., Lv, C., Tang, L., Feng, S., Shi, J., & Bai, M. (2025). Phylogeny and Specific Determination of Gloydius halys-intermedius Complex Based on Complete Mitochondrial Genes. Genes, 16(3), 276. https://doi.org/10.3390/genes16030276






