Clinical Implications of Cell-Free DNA in Managing BRAF V600E Mutation-Positive Colorectal Cancer
Abstract
1. Introduction
1.1. Real-Time Evaluation of Treatment Response
1.2. Minimal Residual Disease (MRD) Evaluation After Curative Resection
2. Materials and Methods
2.1. Study Population and Design
- Cohort 1 (Stage IV cases): Prediction of treatment response using cfDNA;
- Cohort 2 (Stage I–III curatively resected cases): Minimal residual disease (MRD) assessment using cfDNA
2.2. Blood Sample Collection and cfDNA Extraction
2.3. Droplet Digital PCR (ddPCR)
2.4. Statistical Analysis
2.5. Ethical Considerations
3. Results
3.1. Cohort 1: Stage IV Cases (n = 14)
3.1.1. First-Line Treatment Response and cfDNA
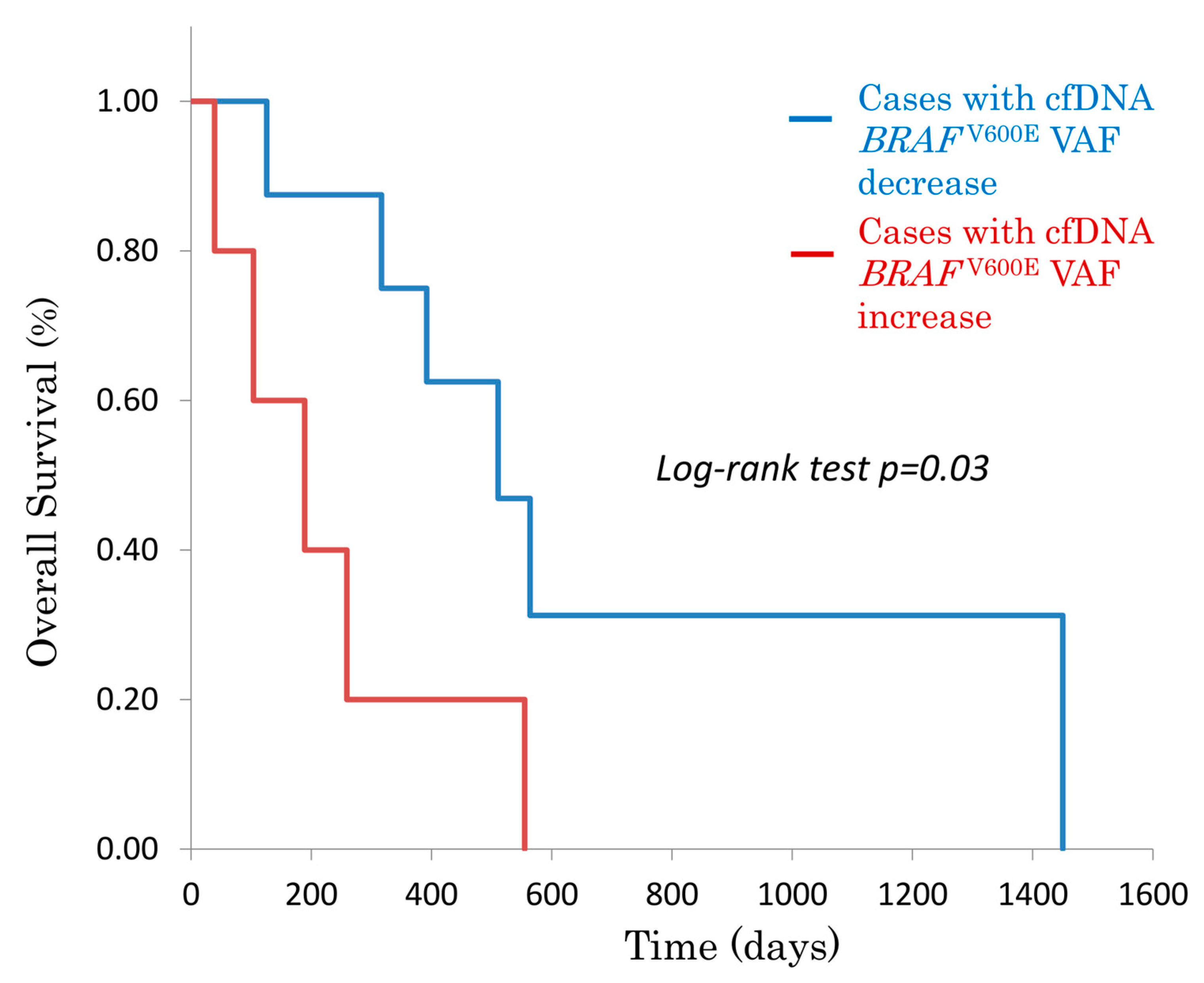
3.1.2. Overall Survival (OS) Based on cfDNA BRAFV600E Status
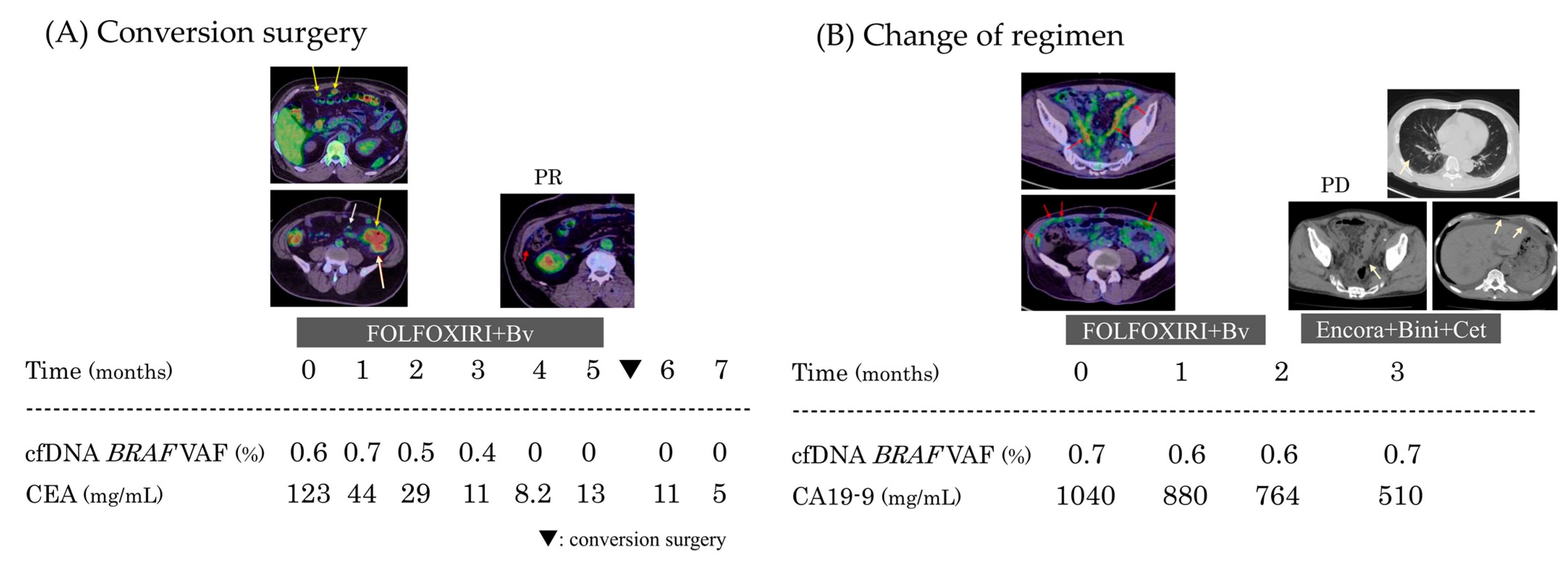
3.1.3. Comparison of cfDNA and Tumor Markers in Clinical Practice
Case 1: Conversion Surgery After Initial Treatment
Case 2: Timing of Regimen Change
3.2. Cohort 2: Stage II–III Curatively Resected Cases (n = 23)
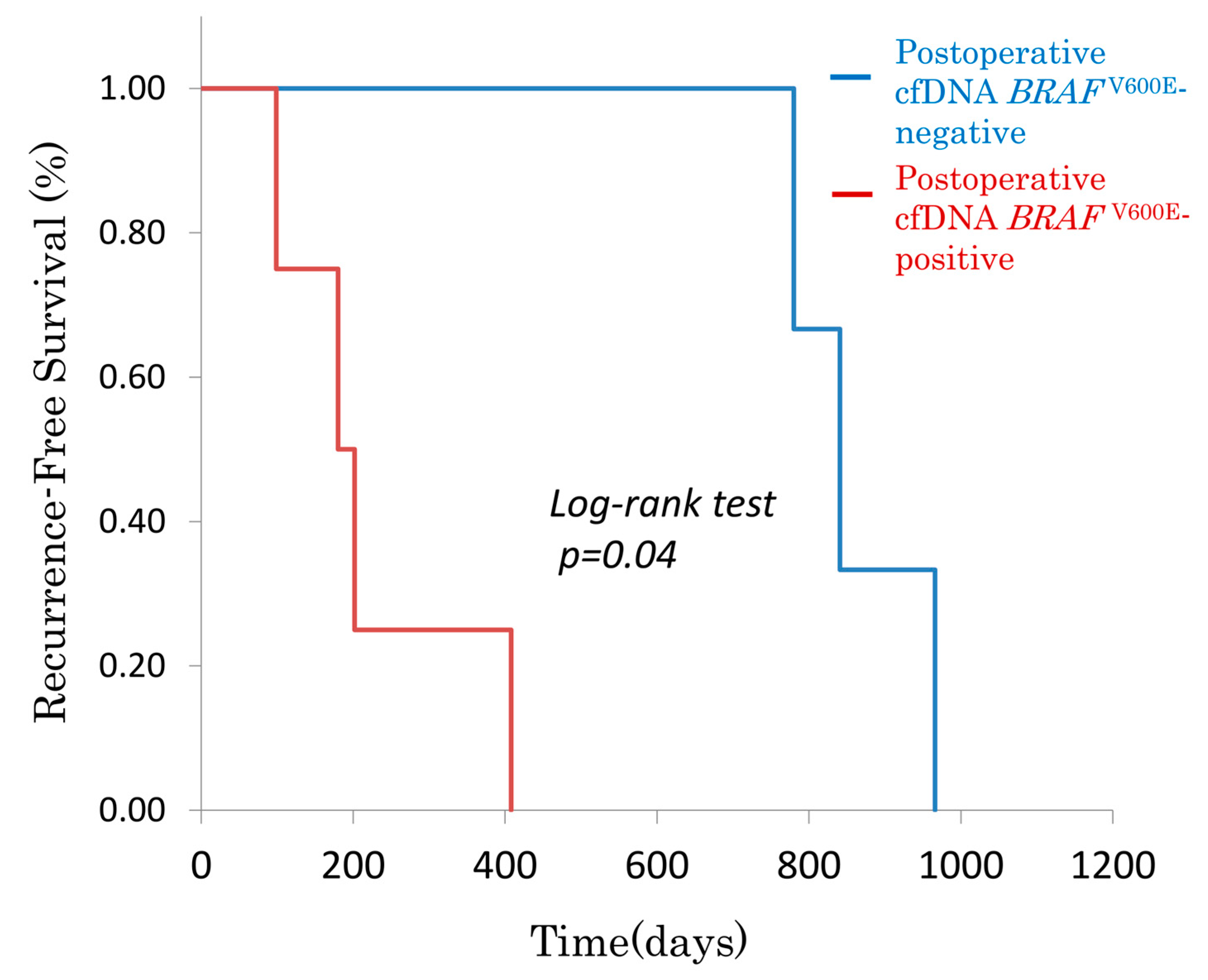
3.2.1. Postoperative cfDNA BRAFV600E and Recurrence-Free Survival (RFS)
3.2.2. Comparison with Conventional Tumor Markers
3.2.3. Postoperative cfDNA BRAFV600E Status and DFS in Recurrent Seven Cases
4. Discussion
Limitation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kawazoe, A.; Shitara, K.; Fukuoka, S.; Kuboki, Y.; Bando, H.; Okamoto, W.; Kojima, T.; Fuse, N.; Yamanaka, T.; Doi, T.; et al. A retrospective observational study of clinicopathological features of KRAS, NRAS, BRAF and PIK3CA mutations in Japanese patients with metastatic colorectal cancer. BMC Cancer 2015, 15, 258. [Google Scholar] [CrossRef]
- Yokota, T.; Ura, T.; Shibata, N.; Takahari, D.; Shitara, K.; Nomura, M.; Kondo, C.; Mizota, A.; Utsunomiya, S.; Muro, K.; et al. BRAF mutation is a powerful prognostic factor in advanced and recurrent colorectal cancer. Br. J. Cancer 2011, 104, 856–862. [Google Scholar] [CrossRef] [PubMed]
- Kopetz, S.; Grothey, A.; Yaeger, R.; Van Cutsem, E.; Desai, J.; Yoshino, T.; Wasan, H.; Ciardiello, F.; Loupakis, F.; Hong, Y.S.; et al. Encorafenib, Binimetinib, and Cetuximab in BRAF V600E-Mutated Colorectal Cancer. N. Engl. J. Med. 2019, 381, 1632–1643. [Google Scholar] [CrossRef]
- Corcoran, R.B.; André, T.; Atreya, C.E.; Schellens, J.H.; Yoshino, T.; Bendell, J.C.; Hollebecque, A.; McRee, A.J.; Siena, S.; Middleton, G.; et al. Combined BRAF, EGFR, and MEK Inhibition in Patients with BRAFV600E-Mutant Colorectal Cancer. Cancer Discov. 2018, 8, 428–443. [Google Scholar] [CrossRef] [PubMed]
- Kadowaki, S.; Kakuta, M.; Takahashi, S.; Takahashi, A.; Arai, Y.; Nishimura, Y.; Yatsuoka, T.; Ooki, A.; Yamaguchi, K.; Matsuo, K.; et al. Prognostic value of KRAS and BRAF mutations in curatively resected colorectal cancer. World J. Gastroenterol. 2015, 21, 1275–1283. [Google Scholar] [CrossRef] [PubMed]
- Formica, V.; Sera, F.; Cremolini, C.; Riondino, S.; Morelli, C.; Arkenau, H.-T.; Roselli, M. KRAS and BRAF Mutations in Stage II and III Colon Cancer: A Systematic Review and Meta-Analysis. J. Natl. Cancer Inst. 2022, 114, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Hoon, D.S.B.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Bronkhorst, A.J.; Ungerer, V.; Holdenrieder, S. The emerging role of cell-free DNA as a molecular marker for cancer management. Biomol. Detect. Quantif. 2019, 17, 100087. [Google Scholar] [CrossRef] [PubMed]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.D.; Knippers, R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar]
- Martins, I.; Ribeiro, I.P.; Jorge, J.; Gonçalves, A.C.; Sarmento-Ribeiro, A.B.; Melo, J.B.; Carreira, I.M. Liquid biopsies: Applications for cancer diagnosis and monitoring. Genes 2021, 12, 349. [Google Scholar] [CrossRef]
- Zmrzljak, U.P.; Košir, R.; Krivokapić, Z.; Radojković, D.; Nikolić, A. Detection of somatic mutations with ddPCR from liquid biopsy of colorectal cancer patients. Genes 2021, 12, 289. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Matsuda, A.; Koizumi, M.; Shinji, S.; Takahashi, G.; Iwai, T.; Takeda, K.; Ueda, K.; Yokoyama, Y.; Hara, K.; et al. Liquid biopsy for the management of patients with colorectal cancer. Digestion 2019, 99, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Nakai, M.; Yamada, T.; Sekiya, K.; Sato, A.; Hankyo, M.; Kuriyama, S.; Takahashi, G.; Kurita, T.; Yanagihara, K.; Yoshida, H.; et al. Use of liquid biopsy to detect PIK3CA mutation in metastatic breast cancer. J. Nippon. Med. Sch. 2022, 89, 66–71. [Google Scholar] [CrossRef]
- Tanaka, K.; Yoshida, Y.; Yamada, T.; Hayashi, T.; Shimaoka, H.; Yoshimura, F.; Kajitani, R.; Munechika, T.; Matsumoto, Y.; Nagano, H.; et al. Oncological evaluation in the perioperative period using cfDNA with BRAF V600E mutation in patients with colorectal cancer. Sci. Rep. 2021, 11, 13263. [Google Scholar] [CrossRef]
- Yamada, T.; Takahashi, G.; Iwai, T.; Takeda, K.; Ueda, K.; Koizumi, M.; Shinji, S.; Yokoyama, Y.; Hara, K.; Matsuda, A.; et al. Emergence of KRAS mutations and acquisition of resistance to EGFR blockade. J. Clin. Oncol. 2017, 35, 3598. [Google Scholar] [CrossRef]
- Van Cutsem, E.; Taieb, J.; Yaeger, R.; Yoshino, T.; Grothey, A.; Maiello, E.; Elez, E.; Dekervel, J.; Ross, P.; Ruiz-Casado, A.; et al. ANCHOR CRC: Results From a Single-Arm, Phase II Study of Encorafenib Plus Binimetinib and Cetuximab in Previously Untreated BRAFV600E-Mutant Metastatic Colorectal Cancer. J. Clin. Oncol. 2023, 41, 2628–2637. [Google Scholar] [CrossRef]
- Kopetz, S.; Yoshino, T.; Kim, T.W.; Yaeger, R.; Desai, J.; Wasan, H.S.; Van Cutsem, E.; Ciardiello, F.; Maughan, T.; Eng, C.; et al. BREAKWATER safety lead-in (SLI): Encorafenib (E) + cetuximab (C) + chemotherapy for BRAFV600E metastatic colorectal cancer (mCRC). J. Clin. Oncol. 2023, 41, 119. [Google Scholar] [CrossRef]
- Kobayashi, S.; Bando, H.; Taketomi, A.; Takamoto, T.; Shinozaki, E.; Shiozawa, M.; Hara, H.; Yamazaki, K.; Komori, K.; Matsuhashi, N.; et al. NEXUS trial: A multicenter phase II clinical study evaluating the efficacy and safety of the perioperative use of encorafenib, binimetinib, and cetuximab in patients with previously untreated surgically resectable BRAF V600E mutant colorectal oligometastases. BMC Cancer 2023, 23, 779. [Google Scholar] [CrossRef]
- Rao, H.; Wu, H.; Huang, Q.; Yu, Z.; Zhong, Z. Clinical Value of Serum CEA, CA24-2 and CA19-9 in Patients with Colorectal Cancer. Clin Lab. 2021, 67. [Google Scholar] [CrossRef]
- Inagaki, C.; Matoba, R.; Yuki, S.; Shiozawa, M.; Tsuji, A.; Inoue, E.; Muro, K.; Ichikawa, W.; Fujii, M.; Sunakawa, Y. The BEETS (JACCRO CC-18) trial: An observational and translational study of BRAF-mutated metastatic colorectal cancer. Future Oncol. 2023, 19, 1165–1174. [Google Scholar] [CrossRef] [PubMed]
- Sawayama, H.; Miyamoto, Y.; Ogawa, K.; Yoshida, N.; Baba, H. Investigation of colorectal cancer in accordance with consensus molecular subtype classification. Ann. Gastroenterol. Surg. 2020, 4, 528–539. [Google Scholar] [CrossRef]
- Ten Hoorn, S.; de Back, T.R.; Sommeijer, D.W.; Vermeulen, L. Clinical value of consensus molecular subtypes in colorectal cancer: A systematic review and meta-analysis. J. Natl. Cancer Inst. 2022, 114, 503–516. [Google Scholar] [CrossRef]
- Stintzing, S.; von der Heyde, E.; Wierecky, J.; Bürkle, D.; Forstbauer, H.; Hübner, G.; Schmidt, B.; Schroeder, J.; Distelrath, A.; Flum, M.; et al. Disease characteristics and clinical practice of BRAF V600E-mutant metastatic colorectal cancer treatment: Baseline analysis of patients enrolled in the BERING CRC study. J. Clin. Oncol. 2023, 41, 34. [Google Scholar] [CrossRef]
- Chen, Y.; Deng, Y.; Ma, Y.; Huang, H.; Liu, Y.; Zhang, Y.; Zhang, H.; Ye, S.; Guo, H.; Wu, M.; et al. Clinical and genomic distinction of class 1/2/3 BRAF-mutant colorectal cancer and differential prognosis. J. Clin. Oncol. 2022, 40, 3533. [Google Scholar] [CrossRef]
| Cohort 1: Stage IV Colorectal Cancer with BRAFV600E Mutation | ||
|---|---|---|
| Age (y/o) | 69 | (59.0–71.5) |
| Sex [M/F] | 10:4 | |
| Primary site [right/left] | 9:5 | |
| Tumor size (mm) 1 | 47.5 | (40.3–55) |
| Histology type [tub/por/muc] 2 | 5:5:4 | |
| Distal metastasis [liver/lung/LN/dissemination/bone] 3 | 6:2:10:1 | |
| Primary tumor resection [yes/no] | 10:4 | |
| Surgical procedure (ileocecal resection 3:right hemicolectomy 4:partial resection 3) | ||
| First-line treatment regimen [mFOLFOX6 + Bv/FOLFOXIRI + Bv/mFOLFOX6] | 5:6:3 | |
| Pre-treatment CEA (ng/mL) | 6:2 | (3.5–18.5) |
| Pre-treatment CA19-9 (U/mL) | 87 | (2.1–2249.4) |
| Pre-treatment cfDNA volume (ng/mL) | 123 | (101.5–145) |
| median | (Interquartile range) | |
| Cohort 2: BRAF V600E Mutation-Positive Colorectal Cancer with Curative Resection | ||
|---|---|---|
| Age (y/o) | 72 | (64.5–82.5) |
| Sex [M/F] | 11:12 | |
| Primary site [right/left] | 18:5 | |
| Tumor size (mm) | 39 | (30–57) |
| pT [1b/2/3/4a] | 2:2:14:5 | |
| pN [0/1a/1b/2a/2b/3] | 11:4:3:3:1:1 | |
| Histology type [tub/por/muc] 1 | 12:5:6 | |
| Surgical procedure [ICR/RHC/partial resection/Sigmoidectomy/LAR] | 10:8:4:1 | |
| Laparoscopic/open | 18:5 | |
| Adjuvant chemotherapy [Yes/No] | 9:14 | |
| regimen [CAPOX 5: FOLFOX 1: UFT/UZEL 3] | ||
| Pre-treatment CEA (ng/mL) | 4.7 | (2.1–8.5) |
| Pre-treatment CA19-9 (U/mL) | 12.7 | (7.8–27.7) |
| Pre-treatment cfDNA volume (ng/mL) | 138 | (82–151) |
| median | (Interquartile range) | |
| Relapse | No Relapse | Relapse | No Relapse | ||
|---|---|---|---|---|---|
| BRAFV600E positive in cfDNA | 4 | CEA ≥5.0 ng/mL | 2 | 3 | |
| BRAFV600E negative in cfDNA | 3 | 16 | CEA <5.0 ng/mL | 5 | 13 |
| p = 0.004 | p = 0.58 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iwai, T.; Yamada, T.; Uehara, K.; Shinji, S.; Matsuda, A.; Yokoyama, Y.; Takahashi, G.; Miyasaka, T.; Yoshida, H. Clinical Implications of Cell-Free DNA in Managing BRAF V600E Mutation-Positive Colorectal Cancer. Genes 2025, 16, 275. https://doi.org/10.3390/genes16030275
Iwai T, Yamada T, Uehara K, Shinji S, Matsuda A, Yokoyama Y, Takahashi G, Miyasaka T, Yoshida H. Clinical Implications of Cell-Free DNA in Managing BRAF V600E Mutation-Positive Colorectal Cancer. Genes. 2025; 16(3):275. https://doi.org/10.3390/genes16030275
Chicago/Turabian StyleIwai, Takuma, Takeshi Yamada, Kay Uehara, Seiichi Shinji, Akihisa Matsuda, Yasuyuki Yokoyama, Goro Takahashi, Toshimitsu Miyasaka, and Hiroshi Yoshida. 2025. "Clinical Implications of Cell-Free DNA in Managing BRAF V600E Mutation-Positive Colorectal Cancer" Genes 16, no. 3: 275. https://doi.org/10.3390/genes16030275
APA StyleIwai, T., Yamada, T., Uehara, K., Shinji, S., Matsuda, A., Yokoyama, Y., Takahashi, G., Miyasaka, T., & Yoshida, H. (2025). Clinical Implications of Cell-Free DNA in Managing BRAF V600E Mutation-Positive Colorectal Cancer. Genes, 16(3), 275. https://doi.org/10.3390/genes16030275






