Systematic Analysis of Stay-Green Genes in Six Ipomoea Species Reveals the Evolutionary Dynamics, Carotenoid and Anthocyanin Accumulation, and Stress Responses of Sweet Potato
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of SGRs
2.2. Property Prediction of SGRs
2.3. Phylogenetic Analysis of SGRs
2.4. Domain Identification, Conserved Motif Analysis, Gene Structure Analysis of SGRs
2.5. Collinearity Analysis and Classification of Gene Duplication
2.6. Cis-Acting Element Analysis of the Promoter of SGRs
2.7. Transcriptome Analysis and Construction of Protein Interaction Network
3. Results
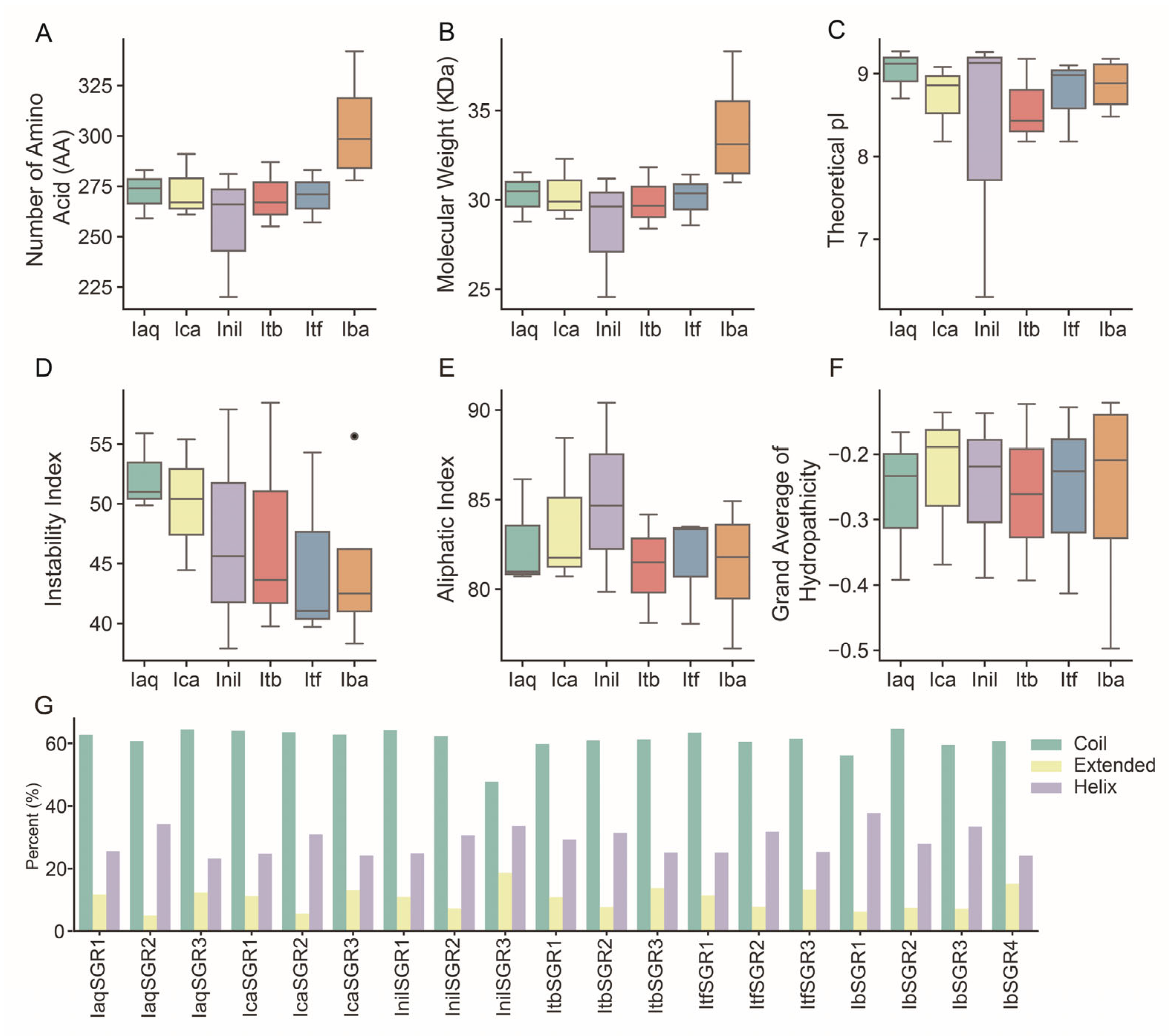
3.1. Identification and Characterization of SGRs in the Six Ipomoea Species
3.2. Evolutionary and Phylogenetic Relationship Analysis of SGRs in the Six Ipomoea Species
3.3. Conserved Domain and Motif Analysis of SGRs in the Six Ipomoea Species
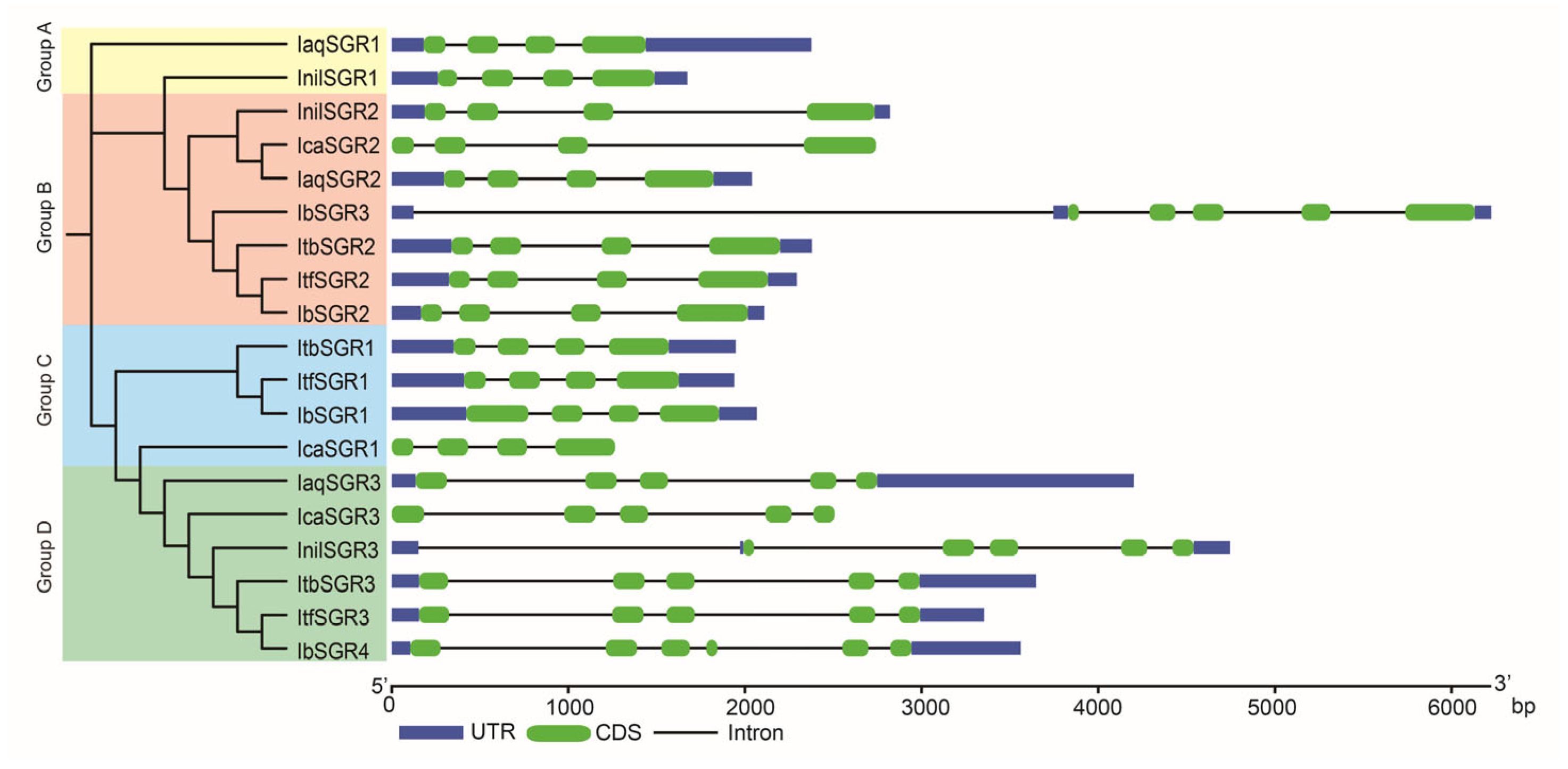
3.4. Gene Structure Analysis of SGRs in the Six Ipomoea Species
3.5. Collinearity Analysis of SGRs in the Genomes of the Six Ipomoea Species
3.6. Cis-Element Analysis in the Promoters of SGRs in the Six Ipomoea Species
3.7. Expression Patterns of IbSGRs
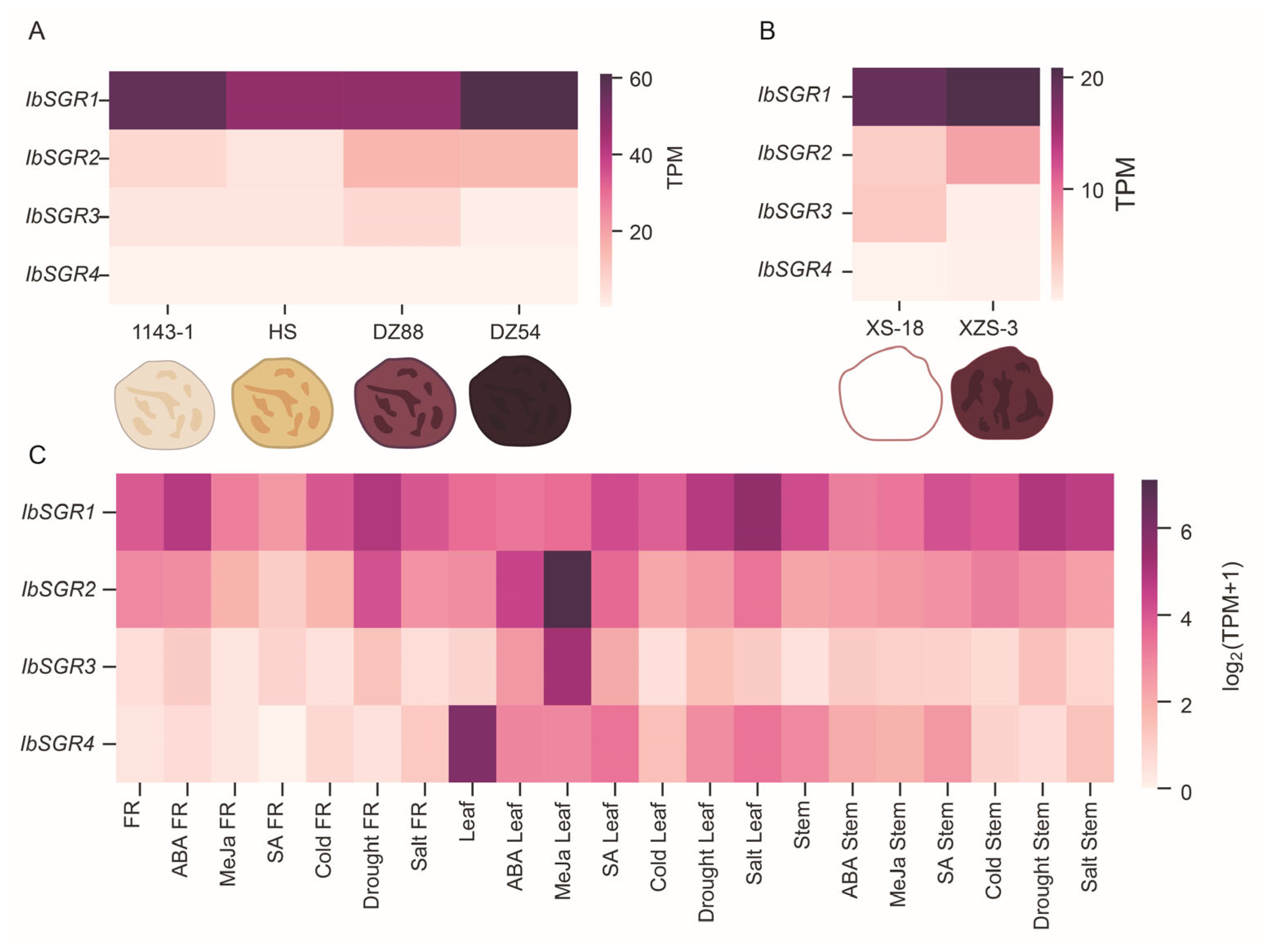
3.7.1. Expression Patterns of IbSGRs in Different Flesh-Colored Sweet Potato Varieties
3.7.2. Expression Patterns of IbSGRs in Different Hormones and Stress Responses
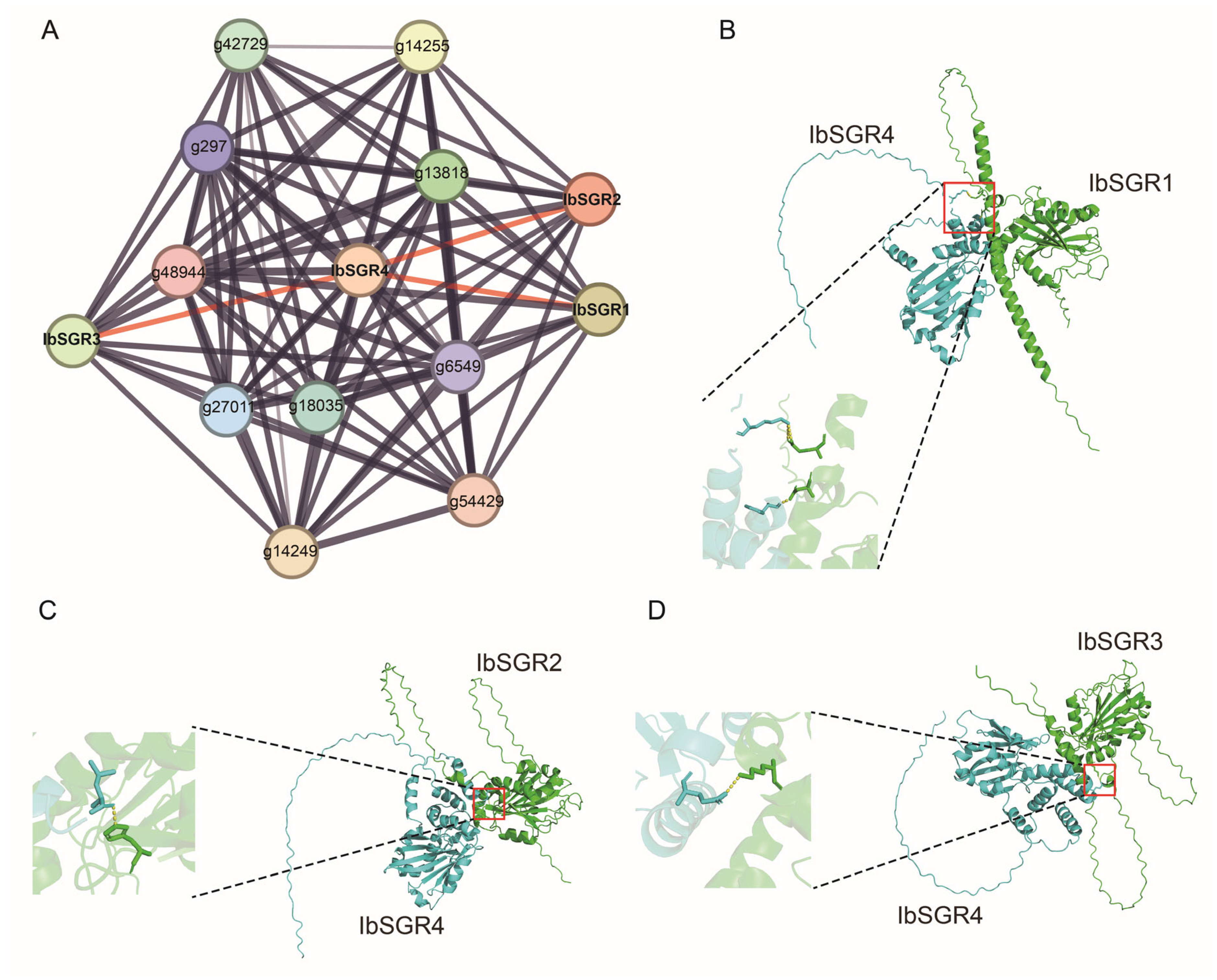
3.8. Protein Interaction Network of IbSGRs in Sweet Potato
4. Discussion
4.1. Evolution of SGRs in the Six Ipomoea Species
4.2. IbSGRs Are Involved in Carotenoid and Anthocyanin Accumulation in Sweet Potato
4.3. IbSGRs Regulate Hormone and Stress Responses in Sweet Potato
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gao, F.; Guo, J.X.; Shen, Y.Y. Advances from chlorophyll biosynthesis to photosynthetic adaptation, evolution and signaling. Plant Stress 2024, 12, 100470. [Google Scholar] [CrossRef]
- Beale, S.I. Enzymes of chlorophyll biosynthesis. Photosynth. Res. 1999, 60, 43–73. [Google Scholar] [CrossRef]
- Hörtensteiner, S.; Kräutler, B. Chlorophyll breakdown in higher plants. Biochim. Biophys. Acta (BBA)-Bioenerg. 2011, 1807, 977–988. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Yu, J.W.; Park, J.S.; Li, J.; Yoo, S.C.; Lee, N.Y.; Lee, S.K.; Jeong, S.W.; Seo, H.S.; Koh, H.J.; et al. The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 2007, 19, 1649–1664. [Google Scholar] [CrossRef] [PubMed]
- Hortensteiner, S. Stay-green regulates chlorophyll and chlorophyll-binding protein degradation during senescence. Trends Plant Sci. 2009, 14, 155–162. [Google Scholar] [CrossRef]
- Luo, J.; Abid, M.; Zhang, Y.; Cai, X.X.; Tu, J.; Gao, P.X.; Wang, Z.P.; Huang, H.W. Genome-wide identification of kiwifruit SGR family members and functional characterization of SGR2 protein for chlorophyll degradation. Int. J. Mol. Sci. 2023, 24, 1993. [Google Scholar] [CrossRef] [PubMed]
- Rong, H.; Tang, Y.Y.; Zhang, H.; Wu, P.Z.; Chen, Y.P.; Li, M.R.; Wu, G.J.; Jiang, H.W. The Stay-Green Rice like (SGRL) gene regulates chlorophyll degradation in rice. J. Plant Physiol. 2013, 170, 1367–1373. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Park, S.Y.; Kim, Y.S.; Wang, S.H.; Yoo, S.C.; Hortensteiner, S.; Paek, N.C. Arabidopsis STAY-GREEN2 is a negative regulator of chlorophyll degradation during leaf senescence. Mol. Plant 2014, 7, 1288–1302. [Google Scholar] [CrossRef] [PubMed]
- Uluisik, S.; Kıyak, A.; Kurt, F.; Filiz, E. STAY-GREEN (SGR) genes in tomato (Solanum lycopersicum): Genome-wide identification, and expression analyses reveal their involvements in ripening and salinity stress responses. Hortic. Environ. Biotechnol. 2022, 63, 557–569. [Google Scholar] [CrossRef]
- Ren, H.Z.; Yu, Y.T.; Huang, C.; Li, D.Y.; Ni, J.L.; Lv, W.Y.; Wei, K.; Wang, L.Y.; Wang, Y.C. Genome-wide identification and characterization of tea SGR family members reveal their potential roles in chlorophyll degradation and stress tolerance. Agronomy 2024, 14, 769. [Google Scholar] [CrossRef]
- Bade, R.G.; Bao, M.L.; Jin, W.Y.; Ma, Y.; Niu, Y.D.; Hasi, A. Genome-wide identification and analysis of the SGR gene family in Cucumis melo L. Genet. Mol. Res. 2016, 15, gmr15048485. [Google Scholar] [CrossRef]
- Ren, G.D.; An, K.; Liao, Y.; Zhou, X.; Cao, Y.J.; Zhao, H.F.; Ge, X.C.; Kuai, B.K. Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in Arabidopsis. Plant Physiol. 2007, 144, 1429–1441. [Google Scholar] [CrossRef]
- Cha, K.W.; Lee, Y.J.; Koh, H.J.; Lee, B.M.; Nam, Y.W.; Paek, N.C. Isolation, characterization, and mapping of the stay green mutant in rice. Theor. Appl. Genet. 2002, 104, 526–532. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.W.; Li, M.R.; Liang, N.T.; Yan, H.B.; Wei, Y.B.; Xu, X.L.; Liu, J.; Xu, Z.F.; Chen, F.; Wu, G.J. Molecular cloning and function analysis of the stay green gene in rice. Plant J. 2007, 52, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Guo, Y.J.; Kuai, B.K. Isolation and characterization of a chlorophyll degradation regulatory gene from tall fescue. Plant Cell Rep. 2011, 30, 1201–1207. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.E.; Han, L.; Pislariu, C.; Nakashima, J.; Fu, C.X.; Jiang, Q.Z.; Quan, L.; Blancaflor, E.B.; Tang, Y.H.; Bouton, J.H.; et al. From model to crop: Functional analysis of a STAY-GREEN gene in the model legume Medicago truncatula and effective use of the gene for alfalfa improvement. Plant Physiol. 2011, 157, 1483–1496. [Google Scholar] [CrossRef]
- Li, Z.P.; Wu, S.X.; Chen, J.Y.; Wang, X.Y.; Gao, J.; Ren, G.D.; Kuai, B.K. NYEs/SGRs-mediated chlorophyll degradation is critical for detoxification during seed maturation in Arabidopsis. Plant J. 2017, 92, 650–661. [Google Scholar] [CrossRef]
- Barry, C.S.; McQuinn, R.P.; Chung, M.Y.; Besuden, A.; Giovannoni, J.J. Amino acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiol. 2008, 147, 179–187. [Google Scholar] [CrossRef]
- Borovsky, Y.; Paran, I. Chlorophyll breakdown during pepper fruit ripening in the chlorophyll retainer mutation is impaired at the homolog of the senescence-inducible stay-green gene. Theor. Appl. Genet. 2008, 117, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.D.; Zhang, J.H.; Li, J.H.; Yang, C.X.; Wang, T.T.; Ouyang, B.; Li, H.X.; Giovannoni, J.; Ye, Z.B. A STAY-GREEN protein SlSGR1 regulates lycopene and β-carotene accumulation by interacting directly with SlPSY1 during ripening processes in tomato. New Phytol. 2013, 198, 442–452. [Google Scholar] [CrossRef]
- Xie, W.Y.; Xue, X.; Wang, Y.; Zhang, G.Y.; Zhao, J.H.; Zhang, H.M.; Wang, G.D.; Li, L.; Wang, Y.Q.; Shan, W.F.; et al. Natural mutation in Stay-Green (OsSGR) confers enhanced resistance to rice sheath blight through elevating cytokinin content. Plant Biotechnol. J. 2024. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.; Tan, J.Y.; Wu, Z.M.; VandenLangenberg, K.; Wehner, T.C.; Wen, C.L.; Zheng, X.Y.; Owens, K.; Thornton, A.; Bang, H.H.; et al. Staygreen, Stay Healthy: A loss-of-susceptibility mutation in the STAYGREEN gene provides durable, broad-spectrum disease resistances for over 50 years of US cucumber production. New Phytol. 2019, 221, 415–430. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.T.; Wei, G.Q.; Xue, J.Y.; Xu, J. CfSGR1 and CfSGR2 from Cryptomeria fortunei exhibit contrasting responses to hormones and abiotic stress in transgenic Arabidopsis. Plant Physiol. Biochem. 2024, 216, 109152. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Gao, J.; Zhu, X.Y.; Song, Y.; Li, Z.P.; Ren, G.D.; Zhou, X.; Kuai, B.K. ABF2, ABF3, and ABF4 promote ABA-mediated chlorophyll degradation and leaf senescence by transcriptional activation of chlorophyll catabolic genes and senescence-associated genes in Arabidopsis. Mol. Plant 2016, 9, 1272–1285. [Google Scholar] [CrossRef] [PubMed]
- Delmas, F.; Sankaranarayanan, S.; Deb, S.; Widdup, E.; Bournonville, C.; Bollier, N.; Northey, J.G.; McCourt, P.; Samuel, M.A. ABI3 controls embryo degreening through Mendel’s I locus. Proc. Natl. Acad. Sci. USA 2013, 110, E3888–E3894. [Google Scholar] [CrossRef]
- Yang, M.M.; Zhu, S.B.; Jiao, B.Z.; Duan, M.; Meng, Q.W.; Ma, N.N.; Lv, W. SlSGRL, a tomato SGR-like protein, promotes chlorophyll degradation downstream of the ABA signaling pathway. Plant Physiol. Biochem. 2020, 157, 316–327. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.Y.; Wang, Y.X.; Fan, Y.; Jiang, Z.C.; Wei, Z.H.; Zhai, H.; He, S.Z.; Zhang, H.; Yang, Y.F.; Zhao, N.; et al. IbNF-YA1 is a key factor in the storage root development of sweet potato. Plant J. 2024, 118, 1991–2002. [Google Scholar] [CrossRef]
- Zhang, X.B.; Tang, C.C.; Jiang, B.Z.; Zhang, R.; Li, M.; Wu, Y.Y.; Yao, Z.F.; Huang, L.F.; Luo, Z.X.; Zou, H.D.; et al. Refining polyploid breeding in sweet potato through allele dosage enhancement. Nat. Plants 2024, 11, 36–48. [Google Scholar] [CrossRef]
- Nguyen, H.C.; Chen, C.C.; Lin, K.H.; Chao, P.Y.; Lin, H.H.; Huang, M.Y. Bioactive compounds, antioxidants, and health benefits of sweet potato leaves. Molecules 2021, 26, 1820. [Google Scholar] [CrossRef]
- Jiang, Z.C.; Wei, Z.H.; Zhang, J.; Zheng, C.X.; Zhu, H.; Zhai, H.; He, S.Z.; Gao, S.P.; Zhao, N.; Zhang, H.; et al. Source-sink synergy is the key unlocking sweet potato starch yield potential. Nat. Commun. 2024, 15, 7260. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.X.; Wang, J.; Dai, W.W.; Du, M.J.; Dai, X.B.; Zhou, Z.L.; He, H.; Yuan, B.; Zhao, D.L.; Cao, Q.H. Comprehensive characterization of nutritional components in sweetpotato (Ipomoea batatas [L]. Lam.) during long-term post-harvest storage. J Plant. Physiol. 2024, 304, 154404. [Google Scholar] [CrossRef] [PubMed]
- Low, J.W.; Thiele, G. Understanding innovation: The development and scaling of orange-fleshed sweetpotato in major African food systems. Agric. Syst. 2020, 179, 102770. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.M.; Li, R.S.; Ren, L.; Gao, X.L.; Zhang, Y.G.; Ma, Z.M.; Ma, D.F.; Luo, Y.H. A comparative metabolomics study of flavonoids in sweet potato with different flesh colors (Ipomoea batatas (L.) Lam). Food Chem. 2018, 260, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.N.; Nusrat, T.; Begum, P.; Ahsan, M. Carotenoids and β-carotene in orange fleshed sweet potato: A possible solution to vitamin A deficiency. Food Chem. 2016, 199, 628–631. [Google Scholar] [CrossRef] [PubMed]
- Li, R.J.; Kang, C.; Song, X.J.; Yu, L.; Liu, D.G.; He, S.Z.; Zhai, H.; Liu, Q.C. A ζ-carotene desaturase gene, IbZDS, increases β-carotene and lutein contents and enhances salt tolerance in transgenic sweetpotato. Plant Sci. 2017, 262, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Kou, M.; Li, C.; Zhang, Y.G. Comparative transcriptome analysis reveals candidate genes involved in anthocyanin biosynthesis in sweetpotato (Ipomoea batatas L.). Plant Physiol. Biochem. 2021, 158, 508–517. [Google Scholar] [CrossRef] [PubMed]
- Khoo, H.E.; Azlan, A.; Tang, S.T.; Lim, S.M. Anthocyanidins and anthocyanins: Colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res. 2017, 61, 1361779. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.B.; Xu, X.; Hu, X.W.; Liu, Y.D.; Cao, H.H.; Chan, H.L.; Gong, Z.H.; Yuan, Y.J.; Luo, Y.Q.; Feng, B.H.; et al. SlMYB72 regulates the metabolism of chlorophylls, carotenoids, and flavonoids in tomato fruit. Plant Physiol. 2020, 183, 854–868. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Y.; Ji, H.Y.; Huang, W.K.; Zhang, Z.H.; Zhu, K.J.; Zhu, S.P.; Chai, L.J.; Ye, J.L.; Deng, X.X. Transcription factor CrWRKY42 coregulates chlorophyll degradation and carotenoid biosynthesis in citrus. Plant Physiol. 2024, 195, 728–744. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.H.; Li, R.J.; Zhao, H.Q.; Zhai, H.; He, S.Z.; Zhang, H.; Zhou, Y.Y.; Zhao, N.; Gao, S.P.; Liu, Q.C. The transcription factor IbNAC29 positively regulates the carotenoid accumulation in sweet potato. Hortic. Res. 2023, 10, uhad010. [Google Scholar] [CrossRef]
- Hoie, M.H.; Kiehl, E.N.; Petersen, B.; Nielsen, M.; Winther, O.; Nielsen, H.; Hallgren, J.; Marcatili, P. NetSurfP-3.0: Accurate and fast prediction of protein structural features by protein language models and deep learning. Nucleic Acids Res. 2022, 50, W510–W515. [Google Scholar] [CrossRef]
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; DeBarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.-H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Chen, C.J.; Wu, Y.; Li, J.W.; Wang, X.; Zeng, Z.H.; Xu, J.; Liu, Y.L.; Feng, J.T.; Chen, H.; He, Y.H.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Xiao, J.P.; Xu, X.Y.; Li, M.X.; Wu, X.J.; Guo, H.C. Regulatory network characterization of anthocyanin metabolites in purple sweetpotato via joint transcriptomics and metabolomics. Front. Plant Sci. 2023, 14, 1030236. [Google Scholar] [CrossRef] [PubMed]
- He, L.H.; Liu, S.F.; Zhang, Y.; Sun, Y.; Tang, R.M.; Wang, W.B.; Cui, H.L.; Li, R.Z.; Jia, X.Y. Transcriptomic and targeted metabolomic analysis identifies genes and metabolites involved in anthocyanin accumulation in tuberous roots of sweetpotato (Ipomoea batatas L.). Plant Physiol. Biochem. 2020, 156, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Z.; He, P.W.; Xu, X.M.; Lü, Z.F.; Cui, P.; George, M.S.; Lu, G.Q. Genome-wide identification and expression analysis of the xyloglucan endotransglucosylase/hydrolase gene family in sweet potato [Ipomoea batatas (L.) Lam]. Int. J. Mol. Sci. 2023, 24, 775. [Google Scholar] [CrossRef]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Fong, J.H.; Marchler-Bauer, A. Protein subfamily assignment using the conserved domain database. BMC Res. Notes 2008, 1, 114. [Google Scholar] [CrossRef]
- Ramola, R.; Friedberg, I.; Radivojac, P. The field of protein function prediction as viewed by different domain scientists. Bioinform. Adv. 2022, 2, vbac057. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.W.; Dou, Y.T.; Pan, L.R.; Xu, L.W.; Peng, S.L. Improving prediction performance of general protein language model by domain-adaptive pretraining on DNA-binding protein. Nat. Commun. 2024, 15, 7838. [Google Scholar] [CrossRef] [PubMed]
- Aubry, S.; Mani, J.; Hortensteiner, S. Stay-green protein, defective in Mendel’s green cotyledon mutant, acts independent and upstream of pheophorbide a oxygenase in the chlorophyll catabolic pathway. Plant Mol. Biol. 2008, 6, 243–256. [Google Scholar] [CrossRef] [PubMed]
- Jiao, B.; Meng, Q.; Lv, W. Roles of stay-green (SGR) homologs during chlorophyll degradation in green plants. Bot. Stud. 2020, 61, 25. [Google Scholar] [CrossRef] [PubMed]
- Frugoli, J.A.; McPeek, M.A.; Thomas, T.L.; McClung, C. R Intron loss and gain during evolution of the catalase gene family in angiosperms. Genetics 1998, 14, 355–365. [Google Scholar] [CrossRef]
- Shimoda, Y.; Ito, H.; Tanaka, A. Arabidopsis STAY-GREEN, mendel’s green cotyledon gene, encodes magnesium-dechelatase. Plant Cell 2016, 28, 2147–2160. [Google Scholar] [CrossRef] [PubMed]
- Biswal, B. Carotenoid catabolism during leaf senescence and its control by light. J. Photochem. Photobiol. B Biol. 1995, 30, 3–13. [Google Scholar] [CrossRef]
- Fukao, Y. Protein-protein interactions in plants. Plant Cell Physiol. 2012, 53, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Dhatterwal, P.; Basu, S.; Mehrotra, S.; Mehrotra, R. Genome wide analysis of W-box element in Arabidopsis thaliana reveals TGAC motif with genes down regulated by heat and salinity. Sci. Rep. 2019, 9, 1681. [Google Scholar] [CrossRef]
- Yuan, Y.; Ren, S.Y.; Liu, X.F.; Su, L.Y.; Wu, Y.; Zhang, W.; Li, Y.; Jiang, Y.D.; Wang, H.H.; Fu, R.; et al. SlWRKY35 positively regulates carotenoid biosynthesis by activating the MEP pathway in tomato fruit. New Phytol. 2022, 234, 164–178. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.J.; Wu, M.; Cao, L.Y.; Yuan, W.J.; Dong, M.F.; Wang, X.H.; Chen, W.C.; Shang, F.D. Characterization of OfWRKY3, a transcription factor that positively regulates the carotenoid cleavage dioxygenase gene OfCCD4 in Osmanthus fragrans. Plant Mol. Biol. 2016, 91, 485–496. [Google Scholar] [CrossRef] [PubMed]
- Su, M.Y.; Zuo, W.F.; Wang, Y.C.; Liu, W.J.; Zhang, Z.Y.; Wang, N.; Chen, X.S. The WKRY transcription factor MdWRKY75 regulates anthocyanins accumulation in apples (Malus domestica). Funct. Plant Biol. 2022, 49, 799–809. [Google Scholar] [CrossRef]
- Yan, H.L.; Pei, X.N.; Zhang, H.; Li, X.; Zhang, X.X.; Zhao, M.H.; Chiang, V.L.; Sederoff, R.R.; Zhao, X.Y. MYB-Mediated regulation of anthocyanin biosynthesis. Int. J. Mol. Sci. 2021, 22, 3103. [Google Scholar] [CrossRef] [PubMed]
- Li, C.X.; Yu, W.J.; Xu, J.R.; Lu, X.F.; Liu, Y.Z. Anthocyanin biosynthesis induced by MYB transcription factors in plants. Int. J. Mol. Sci. 2022, 23, 11701. [Google Scholar] [CrossRef] [PubMed]
- Jian, W.; Cao, H.H.; Yuan, S.; Liu, Y.D.; Lu, J.F.; Lu, W.; Li, N.; Wang, J.H.; Zou, J.; Tang, N.; et al. SlMYB75, an MYB-type transcription factor, promotes anthocyanin accumulation and enhances volatile aroma production in tomato fruits. Hortic. Res. 2019, 6, 22. [Google Scholar] [CrossRef]
- Vimolmangkang, S.; Han, Y.P.; Wei, G.C.; Korban, S.S. An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biol. 2013, 13, 176. [Google Scholar] [CrossRef]
- Liu, Y.; Feng, X.; Jin, H.B.; Zhang, Y.T.; Tong, X.R.; Zhu, P.F. BoMYBL2b, an R3-MYB transcription factor, inhibits anthocyanin accumulation via directly repressing BoDFR1 gene transcription in kale. Plant Physiol. Biochem. 2024, 216, 109140. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.H.; Wang, X.Q.; Yu, C.Y.; Wang, C.T.; Jin, Y.L.; Zhang, H.X. MYB transcription factor PdMYB118 directly interacts with bHLH transcription factor PdTT8 to regulate wound-induced anthocyanin biosynthesis in poplar. BMC Plant Biol. 2020, 20, 173. [Google Scholar] [CrossRef]
- Abdelrahman, M.; El-Sayed, M.; Jogaiah, S.; Burritt, D.J.; Tran, L.P. The “STAY-GREEN” trait and phytohormone signaling networks in plants under heat stress. Plant Cell Rep. 2017, 36, 1009–1025. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Kim, D.; Kim, Y.S.; Hortensteiner, S.; Paek, N.C. Arabidopsis STAYGREEN-LIKE (SGRL) promotes abiotic stress-induced leaf yellowing during vegetative growth. FEBS Lett. 2014, 588, 3830–3837. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zuo, Z.; Ma, H.; Li, L.; Qian, J.; Zhang, M.; Li, X.; Sheng, Y.; Wang, Y. Systematic Analysis of Stay-Green Genes in Six Ipomoea Species Reveals the Evolutionary Dynamics, Carotenoid and Anthocyanin Accumulation, and Stress Responses of Sweet Potato. Genes 2025, 16, 266. https://doi.org/10.3390/genes16030266
Zuo Z, Ma H, Li L, Qian J, Zhang M, Li X, Sheng Y, Wang Y. Systematic Analysis of Stay-Green Genes in Six Ipomoea Species Reveals the Evolutionary Dynamics, Carotenoid and Anthocyanin Accumulation, and Stress Responses of Sweet Potato. Genes. 2025; 16(3):266. https://doi.org/10.3390/genes16030266
Chicago/Turabian StyleZuo, Zhidan, Huihui Ma, Longteng Li, Jialin Qian, Minghui Zhang, Xiang Li, Yeshun Sheng, and Yuxin Wang. 2025. "Systematic Analysis of Stay-Green Genes in Six Ipomoea Species Reveals the Evolutionary Dynamics, Carotenoid and Anthocyanin Accumulation, and Stress Responses of Sweet Potato" Genes 16, no. 3: 266. https://doi.org/10.3390/genes16030266
APA StyleZuo, Z., Ma, H., Li, L., Qian, J., Zhang, M., Li, X., Sheng, Y., & Wang, Y. (2025). Systematic Analysis of Stay-Green Genes in Six Ipomoea Species Reveals the Evolutionary Dynamics, Carotenoid and Anthocyanin Accumulation, and Stress Responses of Sweet Potato. Genes, 16(3), 266. https://doi.org/10.3390/genes16030266





