Transcriptome Analysis of Environmental Adaptation of Largemouth Bass (Micropterus salmonides)
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. cDNA Library Preparation and Sequencing
2.3. Transcriptome Analysis
2.4. Analysis of Gene Expression Levels and Alternative Splicing Genes
2.5. Analysis of Gene Expression Differences
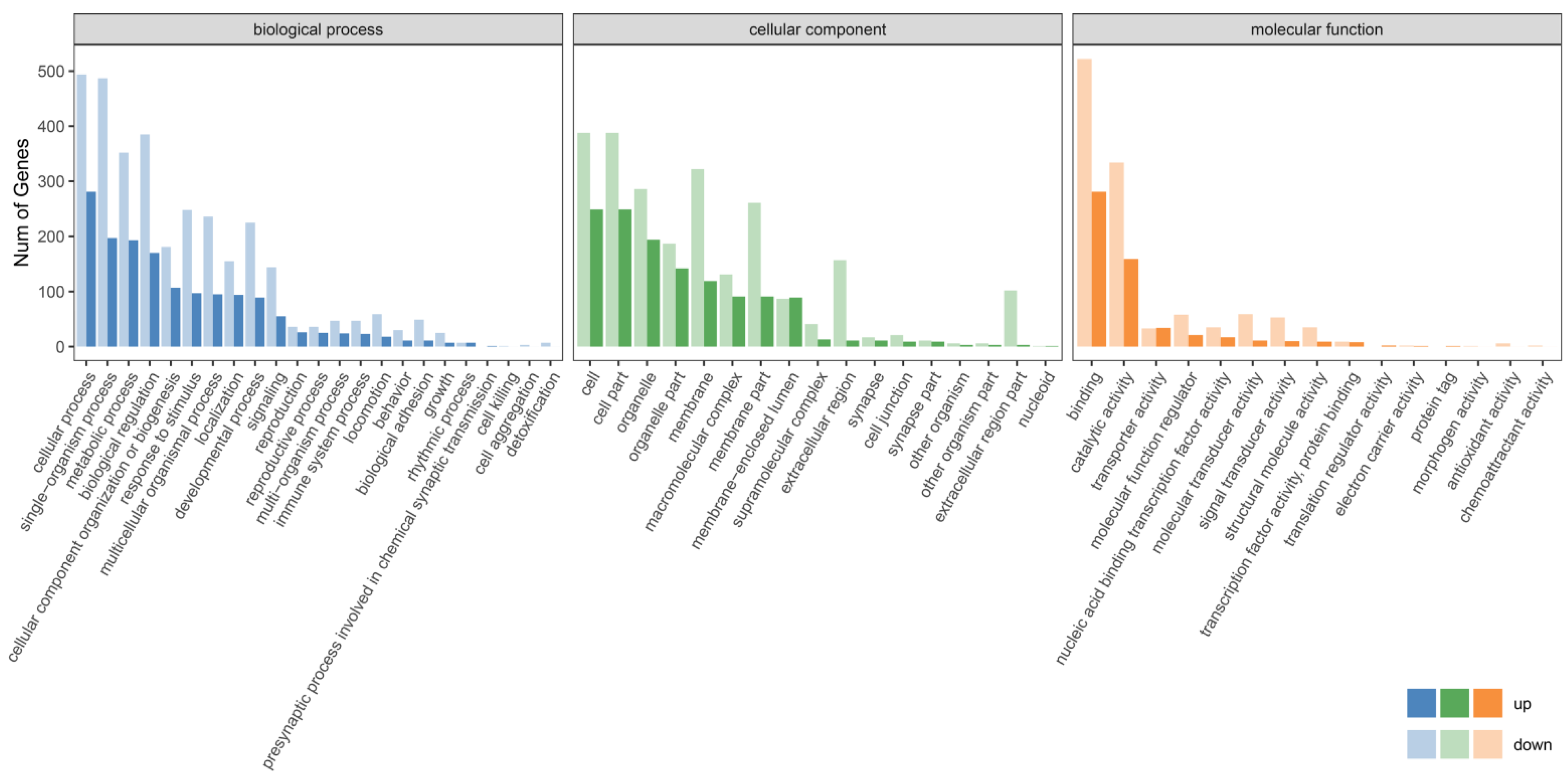
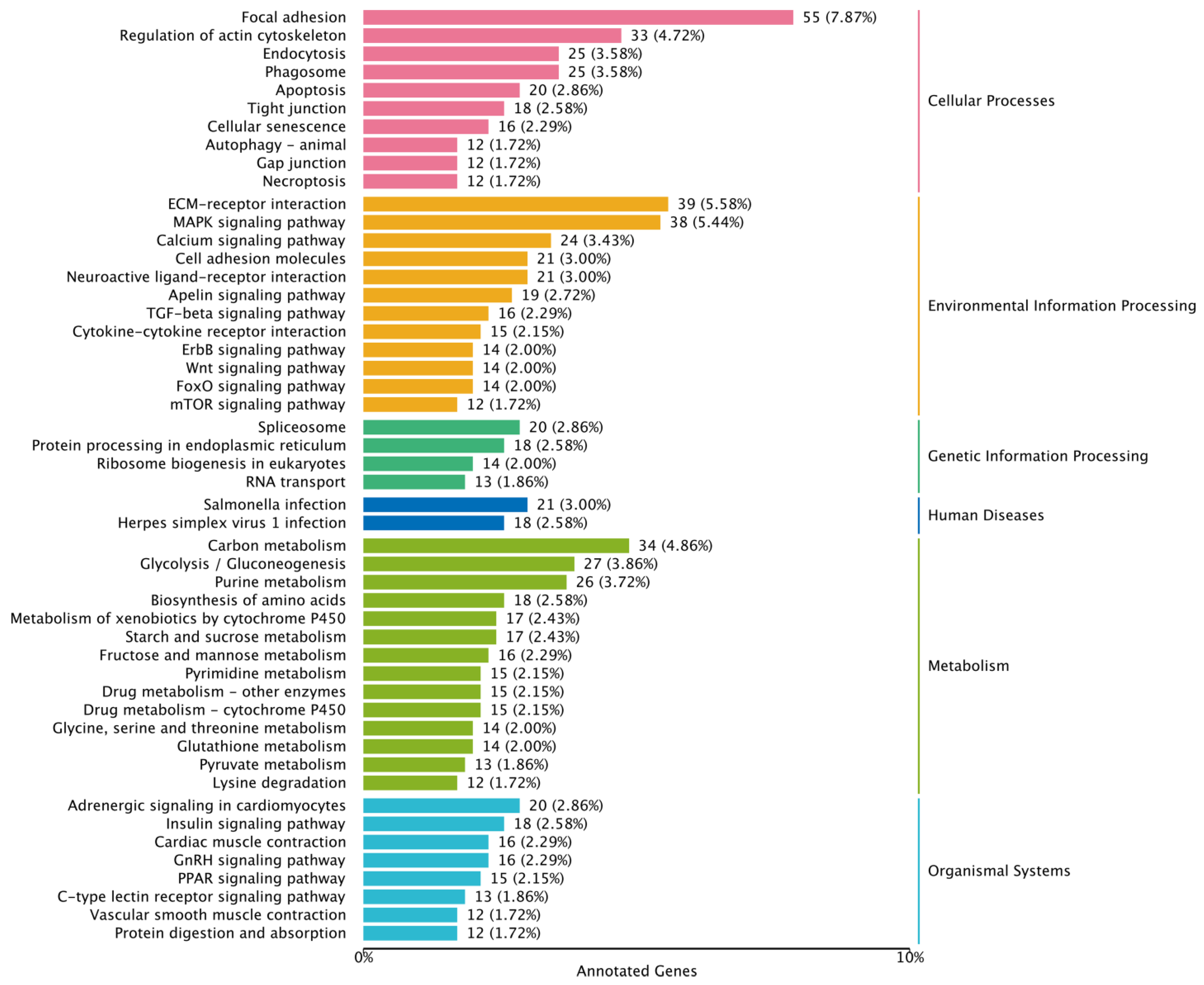
2.6. Functional Analysis of Differentially Expressed Genes
3. Results
3.1. Statistics of High-Throughput Sequencing Data
3.2. Alternative Splicing (AS) Analysis
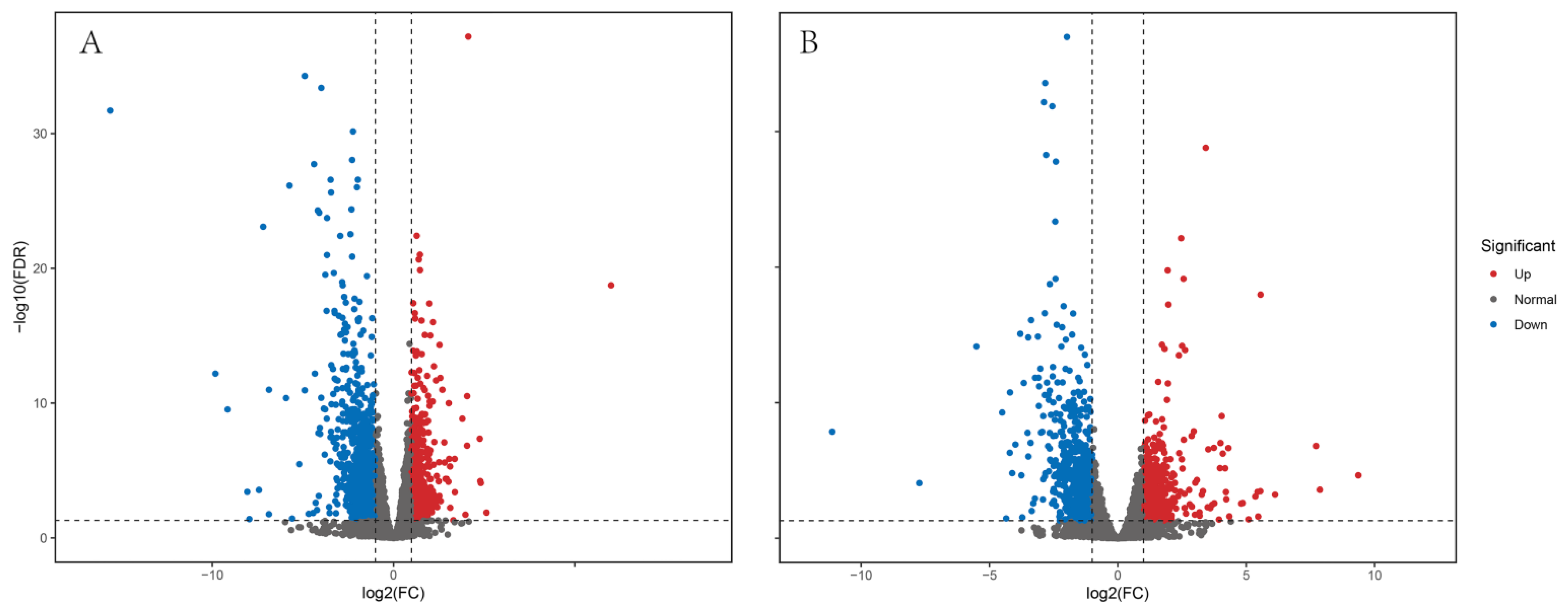
3.3. Differentially Expressed Gene (DEG) Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sun, C.; Li, J.; Dong, J.; Niu, Y.; Hu, J.; Lian, J.; Li, W.; Li, J.; Tian, Y.; Shi, Q.; et al. Chromosome-level genome assembly for the largemouth bass Micropterus salmoides provides insights into adaptation to fresh and brackish water. Mol. Ecol. Resour. 2021, 21, 301–315. [Google Scholar] [CrossRef]
- Lin, Z.; Cai, Z.; Li, L.; Wei, Y.; Ling, Q. c-Jun N-terminal kinase 1/P53 signaling mediates intrinsic apoptosis of largemouth bass (Micropterus salmoides) hepatocytes under heat stress. Sci. Total Environ. 2024, 947, 174664. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhao, J.; An, N.; Li, D.C.; Huang, M.M.; Fei, H. Updates on infectious diseases of largemouth bass: A major review. Fish Shellfish Immunol. 2024, 154, 109976. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, J.; Ao, H.; Liu, L.; Chen, Y. Effects of extruded and pelleted diets with different protein levels on growth performance and nutrient retention of largemouth bass (Micropterus salmoides). Aquac. Rep. 2023, 29, 101479. [Google Scholar] [CrossRef]
- Wang, K.-W.; Liu, Q.-Q.; Zhu, J.; Deng, X.; Luo, L.; Lin, S.-M.; Qin, C.-J.; Chen, Y.-J. Transcriptome analysis provides insights into the molecular mechanism of liver inflammation and apoptosis in juvenile largemouth bass Micropterus salmoides fed low protein high starch diets. Comp. Biochem. Physiol. Part D Genom. Proteom. 2023, 45, 101047. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Liu, Y.; Bao, X.; Yue, Y.; Tong, B.; Yang, X.; Yu, H.; Yang, Y.; Liu, Y.; Yu, Y. Potential of Chinese Yam (Dioscorea polystachya Turczaninow) By-Product as a Feed Additive in Largemouth bass (Micropterus salmoides): Turning Waste into Valuable Resources. Aquac. Nutr. 2023, 2023, 9983499. [Google Scholar] [CrossRef]
- Wang, M.; Wang, Y.; Li, X.; Yin, Y.; Zhang, X.; Wu, S.; Wang, H.; Zhao, Y. Effects of Dietary Ursolic Acid on Growth Performance and Intestinal Health of Largemouth bass (Micropterus salmoides). Animals 2024, 14, 2492. [Google Scholar] [CrossRef]
- Li, B.Y.; Qin, J.C.; Shen, Y.F.; Yang, F.; Wang, T.; Ling, F.; Wang, G.X. A therapeutic agent of ursolic acid demonstrates potential application in aquaculture. Virus Res. 2023, 323, 198965. [Google Scholar] [CrossRef]
- Yang, M.; Jiang, D.; Lai, W.; Chen, K.; Zhang, L.; Lu, L.; Xu, Y.; Liu, Y.; Khan, M.S.; Jiang, J. Hydrolysable tannin improves growth performance and liver health of largemouth bass (Micropterus salmoides) fed high soybean meal diets. Int. J. Biol. Macromol. 2024, 276, 133773. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, M.; Li, N.; Dong, Z.; Cai, L.; Wu, B.; Xie, J.; Liu, L.; Ren, L.; Shi, B. New insights into β-glucan-enhanced immunity in largemouth bass Micropterus salmoides by transcriptome and intestinal microbial composition. Front. Immunol. 2022, 13, 1086103. [Google Scholar] [CrossRef]
- Zhao, F.; Huo, X.; Wang, P.; Liu, Q.; Yang, C.; Su, J. The Combination of β-Glucan and Astragalus Polysaccharide Effectively Resists Nocardia seriolae Infection in Largemouth bass (Micropterus salmoides). Microorganisms 2023, 11, 2529. [Google Scholar] [CrossRef]
- Xv, Z.; Chen, S.; Song, G.; Hu, H.; Lin, S.; Long, Y. Biochemical, histological and transcriptomic analyses for the immunological organs provide insights into heat stress-induced disease susceptibility in Largemouth bass. Sci. Total Environ. 2024, 912, 168758. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, Y.; Wang, Z.; Luo, T.; Huang, J.; Shao, J. Analysis of Differential Alternative Splicing in Largemouth bass After High Temperature Exposure. Animals 2024, 14, 3005. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Li, Z.; Yan, H.; Shi, L.; Yang, H.; Liu, Q.; Song, K.; Hu, Y.; Wang, B.; Yang, S.; et al. Cold temperature delays ovarian development of largemouth bass by inhibiting sex hormone release, angiogenesis, apoptosis and autophagy during out-of-season reproduction. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2024, 301, 111795. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J.; Tan, Y.; Wang, X.; Chen, H.; Yu, H.; Chen, F.; Yan, X.; Sun, J.; Luo, J.; et al. Kidney transcriptome analysis reveals the molecular responses to salinity adaptation in largemouth bass (Micropterus salmoides). Comp. Biochem. Physiol. Part D Genom. Proteom. 2025, 53, 101362. [Google Scholar] [CrossRef]
- Yan, D.; Gan, L.; Dong, X.; Tie, H.; Luo, C.; Wang, Z.; Jiang, H.; Chen, J.; An, M.; Qin, C.; et al. The impact of crowding stress on growth and intestinal integrity in largemouth bass (Micropterus salmoides): Insights into ER stress, autophagy and apoptosis. Fish Shellfish Immunol. 2024, 154, 109955. [Google Scholar] [CrossRef]
- Song, Z.; Ye, W.; Tao, Y.; Zheng, T.; Qiang, J.; Li, Y.; Liu, W.; Xu, P. Transcriptome and 16S rRNA Analyses Reveal That Hypoxic Stress Affects the Antioxidant Capacity of Largemouth bass (Micropterus salmoides), Resulting in Intestinal Tissue Damage and Structural Changes in Microflora. Antioxidants 2022, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Tang, X.; Huang, R.; Liu, Q.; Liao, L.; Hu, Y.; He, K.; Zhang, X.; Guo, J.; Chen, S.; et al. Acute hypoxia promotes the liver angiogenesis of largemouth bass (Micropterus salmoides) by HIF—Dependent pathway. Fish Shellfish Immunol. 2022, 131, 264–273. [Google Scholar] [CrossRef]
- Zhao, L.L.; Liao, L.; Yan, H.X.; Tang, X.H.; He, K.; Liu, Q.; Luo, J.; Du, Z.J.; Chen, S.Y.; Zhang, X.; et al. Physiological responses to acute hypoxia in the liver of largemouth bass by alteration of mitochondrial function and Ca2+ exchange. Aquat. Toxicol. 2023, 256, 106436. [Google Scholar] [CrossRef]
- Sun, J.L.; Zhao, L.L.; Liao, L.; Tang, X.H.; Cui, C.; Liu, Q.; He, K.; Ma, J.D.; Jin, L.; Yan, T.; et al. Interactive effect of thermal and hypoxia on largemouth bass (Micropterus salmoides) gill and liver: Aggravation of oxidative stress, inhibition of immunity and promotion of cell apoptosis. Fish Shellfish Immunol. 2020, 98, 923–936. [Google Scholar] [CrossRef]
- Yu, H.; He, Y.; Zhang, J.; Zhang, Z.; Zhang, X. Hepatic transcriptome analysis reveals the metabolic strategies of largemouth bass (Micropterus salmoides) under different dissolved oxygen condition. Comp. Biochem. Physiol. Part D Genom. Proteom. 2023, 45, 101032. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Yan, H.; Cheng, L.; He, K.; Liu, Q.; Luo, J.; Luo, W.; Zhang, X.; Yan, T.; Du, Z.; et al. Metabolic response provides insights into the mechanism of adaption to hypoxia in largemouth bass (Micropterus salmoides) under intermittent hypoxic conditions. Ecotoxicol. Environ. Saf. 2022, 242, 113957. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.L.; Zhao, L.L.; He, K.; Liu, Q.; Luo, J.; Zhang, D.M.; Liang, J.; Liao, L.; Ma, J.D.; Yang, S. MicroRNA regulation in hypoxic environments: Differential expression of microRNAs in the liver of largemouth bass (Micropterus salmoides). Fish Physiol. Biochem. 2020, 46, 2227–2242. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.L.; Zhao, L.L.; Wu, H.; Liu, Q.; Liao, L.; Luo, J.; Lian, W.Q.; Cui, C.; Jin, L.; Ma, J.D.; et al. Acute hypoxia changes the mode of glucose and lipid utilization in the liver of the largemouth bass (Micropterus salmoides). Sci. Total Environ. 2020, 713, 135157. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, H.; Ge, J.; Luo, J.; He, K.; Yan, H.; Zhang, X.; Tahir, R.; Luo, W.; Li, Z.; et al. Enhance energy supply of largemouth bass (Micropterus salmoides) in gills during acute hypoxia exposure. Fish Physiol. Biochem. 2022, 48, 1649–1663. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Florea, L.; Song, L.; Salzberg, S.L. Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Research 2013, 2, 188. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Middaugh, C.R.; Alfermann, T.; Strickland, P.A.; Nguyen, P. A Regional Evaluation of Suwannee bass and Largemouth bass Exploitation in North Florida Rivers. North Am. J. Fish. Manag. 2016, 36, 958–963. [Google Scholar] [CrossRef]
- Zhou, F.; Qi, M.; Li, J.; Huang, Y.; Chen, X.; Liu, W.; Yao, G.; Meng, Q.; Zheng, T.; Wang, Z.; et al. Comparative Transcriptomic Analysis of Largemouth bass (Micropterus salmoides) Livers Reveals Response Mechanisms to High Temperatures. Genes 2023, 14, 2096. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Ye, W.; Tao, Y.; Li, Y.; Lu, S.; Xu, P.; Qiang, J. Transport Stress Induces Oxidative Stress and Immune Response in Juvenile Largemouth bass (Micropterus salmoides): Analysis of Oxidative and Immunological Parameters and the Gut Microbiome. Antioxidants 2023, 12, 157. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Tang, G.; Xiong, C.; Han, S.; Yang, C.; He, K.; Liu, Q.; Luo, J.; Luo, W.; Wang, Y.; et al. Chronic chlorpyrifos exposure induces oxidative stress, apoptosis and immune dysfunction in largemouth bass (Micropterus salmoides). Environ. Pollut. 2021, 282, 117010. [Google Scholar] [CrossRef] [PubMed]
- Kijewska, A.; Malachowicz, M.; Wenne, R. Alternatively spliced variants in Atlantic cod (Gadus morhua) support response to variable salinity environment. Sci. Rep. 2018, 8, 11607. [Google Scholar] [CrossRef]
- Yu, Z.; Huang, X.; Wen, S.; Cao, H.; Wang, N.; Shen, S.; Ding, M. Alternative Splicing under Cold Stress in Paper Mulberry. Plants 2023, 12, 3950. [Google Scholar] [CrossRef] [PubMed]
- Lei, C.; Song, H.; Song, H.; Zhu, T.; Du, J.; Li, S. RNA-seq and whole-genome re-sequencing reveal Micropterus salmoides growth-linked gene and selection signatures under carbohydrate-rich diet and varying temperature. Sci. Rep. 2024, 14, 25184. [Google Scholar] [CrossRef]
- Zhao, L.L.; Wu, H.; Sun, J.L.; Liao, L.; Cui, C.; Liu, Q.; Luo, J.; Tang, X.H.; Luo, W.; Ma, J.D.; et al. MicroRNA-124 regulates lactate transportation in the muscle of largemouth bass (Micropterus salmoides) under hypoxia by targeting MCT1. Aquat. Toxicol. 2020, 218, 105359. [Google Scholar] [CrossRef]
- Ding, J.; Liu, C.; Luo, S.; Zhang, Y.; Gao, X.; Wu, X.; Shen, W.; Zhu, J. Transcriptome and physiology analysis identify key metabolic changes in the liver of the large yellow croaker (Larimichthys crocea) in response to acute hypoxia. Ecotoxicol. Environ. Saf. 2020, 189, 109957. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Tian, F.; Qi, D.; Qi, H.; Wang, Y.; Xu, S.; Zhao, K. Physiological, metabolomic, and transcriptomic reveal metabolic pathway alterations in Gymnocypris przewalskii due to cold exposure. BMC Genom. 2023, 24, 545. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Sun, J.; Quan, J.; Li, L.; Zhao, G.; Lu, J. Effect of selenium nanoparticles on alternative splicing in heat-stressed rainbow trout primary hepatocytes. Comp. Biochem. Physiol. Part D Genom. Proteom. 2023, 45, 101042. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Z.; Quan, J.; Lu, J.; Zhao, G.; Pan, Y. Effect of Selenium Nanoparticles on Alternative Splicing of Rainbow Trout Head Kidney under Heat Stress. Mar. Biotechnol. 2024, 27, 16. [Google Scholar] [CrossRef]
- Hussain, S.S.; Abbas, M.; Abbas, S.; Wei, M.; El-Sappah, A.H.; Sun, Y.; Li, Y.; Ragauskas, A.J.; Li, Q. Alternative splicing: Transcriptional regulatory network in agroforestry. Front. Plant Sci. 2023, 14, 1158965. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Du, J.; Li, S.; Lei, C.; Zhu, T.; Han, L.; Song, H. Chronic heat stress is capable of reducing the growth performance, causing damage to the liver structure, and altering the liver glucose metabolism and lipid metabolism in largemouth bass (Micropterus salmoides L.). Fish Physiol. Biochem. 2025, 51, 24. [Google Scholar] [CrossRef]
- Kabir, J.; Lobo, M.; Zachary, I. Staurosporine induces endothelial cell apoptosis via focal adhesion kinase dephosphorylation and focal adhesion disassembly independent of focal adhesion kinase proteolysis. Biochem. J. 2002, 367, 145–155. [Google Scholar] [CrossRef]
- Vale, C.; Botana, L.M. Marine toxins and the cytoskeleton: Okadaic acid and dinophysistoxins. FEBS J. 2008, 275, 6060–6066. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Hu, X.; Lian, Z.; Luo, Z.; Lv, A.; Tan, J. Focal adhesion signaling pathway involved in skin immune response of tongue sole Cynoglossus semilaevis to Vibrio vulnificus infection. Fish Shellfish Immunol. 2023, 135, 108651. [Google Scholar] [CrossRef] [PubMed]
- Kuo, J.C.; Han, X.; Yates, J.R., 3rd; Waterman, C.M. Isolation of focal adhesion proteins for biochemical and proteomic analysis. Methods Mol. Biol. 2012, 757, 297–323. [Google Scholar]
- Wei, X.; Zhang, Y.; Li, C.; Ai, K.; Li, K.; Li, H.; Yang, J. The evolutionarily conserved MAPK/Erk signaling promotes ancestral T-cell immunity in fish via c-Myc-mediated glycolysis. J. Biol. Chem. 2020, 295, 3000–3016. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Yu, M.; Chu, X.; Li, W.; Yin, X.; Chen, L. Cold-induced retrotransposition of fish LINEs. J. Genet. Genom. 2017, 44, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Wang, S.; Liu, Y.; Chu, P.; Han, B.; Ning, X.; Wang, T.; Yin, S. Acute cold stress leads to zebrafish ovarian dysfunction by regulating miRNA and mRNA. Comp. Biochem. Physiol. Part D Genom. Proteom. 2023, 48, 101139. [Google Scholar] [CrossRef] [PubMed]
- Fei, H.; Yi, S.F.; Zhang, H.M.; Cheng, Y.; Zhang, Y.Q.; Yu, X.; Qian, S.C.; Huang, M.M.; Yang, S. Transcriptome and 16S rRNA analysis revealed the response of largemouth bass (Micropterus salmoides) to Rhabdovirus infection. Front. Immunol. 2022, 13, 973422. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Ouyang, X.; Liao, Z.; Huang, T.; Tong, G.; Tan, H.; Zhou, M.; Lu, X.; Wei, X.; Yang, X.; et al. Comprehensive transcriptomic, proteomic, and intestinal microbiota analyses of largemouth bass (Micropterus salmoides) intestines reveal new insights into immune responses to Aeromonas hydrophila infection. Fish Shellfish Immunol. 2025, 156, 110057. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, L.; Zhou, F.; Li, J.; Wu, X.; Zhong, X.; Lv, H.; Yi, S.; Gao, Q.; Yang, Z.; et al. Integrated comparative transcriptome and weighted gene co-expression network analysis provide valuable insights into the response mechanisms of crayfish (Procambarus clarkii) to copper stress. J. Hazard. Mater. 2023, 448, 130820. [Google Scholar] [CrossRef] [PubMed]



| Group | Sample | Total Reads | ReadSum | BaseSum | GC (%) | Q30 (%) |
|---|---|---|---|---|---|---|
| Female group in Huzhou (HZ-F) | HZ-F1 | 52,980,370 | 26,490,185 | 7,930,807,418 | 50.90 | 95.29 |
| HZ-F2 | 52,141,432 | 26,070,716 | 7,799,815,066 | 50.84 | 95.43 | |
| HZ-F3 | 42,252,350 | 21,126,175 | 6,324,348,192 | 51.19 | 95.16 | |
| Male group in Huzhou (HZ-M) | HZ-M1 | 46,796,218 | 23,398,109 | 7,004,969,356 | 50.95 | 94.85 |
| HZ-M2 | 44,605,728 | 22,302,864 | 6,677,397,946 | 51.19 | 95.15 | |
| HZ-M3 | 49,471,502 | 24,735,751 | 7,406,998,266 | 51.00 | 94.88 | |
| Female group in Xingtai (XT-F) | XT-F1 | 53,488,148 | 26,744,074 | 8,001,807,860 | 50.50 | 95.37 |
| XT-F2 | 48,387,164 | 24,193,582 | 7,243,791,296 | 50.72 | 95.08 | |
| XT-F3 | 48,131,874 | 24,065,937 | 7,199,491,898 | 50.62 | 95.23 | |
| Male group in Xingtai (XT-M) | XT-M1 | 51,856,652 | 25,928,326 | 7,759,384,482 | 50.71 | 95.48 |
| XT-M2 | 40,941,278 | 20,470,639 | 6,128,805,602 | 50.35 | 94.91 | |
| XT-M3 | 49,290,250 | 24,645,125 | 7,374,428,542 | 50.32 | 95.60 |
| DEG Set | DEG Number | Upregulated | Downregulated |
|---|---|---|---|
| HZ-F vs. XT-F | 1678 | 541 | 1137 |
| HZ-M vs. XT-M | 1287 | 542 | 745 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Li, H.; Li, C.; Tang, W.; Wang, Y.; Hou, H. Transcriptome Analysis of Environmental Adaptation of Largemouth Bass (Micropterus salmonides). Genes 2025, 16, 267. https://doi.org/10.3390/genes16030267
Wang Y, Li H, Li C, Tang W, Wang Y, Hou H. Transcriptome Analysis of Environmental Adaptation of Largemouth Bass (Micropterus salmonides). Genes. 2025; 16(3):267. https://doi.org/10.3390/genes16030267
Chicago/Turabian StyleWang, Yuao, Huan Li, Chuan Li, Weibin Tang, Yanchao Wang, and Hongxia Hou. 2025. "Transcriptome Analysis of Environmental Adaptation of Largemouth Bass (Micropterus salmonides)" Genes 16, no. 3: 267. https://doi.org/10.3390/genes16030267
APA StyleWang, Y., Li, H., Li, C., Tang, W., Wang, Y., & Hou, H. (2025). Transcriptome Analysis of Environmental Adaptation of Largemouth Bass (Micropterus salmonides). Genes, 16(3), 267. https://doi.org/10.3390/genes16030267




