Insights into Phylogeny, Taxonomy, Origins and Evolution of Crataegus and Mespilus, Based on Comparative Chloroplast Genome Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and DNA Extraction
2.2. Chloroplast Genome Sequencing, Assembly and Annotation
2.3. Molecular Marker Identification and Sequence Divergence Analysis
2.4. Phylogenetic Analysis
2.5. Divergence Time Estimation
2.6. Ancestral Area Reconstruction
3. Results
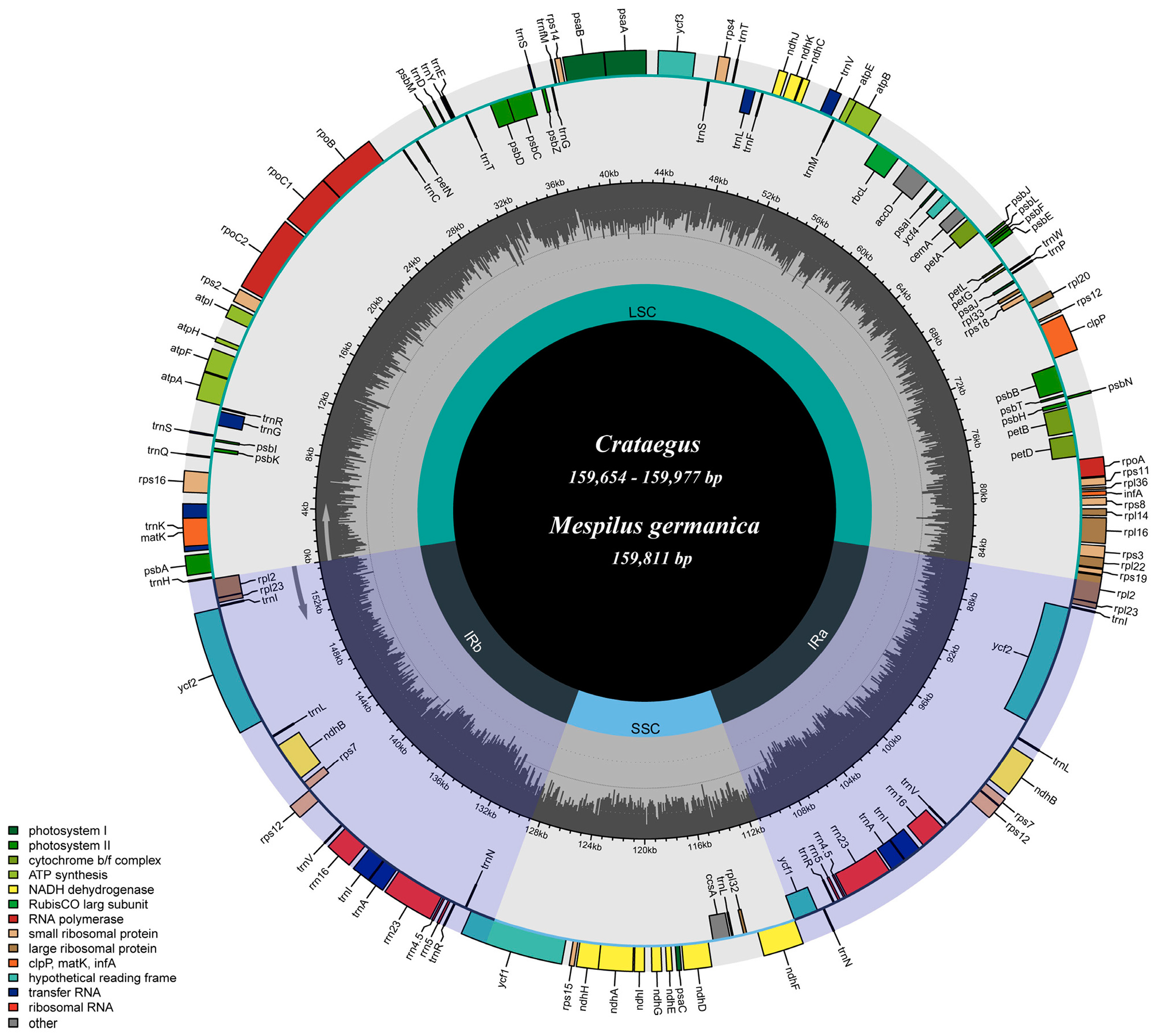
3.1. Basic Characteristics of the Chloroplast Genome
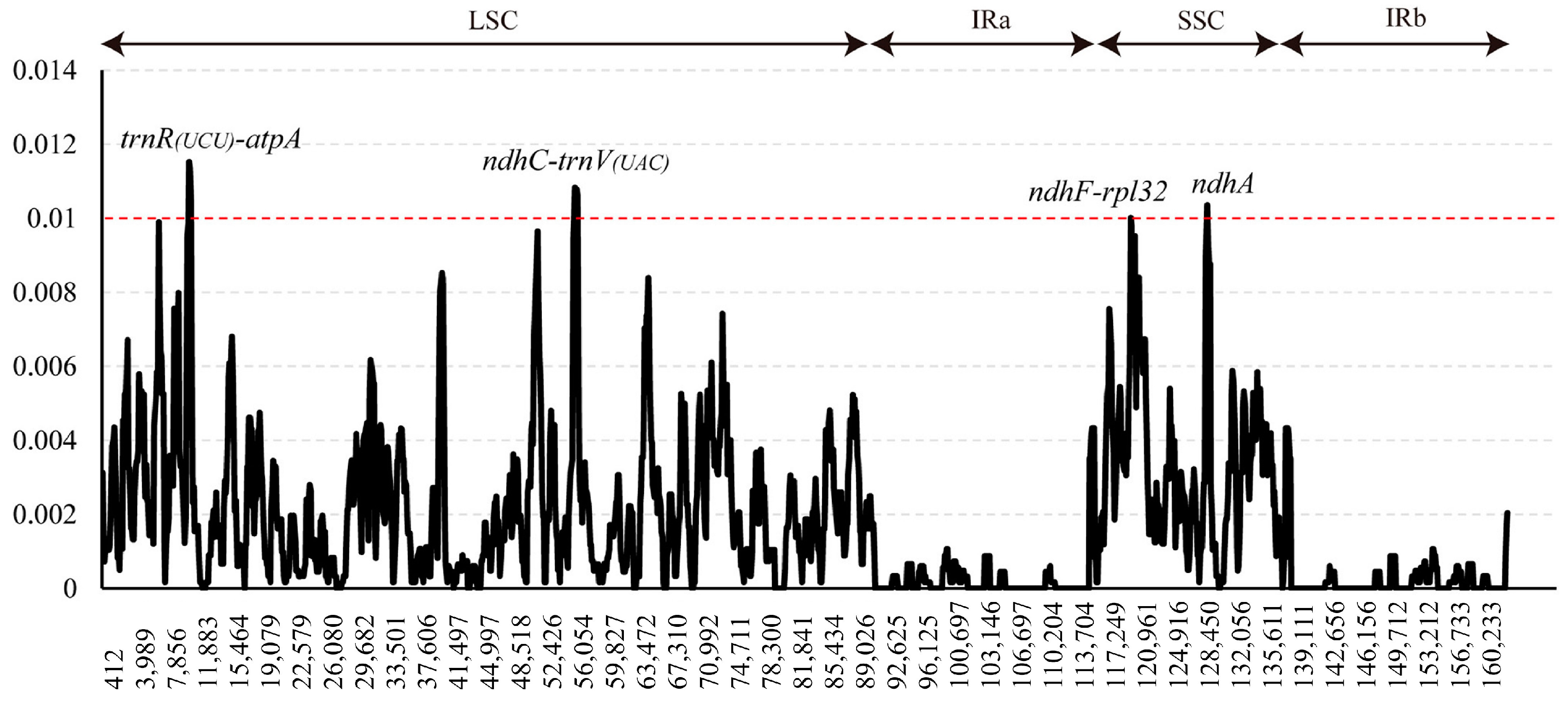
3.2. Sequence Divergence and Hotspots
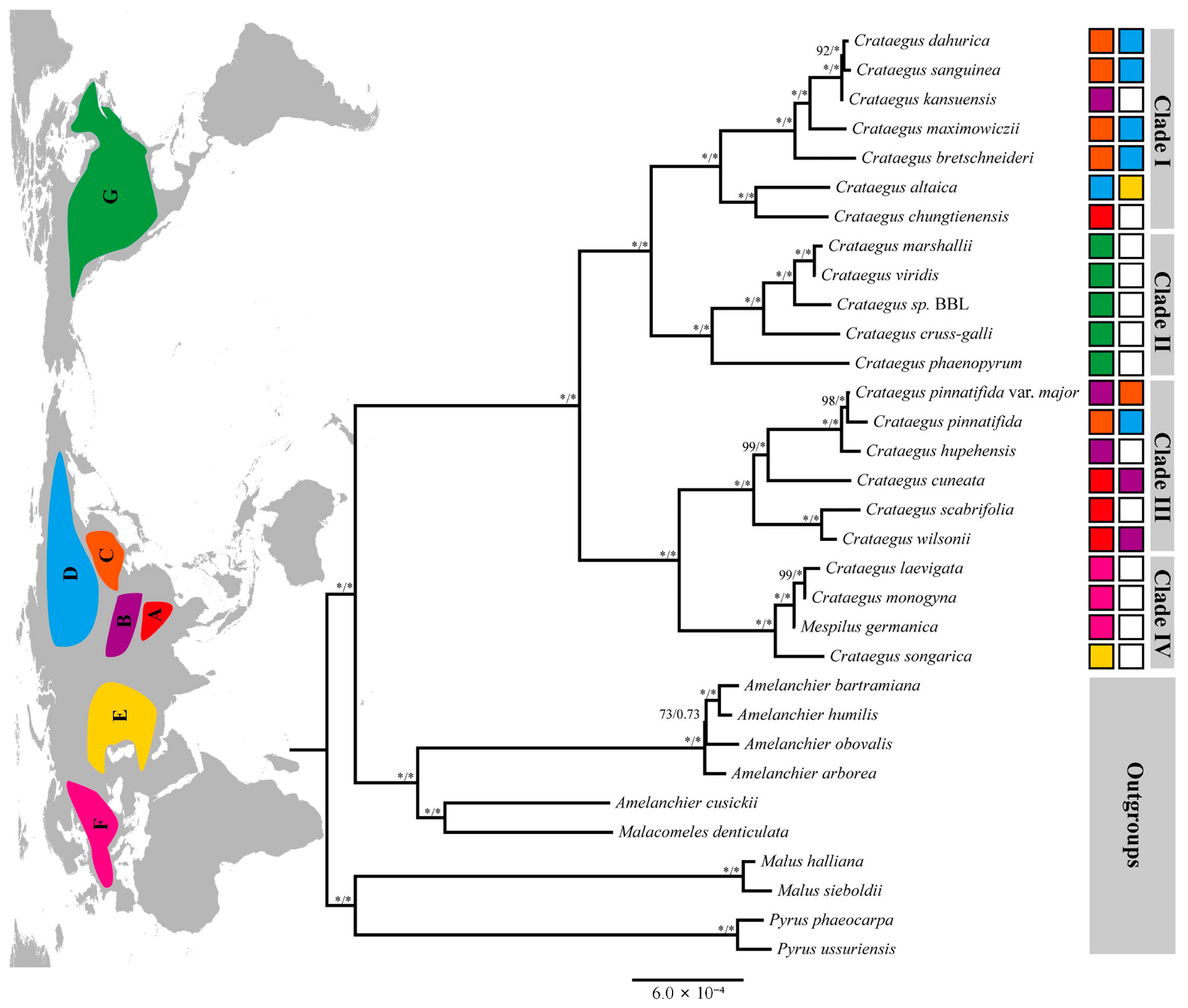
3.3. Phylogenetic Analysis
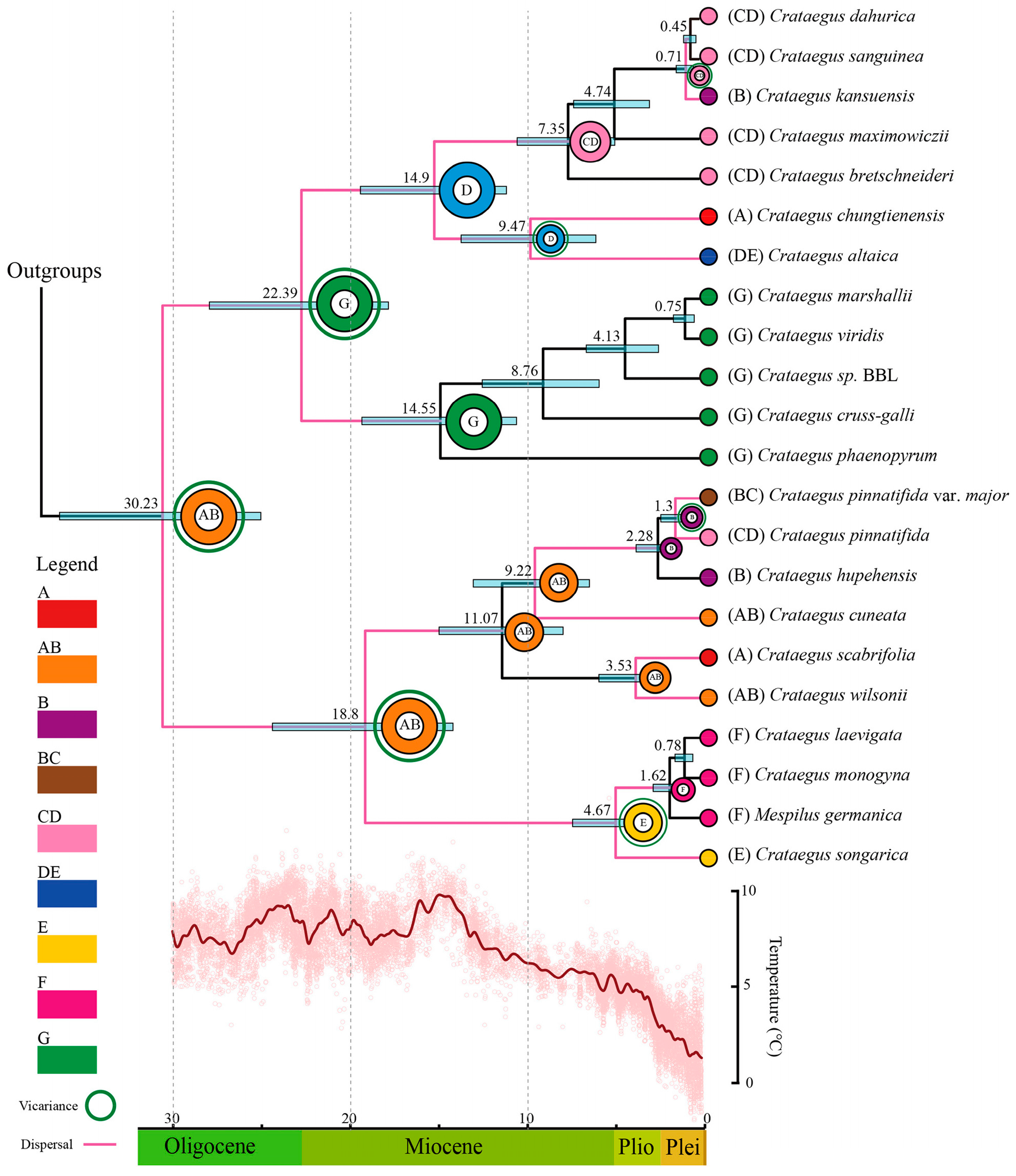
3.4. Divergence Time Estimation
3.5. Biogeographic History
4. Discussion
4.1. Features of the Crataegus and Mespilus Chloroplast Genomes
4.2. Potential Highly Variable Chloroplast Barcodes
4.3. Phylogenetic Relationships
4.4. Origin and Spread of Crataegus
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Christensen, K.I. Revision of Crataegus sect. Crataegus and Nothosect. Crataeguineae (Rosaceae-Maloideae) in the old world. Syst. Bot. Monogr. 1992, 35, 1–199. [Google Scholar] [CrossRef]
- Özcan, M.; Hacıseferoğulları, H.; Marakoğlu, T.; Arslan, D. Hawthorn (Crataegus spp.) fruit: Some physical and chemical properties. J. Food Eng. 2005, 69, 409–413. [Google Scholar] [CrossRef]
- Xu, J.Y.; Zhao, Y.H.; Zhang, X.; Zhang, L.J.; Hou, Y.L.; Dong, W.X. Transcriptome analysis and ultrastructure observation reveal that hawthorn fruit softening is due to Cellulose/Hemicellulose degradation. Front. Plant Sci. 2016, 7, 1524. [Google Scholar] [CrossRef]
- Liu, P.; Yang, B.; Kallio, H. Characterization of phenolic compounds in Chinese hawthorn (Crataegus pinnatifida Bge. Var. Major) fruit by high performance liquid chromatography-electrospray ionization mass spectrometry. Food Chem. 2010, 121, 1188–1197. [Google Scholar]
- Zheng, G.; Deng, J.; Wen, L.; You, L.; Zhao, Z.; Zhou, L. Release of phenolic compounds and antioxidant capacity of Chinese hawthorn “Crataegus pinnatifida” during in vitro digestion. J. Func. Foods 2018, 40, 76–85. [Google Scholar] [CrossRef]
- Brown, J.A.; Beatty, G.E.; Finlay, C.; Montgomery, W.I.; Tosh, D.G.; Provan, J. Genetic analyses reveal high levels of seed and pollen flow in hawthorn (Crataegus monogyna Jacq.), a key component of hedgerows. Tree Genet. Genomes 2016, 12, 58. [Google Scholar] [CrossRef]
- Betancourt-Olvera, M.; Nieto-Angel, R.; Urbano, B.; Gonzalez-Andres, F. Analysis of the biodiversity of hawthorn (Crataegus spp.) from the morphological, molecular, and ethnobotanical approaches, and implications for genetic resource conservation in scenery of increasing cultivation: The case of Mexico. Genet. Resour. Crop Evol. 2018, 65, 897–916. [Google Scholar] [CrossRef]
- Phipps, J.B. Biogeographic, taxonomic, and cladistic relationships between East Asiatic and North American Crataegus. Ann. Mo. Bot. Gard. 1983, 70, 667–700. [Google Scholar] [CrossRef]
- Evans, R.C.; Campbell, C.S. The origin of the apple subfamily (Maloideae; Rosaceae) is clarified by DNA sequence data from duplicated GBSSI genes. Am. J. Bot. 2002, 89, 1478–1484. [Google Scholar] [CrossRef]
- Lo, E.Y.Y.; Stefanović, S.; Christensen, K.I.; Dickinson, T.A. Evidence for genetic association between East Asian and western North American Crataegus L. (Rosaceae) and rapid divergence of the eastern North American lineages based on multiple DNA sequences. Mol. Phylogenet. Evol. 2009, 51, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Lo, E.Y.Y.; Stefanović, S.; Dickinson, T.A. Molecular reappraisal of relationships between Crataegus and Mespilus (Rosaceae, Pyreae)–Two genera or one? Syst. Bot. 2007, 32, 596–616. [Google Scholar]
- Zhao, H.C.; Feng, B.T. China Fruit-Plant Monograph of Hawthorn (Crataegus) Flora; Zhongguo Linye Press: Beijing, China, 1996. [Google Scholar]
- Dong, W.X.; Li, Z.X. The Science and Practice of Chinese Fruit Tree: Hawthorn; Shanxi Science Press: Xi’an, China, 2015. [Google Scholar]
- Du, X.; Zhang, X.; Bu, H.; Zhang, T.; Lao, Y.; Dong, W. Molecular Analysis of Evolution and Origins of Cultivated Hawthorn (Crataegus spp.) and Related Species in China. Front. Plant Sci. 2019, 10, 443. [Google Scholar] [CrossRef] [PubMed]
- Zarrei, M.; Stefanovic’, S.; Dickinson, T.A. Reticulate evolution in North American black-fruited hawthorns (Crataegus section Douglasia; Rosaceae): Evidence from nuclear ITS2 and plastid sequences. Ann. Bot. 2014, 114, 253–269. [Google Scholar] [CrossRef] [PubMed]
- Park, I.; Choi, B.; Weiss-Schneeweiss, H.; So, S.; Myeong, H.H.; Jang, T.S. Comparative Analyses of Complete Chloroplast Genomes and Karyotypes of Allotetraploid Iris koreana and Its PutativeDiploid Parental Species (Iris Series Chinenses, Iridaceae). Int. J. Mol. Sci. 2022, 23, 10929. [Google Scholar] [CrossRef]
- Phipps, J.B. Studies in Mespilus, Crataegus, and ×Crataemespilus (Rosaceae), I. Differentiation of Mespilus and Crataegus, expansion of ×Crataemespilus, with supplementary observations on differences between the Crataegus and Amelanchier clades. Phytotaxa 2016, 257, 201–229. [Google Scholar] [CrossRef][Green Version]
- Lu, R.S.; Li, P.; Qiu, Y.X. The complete chloroplast genomes of three cardiocrinum (Liliaceae) species: Comparative genomic and phylogenetic analyses. Front. Plant Sci. 2017, 7, 2054. [Google Scholar] [CrossRef]
- Roy, N.S.; Jeong, U.; Na, M.; Choi, I.; Cheong, E.J. Genomic analysis and a consensus chloroplast genome sequence of Prunus yedoensis for DNA marker development. Hortic. Environ. Biotechnol. 2020, 61, 859–867. [Google Scholar] [CrossRef]
- Liu, Q.; Li, X.; Li, M.; Xu, W.; Heslop-Harrison, J.S. Comparative chloroplast genome analyses of Avena: Insights into evolutionary dynamics and phylogeny. BMC Plant Biol. 2020, 20, 406. [Google Scholar] [CrossRef] [PubMed]
- McNeal, J.R.; Leebens-Mack, J.H.; Arumuganathan, K.; Kuehl, J.; Boore, J.L.; DePamphilis, C.W. Using partial genomic fosmid libraries for sequencing complete organellar genomes. Biotechniques 2006, 41, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Wicke, S.; Schneeweiss, G.M.; DePamphilis, C.W.; Muller, K.F.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef]
- Mehmetoglu, E.; Kaymaz, Y.; Ates, D.; Kahraman, A.; Tanyolac, M.B. The complete chloroplast genome sequence of Cicer echinospermum, genome organization and comparison with related species. Sci. Hortic. 2022, 296, 110912. [Google Scholar] [CrossRef]
- Wang, J.; Li, Y.; Li, C.; Yan, C.; Shan, S. Twelve complete chloroplast genomes of wild peanuts: Great genetic resources and a better understanding of Arachis phylogeny. BMC Plant Biol. 2019, 19, 504. [Google Scholar] [CrossRef] [PubMed]
- Saina, J.K.; Gichira, A.W.; Li, Z.; Hu, G.; Wang, Q.; Liao, K. The complete chloroplast genome sequence of Dodonae aviscosa: Comparative and phylogenetic analyses. Genetica 2018, 146, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Li, J.L.; Wang, S.; Yu, J.; Wang, L.; Zhou, S.L. A modified CTAB protocol for plant DNA extraction. Chin. Bull. Bot. 2013, 48, 72–78. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Brozynska, M.; Furtado, A.; Henry, R.J. Direct chloroplast sequencing: Comparison of sequencing platforms and analysis tools for whole chloroplast barcoding. PLoS ONE 2014, 9, e110387. [Google Scholar] [CrossRef]
- Huang, D.I.; Cronk, Q. Plann: A command-line application for annotating plastome sequences. Appl. Plant Sci. 2015, 3, 1500026. [Google Scholar] [CrossRef] [PubMed]
- Conant, G.C.; Wolfe, K.H. GenomeVx: Simple web-based creation of editable circular chromosome maps. Bioinformatics 2008, 24, 861–862. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Albert, F.M.; Juan Carlos, S.D.; Sara, G.R.; Pablo, L.; Ramos-Onsins, S.E.; Alejandro, S.G. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, R.; Vaughan, T.G.; BaridoSottani, J.; Duchene, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kuhnert, D.; De Maio, N.; et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef]
- Xiang, Y.Z.; Huang, C.H.; Hu, Y.; Wen, J.; Li, S.S.; Yi, T.S.; Chen, H.Y.; Xiang, J.; Ma, H. Evolution of Rosaceae Fruit Types Based on Nuclear Phylogeny in the Context of Geological Times and Genome Duplication. Mol. Biol. Evol. 2017, 34, 262–281. [Google Scholar] [CrossRef]
- Zhang, S.D.; Jin, J.J.; Chen, S.Y.; Chase, M.W.; Soltis, D.E.; Li, H.T.; Yang, J.B.; Li, D.Z.; Yi, T.S. Diversification of Rosaceae since the Late Cretaceous based on plastid phylogenomics. New Phytol. 2017, 214, 1355–1367. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J. TreeAnnotator v2. 1.2; University of Edinburgh, Institute of Evolutionary Biology: Edinburgh, Scotland, 2014. [Google Scholar]
- Yu, Y.; Harris, A.J.; Blair, C.; He, X. RASP (Reconstruct Ancestral State in Phylogenies): A tool for historical biogeography. Mol. Phylogenet. Evol. 2015, 87, 46–49. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, J.; Qiao, P.; Wang, J.; Wang, M.; Du, Y.; Xiong, F.; Luo, J.; Yuan, Q.; Dong, W.; et al. Evolutionary history of genus Coptis and its dynamic changes in the potential suitable distribution area. Front. Plant Sci. 2022, 13, 1003368. [Google Scholar] [CrossRef] [PubMed]
- Zachos, J.C.; Dickens, G.R.; Zeebe, R.E. An early Cenozoic perspective on greenhouse warming and carbon-cycle dynamics. Nature 2008, 451, 279–283. [Google Scholar] [CrossRef]
- Oliver, M.J.; Murdock, A.G.; Mishler, B.D.; Kuehl, J.V.; Boore, J.L.; Mandoli, D.F.; Everett, K.D.; Wolf, P.G.; Duffy, A.M.; Karol, K.G. Chloroplast genome sequence of the moss Tortula ruralis: Gene content, polymorphism, and structural arrangement relative to other green plant chloroplast genomes. BMC Genom. 2010, 11, 143. [Google Scholar] [CrossRef]
- Jansen, R.K.; Cai, Z.; Raubeson, L.A.; Daniell, H.; Depamphilis, C.W.; Leebens-Mack, J.; Kai, F.M.; Guisinger-Bellian, M.; Haberle, R.C.; Hansen, A.K. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA 2007, 104, 19369–19374. [Google Scholar] [CrossRef]
- Perry, A.S.; Wolfe, K.H. Nucleotide substitution rates in legume chloroplast DNA depend on the presence of the inverted repeat. J. Mol. Evol. 2002, 55, 501–508. [Google Scholar] [CrossRef]
- Zhu, A.D.; Guo, W.H.; Gupta, S.; Fan, W.S.; Mower, J.P. Evolutionary dynamics of the plastid inverted repeat: The effects of expansion, contraction, and loss on substitution rates. New Phytol. 2016, 209, 1747–1756. [Google Scholar] [CrossRef]
- Li, F.W.; Kuo, L.Y.; Pryer, K.M.; Rothfels, C.J. Genes translocated into the plastid inverted repeat show decelerated substitution rates and elevated GC content. Genome Biol. Evol. 2016, 8, 2452–2458. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.S.; Chaw, S.M. Evolutionary stasis in cycad plastomes and the first case of plastome GC-Biased gene conversion. Genome Biol. Evol. 2015, 7, 2000–2009. [Google Scholar] [CrossRef]
- Asaf, S.; Waqas, M.; Khan, A.L.; Khan, M.A.; Kang, S.M.; Imran, Q.M.; Shahzad, R.; Bilal, S.; Yun, B.W.; Lee, I.J. The complete chloroplast genome of wild rice (Oryza minuta) and its comparison to related species. Front. Plant Sci. 2017, 8, 304. [Google Scholar] [CrossRef] [PubMed]
- Emami, A.; Shabanian, N.; Rahmani, M.; Khadivi, A.; Mohammad-Panah, N. Genetic characterization of the Crataegus genus: Implications for in situ conservation. Sci. Hortic. 2018, 231, 56–65. [Google Scholar] [CrossRef]
- Erfani-Moghadam, J.; Mozafari, M.; Fazeli, A. Genetic variation of some hawthorn species based on phenotypic characteristics and RAPD marker. Biotechnol. Biotechnol. Equip. 2016, 30, 247–253. [Google Scholar] [CrossRef]
- Dong, W.P.; Liu, J.; Yu, J.; Wang, L.; Zhou, S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE 2012, 7, e35071. [Google Scholar] [CrossRef]
- Hu, G.; Cheng, L.; Huang, W.; Cao, Q.; Zhou, L.; Jia, W.; Lan, Y. Chloroplast genomes of seven species of Coryloideae (Betulaceae): Structures and comparative analysis. Genome 2020, 63, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Raman, G.; Park, K.T.; Kim, J.H.; Park, S.J. Characteristics of the completed chloroplast genome sequence of Xanthium spinosum: Comparative analyses, identification of mutational hotspots and phylogenetic implications. BMC Genom. 2020, 21, 855. [Google Scholar] [CrossRef]
- Wu, X.; Luo, D.; Zhang, Y.; Yang, C.; Crabbe, M.J.C.; Zhang, T.; Li, G. Comparative Genomic and Phylogenetic Analysis of Chloroplast Genomes of Hawthorn (Crataegus spp.) in Southwest China. Front. Genet. 2022, 13, 900357. [Google Scholar] [CrossRef]
- Yang, Z.; Zhao, T.; Ma, Q.; Liang, L.; Wang, G. Comparative genomics and phylogenetic analysis revealed the chloroplast genome variation and interspecific relationships of Corylus (Betulaceae) species. Front. Plant Sci. 2018, 9, 927. [Google Scholar] [CrossRef]
- Guo, T.J.; Jiao, P.J. Hawthorn (Crataegus) resources in China. HortScience 1995, 30, 1132–1134. [Google Scholar] [CrossRef]
- Campbell, C.S.; Donoghue, M.J.; Wojciechowski, B.M.F. Phylogenetic relationships in Maloideae (Rosaceae): Evidence from sequences of the internal transcribed spacers of nuclear ribosomal DNA and its congruence with morphology. Am. J. Bot. 1995, 82, 903–918. [Google Scholar] [CrossRef]
- Talent, N.; Eckenwalder, J.E.; Lo, E.; Christensen, K.I.; Dickinson, T.A. (1847) Proposal to conserve the name Crataegus against Mespilus (Rosaceae). Taxon 2008, 57, 1007–1008. [Google Scholar] [CrossRef]
- Dnmez, E.O. Pollen morphology in Turkish Crataegus (Rosaceae). Bot. Helv. 2008, 118, 59–70. [Google Scholar] [CrossRef]
- Zhang, Y.; Dai, H.Y.; Zhang, Q.J.; Li, H.; Zhang, Z.H. Assessment of genetic relationship in Crataegus genus by the apple SSR primers. J. Fruit Sci. 2008, 25, 521–525. [Google Scholar]
- Sun, J.; Sun, R.; Liu, H.; Chang, L.; Li, S.; Zhao, M.; Shennan, C.; Lei, J.; Dong, J.; Zhong, C.; et al. Complete chloroplast genome sequencing of ten wild Fragaria species in China provides evidence for phylogenetic evolution of Fragaria. Genomics 2021, 113, 1170–1179. [Google Scholar] [CrossRef]
- Wu, L.W.; Cui, Y.X.; Wang, Q.; Xu, Z.C.; Wang, Y.; Lin, Y.L.; Song, J.Y.; Yao, H. Identification and phylogenetic analysis of five Crataegus species (Rosaceae) based on complete chloroplast genomes. Planta 2021, 254, 14. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.F.; Zhang, Z.H.; Dai, H.Y.; Zhang, Y.; Chang, L.L. Genetic Relationships of Some Hawthorns (Crataegus spp.) Derived from cp DNA PCR-RFL. J. Shenyang Agric. Univ. 2008, 39, 664–668. [Google Scholar]
- Fehrer, J.; Gemeinholzer, B.; Chrtek, J.J.; Brautigam, S. Incongruent plastid and nuclear DNA phylogenies reveal ancient intergeneric hybridization in Pilosella hawkweeds (Hieracium, Cichorieae, Asteraceae). Mol. Phylogenet. Evol. 2007, 42, 347–361. [Google Scholar] [CrossRef]
- Paun, O.; Forest, F.; Fay, M.F.; Chase, M.W. Hybrid speciation in angiosperms: Parental divergence drives ploidy. New Phytol. 2009, 182, 507–518. [Google Scholar] [CrossRef]
- Du, F.K.; Peng, X.L.; Liu, J.Q.; Lascoux, M.; Hu, F.S.; Petit, R.J. Direction and extent of organelle DNA introgression between two spruce species in the Qinghai-Tibetan Plateau. New Phytol. 2011, 192, 1024–1033. [Google Scholar] [CrossRef] [PubMed]
- Acosta, M.C.; Premoli, A.C. Evidence of chloroplast capture in South American Nothofagus (subgenus Nothofagus, Nothofagaceae). Mol. Phylogenet. Evol. 2010, 54, 235–242. [Google Scholar] [CrossRef]
- Zhao, T.; Wang, G.; Ma, Q.; Liang, L.; Yang, Z. Multilocus data reveal deep phylogenetic relationships and intercontinental biogeography of the Eurasian-North American genus Corylus (Betulaceae). Mol. Phylogenet. Evol. 2020, 142, 106658. [Google Scholar] [CrossRef]
- Hokanson, K.E.; Smith, M.J.; Connor, A.M.; Luby, J.J.; Hancock, J.F. Relationships among subspecies of New World octoploid strawberry species, Fragaria virginiana and Fragaria chiloensis, based on simple sequence repeat marker analysis. Can. J. Bot. 2006, 84, 1829–1841. [Google Scholar] [CrossRef]
- Zarei, A.; Erfani-Moghadam, J.; Mozaffari, M. Phylogenetic analysis among some pome fruit trees of Rosaceae family using RAPD markers. Biotechnol. Biotechnol. Equip. 2017, 31, 289–298. [Google Scholar] [CrossRef]
- Xiang, X.G.; Wang, W.; Li, R.Q.; Lin, L.; and Liu, Y.; Zhou, Z.K.; Li, Z.Y.; Chen, Z.D. Large-scale phylogenetic analyses reveal fagalean diversification promoted by the interplay of diaspores and environments in the Paleogene. Perspect. Plant Ecol. Evol. Syst. 2014, 16, 101–110. [Google Scholar] [CrossRef]
- Liu, Z.H.; Pagani, M.; Zinniker, D.; DeConto, R.; Huber, M.; Brinkhuis, H.; Shah, S.R.; Leckie, R.M.; Pearson, A. Global cooling during the Eocene-Oligocene climate transition. Science 2009, 323, 1187–1190. [Google Scholar] [CrossRef] [PubMed]
- Marincovich, L.; Gladenkov, A.Y. Evidence for an early opening of the Bering Strait. Nature 1999, 397, 149–151. [Google Scholar] [CrossRef]
- Jiang, D.; Klaus, S.; Zhang, Y.P.; Hillis, D.M.; Li, J.T. Asymmetric biotic interchange across the Bering land bridge between Eurasia and North America. Natl. Sci. Rev. 2019, 6, 739–745. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Mao, X.; Ding, Z.; Lv, B.; Chen, F. Study on the relation between loess paleoclimate trend and uplift Tibetan Plateau. Quat. Sci. 2009, 29, 988–999. [Google Scholar]
- Gong, H.; Zhang, Y.; Huang, L. Paleoenvironment significance of grain-size composition of Neogene Red Clay in Linxia Basin, Gansu province. Acta Sedimentol. Sin. 2005, 23, 260. [Google Scholar]
- Wang, W.T. On some distribution patterns and some migration routes found in the eastern Asiatic region. Acta Phys. Sin. 1992, 30, 1–24. [Google Scholar]
- Lu, H.; Wang, X.; An, Z.; Miao, X.D.; Zhu, R.X.; Ma, H.Z.; Li, Z.; Tan, H.B.; Wang, X.Y. Geomorphologic evidence of phased uplift of the northeastern Qinghai-Tibet Plateau since 14 million years ago. Sci. China Ser. D 2004, 47, 822–833. [Google Scholar] [CrossRef]
- Li, J.J. The Qinghai-Tibet Plateau Uplifting and Environmental Evolution in Asia: Article Collection of Academician Li Ji-Jun; Science Press: Beijing, China, 2006. [Google Scholar]
- Liu, X.D.; Dong, B.W. Influence of the Tibetan Plateau uplift on the Asian monsoon-arid environment evolution. Chin. Sci. Bull. 2013, 58, 4277–4291. [Google Scholar] [CrossRef]
- Jacques, F.; Guo, S.X.; Tao, S.; Xing, Y.W.; Huang, Y.J.; Liu, Y.S.; Ferguson, D.K.; Zhou, Z.K. Quantitative reconstruction of the Late Miocene monsoon climates of southwest China: A case study of the Lincang flora from Yunnan Province. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2011, 304, 318–327. [Google Scholar] [CrossRef]
- Su, T.; Liu, Y.S.; Jacques, F.; Huang, Y.J.; Xing, Y.W.; Zhou, Z.K. The intensification of the East Asian winter monsoon contributed to the disappearance of Cedrus (Pinaceae) in southwestern China. Quat. Res. 2013, 80, 316–325. [Google Scholar] [CrossRef]
| Taxon | Identification Code | Biogeographic Region | Collection Site | Usage | Main Characteristics |
|---|---|---|---|---|---|
| C. bretschneideri | FSZ | North-eastern China, Mongolia–Siberia | Shenyang | Fresh eating, processing, medicine, ornamental plant, afforestation | Fruit, small, prismatic and oblate, peel, bright red |
| C. pinnatifida | SLH | North-eastern China, Mongolia–Siberia | Shenyang | Medicine, ornamental plant, afforestation | Fruit, subglobose or pear-shaped, peel, dark red |
| C. pinnatifida var. major | SDDJX | Central Plains, Qinling Mountains of China and North-eastern China | Shenyang | Fresh eating, processing, medicine, afforestation | Fruit, large, broad obovate, peel, dark red or purple red |
| C. maximowiczii | MSZ | North-eastern China, Mongolia–Siberia | Shenyang | Fresh eating, ornamental plant, afforestation, timber | Fruit, spherical, pilose when young, glabrous after, peel, red |
| C. hupehensis | HBSZ1H | Central Plains and Qinling Mountains of China | Shenyang | Fresh eating, processing, medicine, ornamental plant, afforestation | Fruit, small, subglobose, peel, bright red |
| C. sanguinea | LNSZ | North-eastern China, Mongolia–Siberia | Shenyang | Fresh eating, medicine, ornamental plant, afforestation | Fruit, extremely small, oblate, peel, blood red and smooth |
| C. kansuensis | GSSZ | Central Plains and Qinling Mountains of China | Shenyang | Fresh eating, processing, medicine, afforestation | Fruit, subspheroidal, peel, red or orange |
| C. scabrifolia | YNSZ | South-western China | Shenyang | Fresh eating, processing, medicine | Fruit, medium size, oblate, peel, stone yellow |
| C. songarica | ZGESZ | Central and Western Asia | Shenyang | Medicine, ornamental plant | Fruit, spherical or oblate, peel, reddish black |
| C. cuneata | XYSZ | Southwest, Central Plains and Qinling Mountains of China | Shenyang | Processing, medicine, rootstock | Fruit, spherical or oblate, peel, red or yellow |
| C. altaica | AETSZ | Central and Western Asia, Mongolia–Siberia | Shenyang | Medicine, ornamental plant | Fruit, small and spherical, peel, golden yellow |
| C. dahurica | GYSZ | North-eastern China, Mongolia–Siberia | Shenyang | Fresh eating, medicine, afforestation, rootstock | Fruit, spherical or oblong, peel, orange-red or orange |
| C. chungtienensis | ZDSA | South-western China | Shenyang | Ornamental plant, afforestation | Fruit, small, spherical or oblong, peel, red |
| C. wilsonii | HZSZ | Southwest, Central Plains and Qinling Mountains of China | Shenyang | Processing, afforestation | Fruit, small, oblong, peel, red |
| C. crussgalli | JJSZ | North America | Shanghai | Ornamental plant, afforestation | Thorn, strong, fruit, small, peel, red |
| C. viridis | LSZ | North America | Shanghai | Ornamental plant, afforestation | Fruit, small, peel, bright red |
| C. phaenopyrum | HSDSZ | North America | Shanghai | Ornamental plant, afforestation | Fruit, small, peel, bright red |
| C. laevigata | HHSZ | Europe | Shanghai | Ornamental plant | Fruit, scarce, peel, dark red |
| C. monogyna | DZSZ1H | Europe | Shanghai | Ornamental plant, afforestation | dunga-runga, peel, dark red |
| M. germanica | OZ | Europe | Shanghai | Fresh eating, processing, afforestation | Dunga-runga, fruit, apple-shaped, peel, brown |
| Species | Plastome Size (bp) | LSC Length (bp) | IR Length (bp) | SSC Length (bp) | Gene Number | Protein-Coding Genes | tRNA Genes | rRNA Genes | Overall GC (%) | GenBank Accession Number |
|---|---|---|---|---|---|---|---|---|---|---|
| C. pinnatifida | 159,676 | 87,744 | 26,384 | 19,164 | 113 | 79 | 4 | 30 | 36.60% | OP963999 |
| C. pinnatifida var. major | 159,656 | 87,749 | 26,384 | 19,139 | 113 | 79 | 4 | 30 | 36.70% | OP964000 |
| C. maximowiczii | 159,800 | 87,778 | 26,384 | 19,254 | 113 | 79 | 4 | 30 | 36.60% | OP964001 |
| C. cuneata | 159,718 | 87,766 | 26,384 | 19,184 | 113 | 79 | 4 | 30 | 36.60% | OP964002 |
| C. sanguinea | 159,851 | 87,844 | 26,384 | 19,239 | 113 | 79 | 4 | 30 | 36.60% | OP964003 |
| C. scabrifolia | 159,744 | 87,759 | 26,383 | 19,219 | 113 | 79 | 4 | 30 | 36.60% | OP964004 |
| C. songarica | 159,871 | 87,880 | 26,389 | 19,213 | 113 | 79 | 4 | 30 | 36.60% | OP964005 |
| C. kansuensis | 159,827 | 87,800 | 26,384 | 19,259 | 113 | 79 | 4 | 30 | 36.60% | OP964006 |
| C. altaica | 159,704 | 87,682 | 26,377 | 19,268 | 113 | 79 | 4 | 30 | 36.60% | OP964007 |
| C. bretschneideri | 159,717 | 87,711 | 26,347 | 19,312 | 113 | 79 | 4 | 30 | 36.60% | OP964008 |
| C. dahurica | 159,852 | 87,847 | 26,384 | 19,237 | 113 | 79 | 4 | 30 | 36.60% | OP964009 |
| C. chungtienensis | 159,835 | 87,803 | 26,384 | 19,264 | 113 | 79 | 4 | 30 | 36.60% | OP964010 |
| C. laevigata | 159,707 | 87,698 | 26,384 | 19,241 | 113 | 79 | 4 | 30 | 36.60% | OP964011 |
| C. monogyna | 159,763 | 87,779 | 26,384 | 19,216 | 113 | 79 | 4 | 30 | 36.60% | OP964012 |
| C. hupehensis | 159,766 | 87,852 | 26,385 | 19,144 | 113 | 79 | 4 | 30 | 36.60% | MW201730 |
| C. wilsonii | 159,779 | 87,773 | 26,383 | 19,240 | 113 | 79 | 4 | 30 | 36.60% | OP964013 |
| C. crussgalli | 159,977 | 88,014 | 26,363 | 19,237 | 113 | 79 | 4 | 30 | 36.60% | OP964014 |
| C. phaenopyrum | 159,953 | 88,061 | 26,331 | 19,230 | 113 | 79 | 4 | 30 | 36.60% | OP964015 |
| C. viridis | 159,654 | 87,704 | 26,365 | 19,220 | 113 | 79 | 4 | 30 | 36.60% | OP964016 |
| M. germanica | 159,811 | 87,804 | 26,396 | 19,215 | 113 | 79 | 4 | 30 | 36.60% | OP964017 |
| Gene Category | Gene Group | Gene Names |
|---|---|---|
| Photosynthesis-related genes | Rubisco | rbcL |
| Photosystem I | psaA, psaB, psaC, psaI, psaJ | |
| Assembly/stability of photosystem I | * ycf3, ycf4 | |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| ATP synthase | atpA, atpB, atpE, * atpF, atpH, atpI | |
| Cytochrome complex | petA, * petB, * petD, petG, petL, petN | |
| Cytochrome synthesis | ccsA | |
| NADPH dehydrogenase | * ndhA, * ndhB, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Transcription- and translation-related genes | Transcription | rpoA, rpoB, rpoC1, rpoC2 |
| Ribosomal proteins | rps2, rps3, rps4, rps7, rps8, rps11, * rps12, rps14, rps15, rps16, rps18, rps19, * rpl2, rpl14, * rpl16, rpl20, rpl22, rpl23, rpl32, rpl33, rpl36 | |
| Translation initiation factor | infA | |
| RNA genes | Ribosomal RNA | rrn5, rrn4.5, rrn16, rrn23 |
| Transfer RNA | * trnA-UGC, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-UCC, * trnG-GCC, trnH-GUG, trnI-CAU, * trnI-GAU, * trnK-UUU, trnL-CAA, * trnL-UAA, trnL-UAG, trnfM-CAUI, trnM-CAU, trnN-GUU, trnP-UGG, trnQ-UUG, trnR-ACG, trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC, * trnV-UAC, trnW-CCA, trnY-GUA | |
| Other genes | RNA processing | matK |
| Carbon metabolism | cemA | |
| Proteolysis | * clpP | |
| Fatty acid synthesis | accD | |
| Genes of unknown function | Conserved reading frames 2 | ycf1, ycf2 |
| Region | Length (bp) | Variable Sites | Parsimony- Informative Sites | Singleton Sites | Nucleotide Diversity | |||
|---|---|---|---|---|---|---|---|---|
| Number | % | Number | % | Number | % | |||
| LSC | 91,171 | 803 | 0.88 | 461 | 0.51 | 342 | 0.38 | 0.00232 |
| SSC | 19,881 | 254 | 1.28 | 159 | 0.80 | 95 | 0.48 | 0.00355 |
| IR | 26,402 | 29 | 0.11 | 17 | 0.06 | 12 | 0.05 | 0.00032 |
| Total | 163,856 | 1115 | 0.68 | 654 | 0.40 | 461 | 0.28 | 0.00180 |
| Marker | Length (bp) | Variable Sites | Parsimony- Informative Sites | Nucleotide Diversity | ||
|---|---|---|---|---|---|---|
| Number | % | Number | % | |||
| trnR(UCU)-atpA | 847 | 36 | 4.25 | 17 | 2.01 | 0.01381 |
| ndhC-trnV(UAC) | 758 | 22 | 2.90 | 12 | 1.58 | 0.01227 |
| ndhF-rpl32 | 1221 | 31 | 0.56 | 18 | 0.49 | 0.00732 |
| ndhA | 2310 | 26 | 1.05 | 17 | 0.66 | 0.00352 |
| trnH-psbA | 297 | 8 | 4.25 | 5 | 2.01 | 0.00781 |
| rbcL | 1428 | 8 | 2.90 | 7 | 1.58 | 0.00188 |
| matK | 1521 | 16 | 2.54 | 10 | 1.47 | 0.00293 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, J.; Wang, Y.; Song, H.; Dong, W.; Dong, N. Insights into Phylogeny, Taxonomy, Origins and Evolution of Crataegus and Mespilus, Based on Comparative Chloroplast Genome Analysis. Genes 2025, 16, 204. https://doi.org/10.3390/genes16020204
Meng J, Wang Y, Song H, Dong W, Dong N. Insights into Phylogeny, Taxonomy, Origins and Evolution of Crataegus and Mespilus, Based on Comparative Chloroplast Genome Analysis. Genes. 2025; 16(2):204. https://doi.org/10.3390/genes16020204
Chicago/Turabian StyleMeng, Jiaxin, Yan Wang, Han Song, Wenxuan Dong, and Ningguang Dong. 2025. "Insights into Phylogeny, Taxonomy, Origins and Evolution of Crataegus and Mespilus, Based on Comparative Chloroplast Genome Analysis" Genes 16, no. 2: 204. https://doi.org/10.3390/genes16020204
APA StyleMeng, J., Wang, Y., Song, H., Dong, W., & Dong, N. (2025). Insights into Phylogeny, Taxonomy, Origins and Evolution of Crataegus and Mespilus, Based on Comparative Chloroplast Genome Analysis. Genes, 16(2), 204. https://doi.org/10.3390/genes16020204





