Product Speculation from Carotenogenic Gene Cluster of Nonlabens spongiae Genome, and Identification of Myxol and Functional Analysis of Each Gene
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome and Gene Clusters from Nonlabens
2.2. Strains and Growth Conditions
2.3. Extraction and Analysis of Carotenoids from N. spongiae
2.4. Extraction of Genomic DNA from N. spongiae
2.5. Cloning of the Carotenogenic Gene Cluster from N. spongiae
2.6. Functional Analysis of cruF, crtY, crtD, crtA-OH, and crtZ Genes from N. spongiae
2.7. Reconstruction of Plasmid for Myxol Production
2.8. Extraction and Analysis of Carotenoids from E. coli Cells
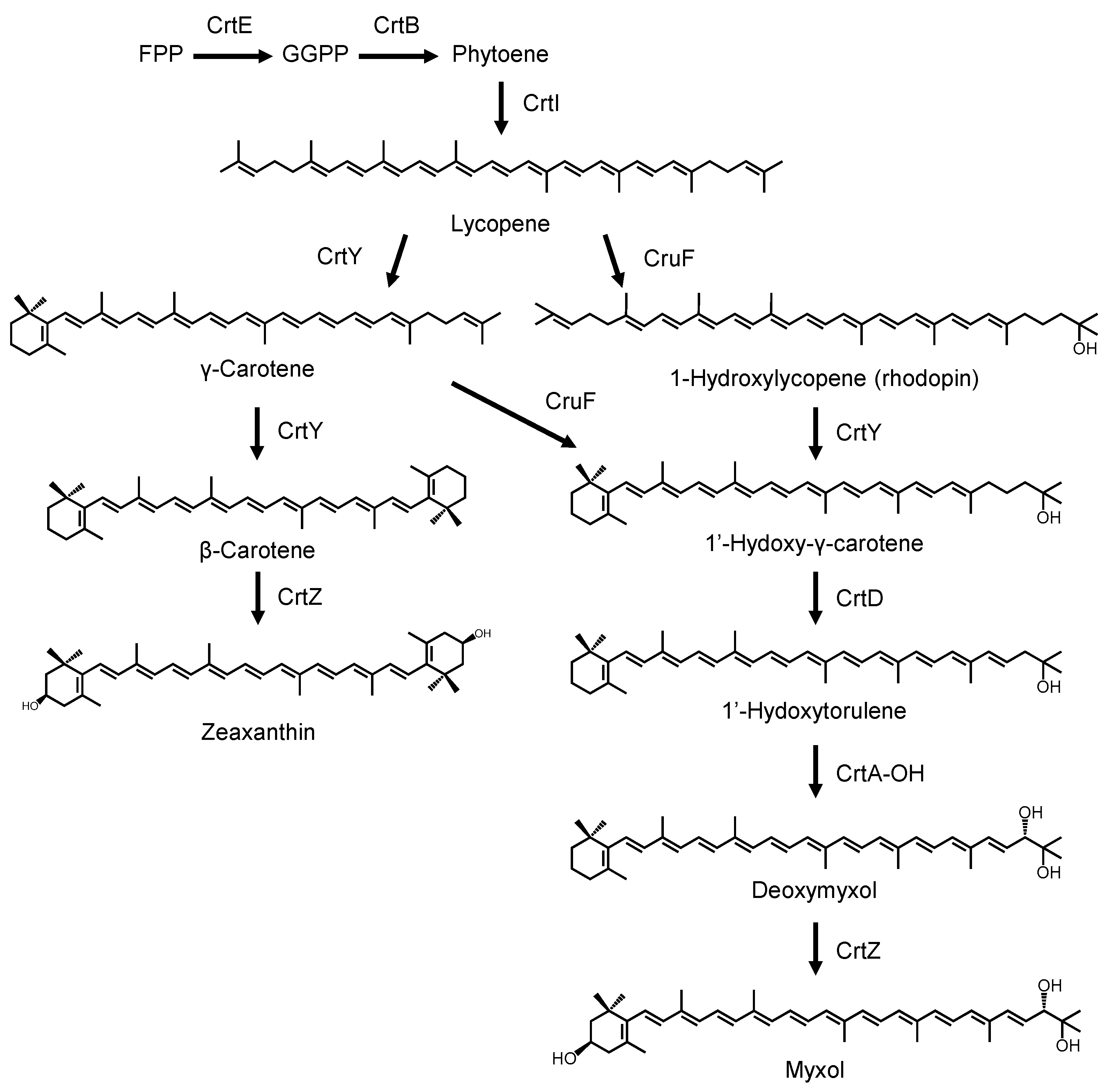
3. Results and Discussion
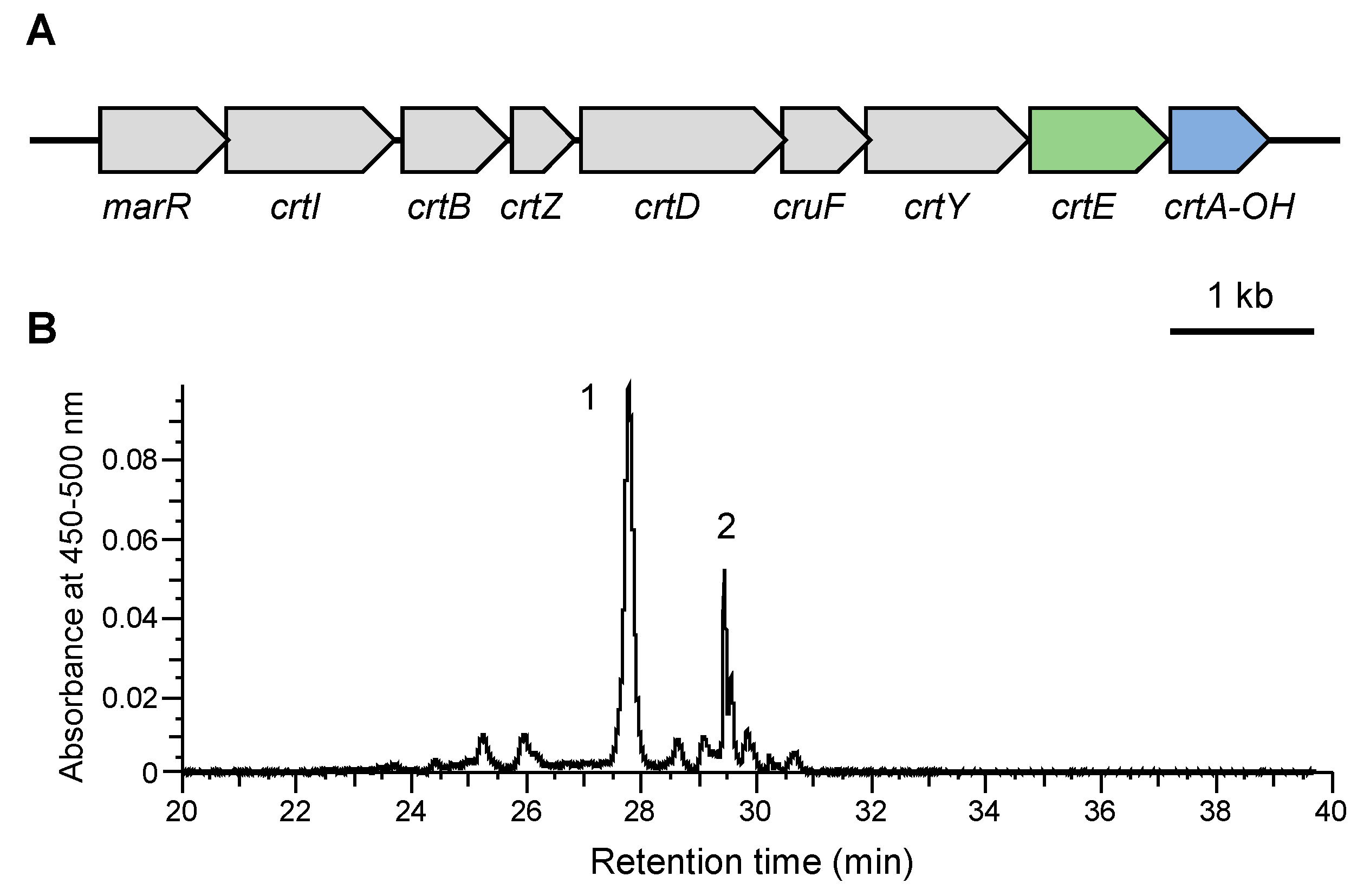
3.1. Analysis of Carotenogenic Gene Cluster in Nonlabens
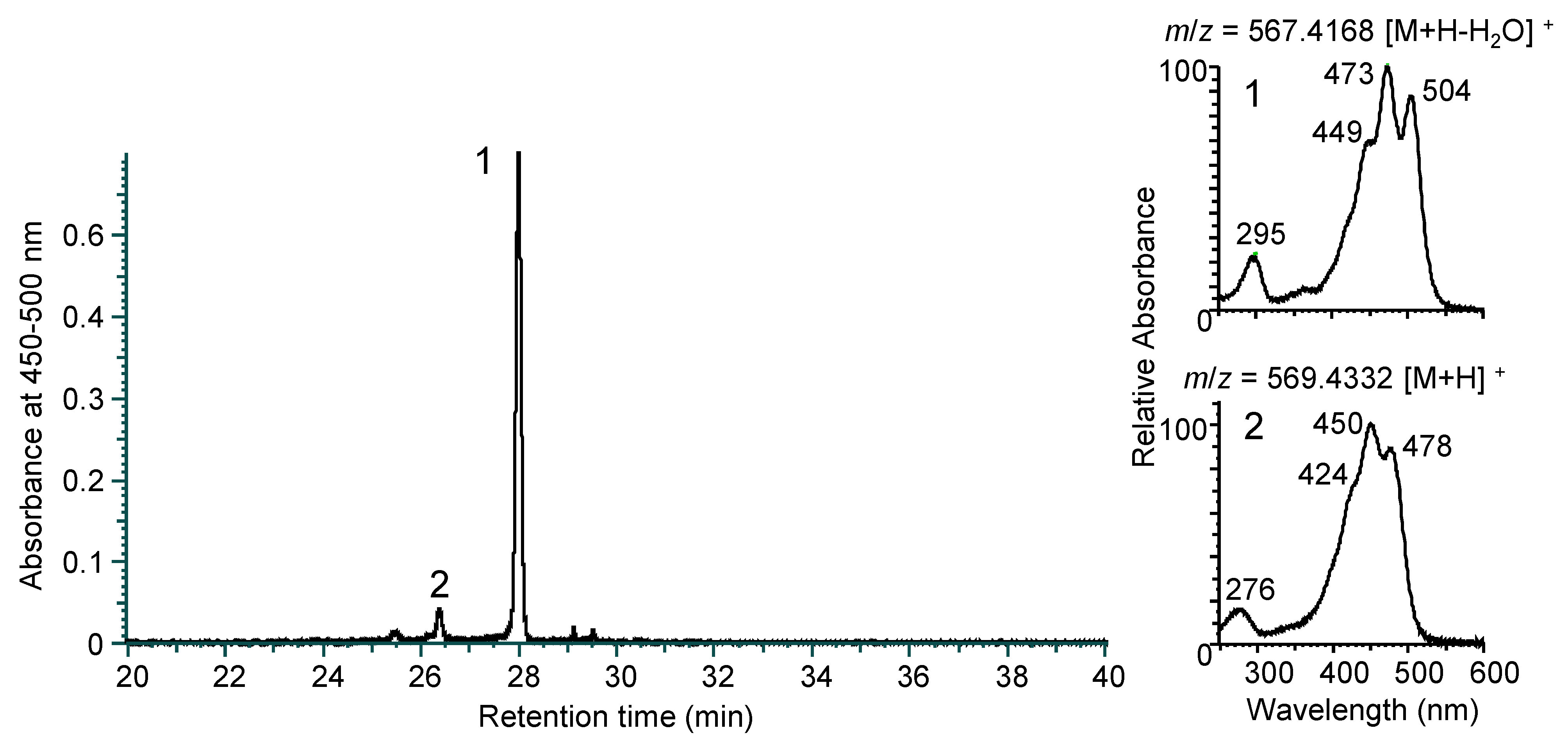
3.2. Identification of Carotenoids from N. spongiae
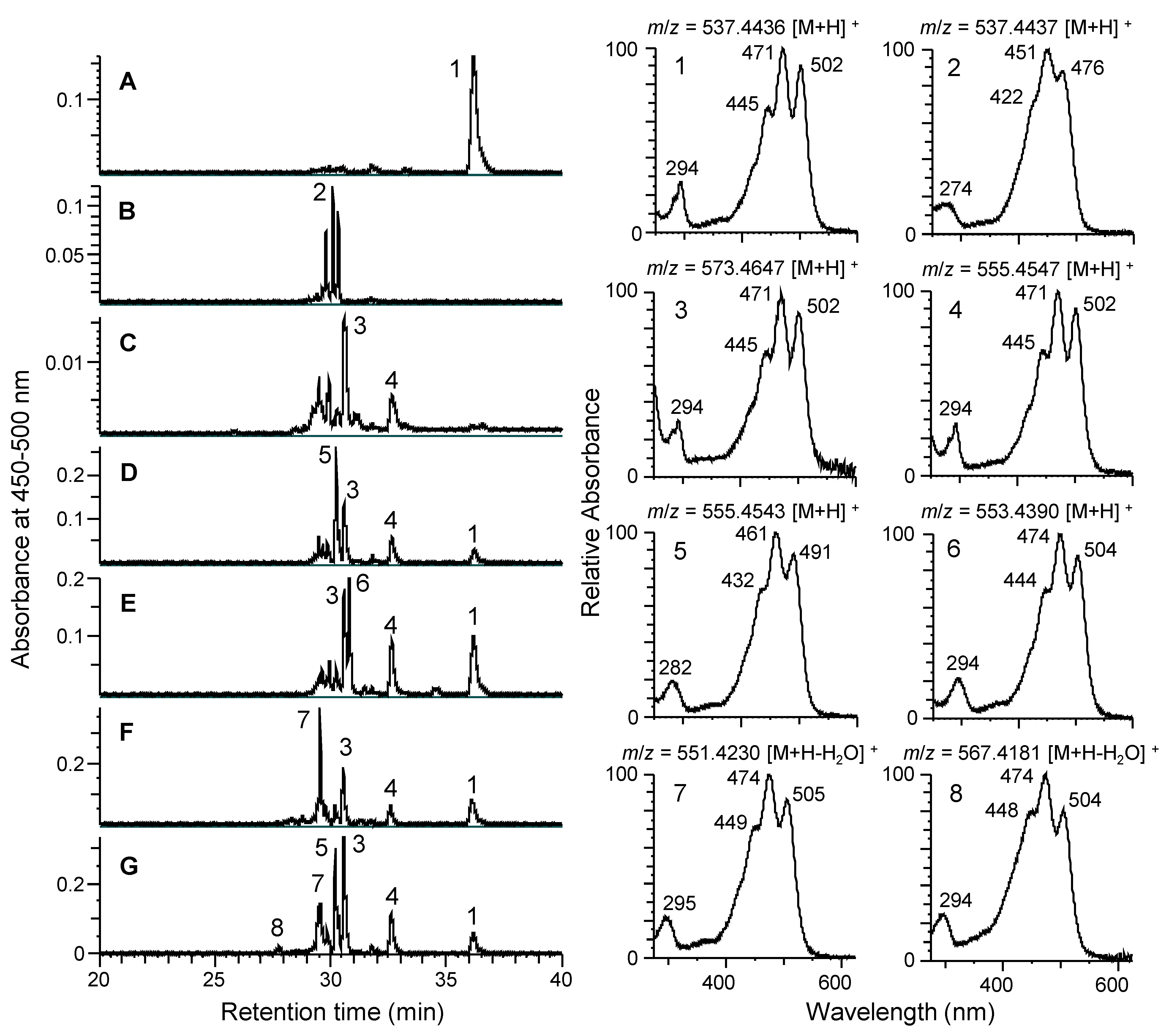
3.3. Functional Analysis of Carotenogenic Genes in Cluster from N. spongiae
3.4. Production of Myxol Using Reconstructed Gene Cluster and crtE from N. spongiae in E. coli
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| APCI | atmospheric-pressure chemical ionization |
| FPP | farnesyl diphosphate |
| JCM | Japan Collection of Microorganisms |
References
- Frank, H.A.; Cogdell, R.J. Carotenoids in photosynthesis. Photochem. Photobiol. 1996, 63, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, H. Carotenoids and the assembly of light-harvesting complexes. In The Photochemistry of Carotenoids; Frank, H.A., Young, A.J., Britton, G., Cogdell, R.J., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherland, 1999; pp. 123–135. [Google Scholar]
- Takaichi, S. Carotenoids in carotenogenic organisms: Distribution, biosynthesis, and functions. In Natural Products: Phytochemistry, Botany and Metabolism of Alkaloids, Phenolics and Terpenes, 2nd ed.; Ramawat, K.G., Merillon, J.M., Eds.; Springer: Berlin/Heidelberg, Germany, 2025; in press. [Google Scholar]
- Maoka, T. Carotenoids as natural functional pigments. J. Nat. Med. 2020, 74, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, G.A.; Alberti, M.; Leach, F.; Hearst, J.E. Nucleotide sequence, organization, and nature of the protein products of the carotenoid biosynthesis gene cluster of Rhodobacter capsulatus. Mol. Gen. Genet. 1989, 216, 254–268. [Google Scholar] [CrossRef] [PubMed]
- Gerjets, T.; Steiger, S.; Sandmann, G. Catalytic properties of the expressed acyclic carotenoid 2-ketolases from Rhodobacter capsulatus and Rubrivivax gelatinosus. Biochim. Biophys. Acta 2009, 1791, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Misawa, N.; Nakagawa, M.; Kobayashi, K.; Yamano, S.; Izawa, Y.; Nakamura, K.; Harashima, K. Elucidation of pathway the Erwinia uredovora carotenoid biosynthetic by functional analysis of gene products expressed in Escherichia coli. J. Bacteriol. 1990, 172, 6704–6712. [Google Scholar] [CrossRef]
- Hannibal, L.; Lorquin, J.; D’ortoli, N.A.; Garcia, N.; Chaintreuil, C.; Masson-Boivin, C.; Dreyfus, B. Isolation and characterization of canthaxanthin biosynthesis genes from the photosynthetic bacterium Bradyrhizobium sp. strain ORS278. J. Bacteriol. 2000, 182, 3850–3853. [Google Scholar] [CrossRef]
- Giraud, E.; Hannibal, L.; Fardoux, J.; Jaubert, M.; Jourand, P.; Dreyfus, B.; Sturgis, J.N.; Verméglio, A. Two distinct crt gene clusters for two different functional classes of carotenoid in Bradyrhizobium. J. Biol. Chem. 2004, 279, 15076–15083. [Google Scholar] [CrossRef]
- Kang, M.; Chhetri, G.; Kim, J.; Kim, I.; Seo, T. Sphingomonas sabuli sp. nov., a carotenoid-producing bacterium isolated from beach sand. Int. J. Syst. Evol. Microbiol. 2021, 71, 004896. [Google Scholar] [CrossRef]
- Serrano, S.; Mendo, S.; Caetano, T. Haloarchaea have a high genomic diversity for the biosynthesis of carotenoids of biotechnological interest. Res. Microbiol. 2022, 173, 103919. [Google Scholar] [CrossRef]
- Hertzberg, S.; Liaaen-Jensen, S. The structure of myxoxanthophyll. Phytochemistry 1969, 8, 1259–1280. [Google Scholar] [CrossRef]
- Takaichi, S.; Mochimaru, M. Carotenoids and carotenogenesis in cyanobacteria: Unique ketocarotenoids and carotenoid glycosides. Cell Mol. Life Sci. 2007, 64, 2607–2619. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, K.; Ebisawa, M.; Yamada, M.; Nagashima, Y.; Suzuki, H.; Maoka, T.; Takaichi, S. Functional lycopene cyclase (CruA) in cyanobacterium, Arthrospira platensis NIES-39, and its role in carotenoid synthesis. Plant Cell Physiol. 2017, 58, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, A.; Miki, W. Isolation of myxol from a marine bacterium Flavobacterium sp. associated with a marine sponge. Fish. Sci. 1995, 61, 684–686. [Google Scholar] [CrossRef]
- Teramoto, M.; Takaichi, S.; Inomata, Y.; Ikenaga, H.; Misawa, N. Structural and functional analysis of a lycopene L-monocyclase gene isolated from a unique marine bacterium that produces myxol. FEBS Lett. 2003, 545, 120–126. [Google Scholar] [CrossRef]
- Choi, S.K.; Matsuda, S.; Hoshino, T.; Peng, X.; Misawa, N. Characterization of bacterial β-carotene 3,3′-hydroxylases, CrtZ, and P450 in astaxanthin biosynthetic pathway and adonirubin production by gene combination in Escherichia coli. Appl. Microbiol. Biotechnol. 2006, 72, 1238–1246. [Google Scholar] [CrossRef]
- Teramoto, M.; Rählert, N.; Misaswa, N.; Sandmann, G. 1-Hydroxy monocyclic carotenoid 3,4-dehydrogenase from a marine bacterium that produces myxol. FEBS Lett. 2004, 570, 184–188. [Google Scholar] [CrossRef]
- Rählert, N.; Fraser, P.D.; Sandmann, G. A crtA-related gene from Flavobacterium P99-3 encodes a novel carotenoid 2-hydroxylase involved in myxol biosynthesis. FEBS Lett. 2009, 583, 1605–1610. [Google Scholar] [CrossRef]
- Graham, J.E.; Bryant, D.A. The biosynthetic pathway for myxol-2 fucoside (myxoxanthophyll) in the cyanobacterium Synechococcus sp. strain PCC 7002. J. Bacteriol. 2009, 191, 3292–3300. [Google Scholar] [CrossRef]
- Umeno, D.; Arnold, F.H. Evolution of a pathway to novel long-chain carotenoids. J. Bacteriol. 2004, 186, 1531–1536. [Google Scholar] [CrossRef]
- Li, L.; Furubayashi, M.; Wang, S.; Maoka, T.; Kawai-Noma, S.; Saito, K.; Umeno, D. Genetically engineered biosynthetic pathways for nonnatural C60 carotenoids using C5-elongases and C50-cyclases in Escherichia coli. Sci. Rep. 2019, 9, 2982. [Google Scholar] [CrossRef]
- Takemura, M.; Kudo, A.; Higuchi, Y.; Maoka, T.; Sahara, T.; Yaoi, K.; Ohdan, K.; Umeno, D.; Misawa, N. Pathway engineering for efficient biosynthesis of violaxanthin in Escherichia coli. Appl. Microbiol. Biotechnol. 2019, 103, 9393–9399. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.C.K.; Tsoi, M.M.Y.; Li, X.; Plakhotnikova, I.; Dobretsov, S.; Wu, M.; Wong, P.K.; Pawlik, J.R.; Qian, P.Y. Stenothermobacter spongiae gen. nov., sp. nov., a novel member of the family Flavobacteriaceae isolated from a marine sponge in the Bahamas, and emended description of Nonlabens tegetincola. Int. J. Syts. Evol. Microbiol. 2006, 56, 181–185. [Google Scholar] [CrossRef][Green Version]
- Yi, H.; Chun, J. Unification of the genera Nonlabens, Persicivirga, Sandarakinotalea and Stenothermobacter into a single emended genus, Nonlabens, and description of Nonlabens agnitus sp. nov. Syst. Appl. Microbiol. 2012, 35, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.K.; Kim, B.K.; Song, J.Y.; Kwak, M.J.; Lee, C.H.; Yoon, J.H.; Oh, T.K.; Kim, J.F. Genomic makeup of the marine flavobacterium Nonlabens (Donghaeana) dokdonensis and identification of a novel class of Rhodopsins. Genome Biol. Evol. 2013, 5, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Kopel, M.; Helbert, W.; Henrissat, B.; Doniger, T.; Banin, E. Draft genome sequence of Nonlabens ulvanivorans, an Ulvan-Degrading Bacterium. Genome Announc. 2014, 2, e00793-14. [Google Scholar] [CrossRef]
- Kumagai, Y.; Yoshizawa, S.; Nakajima, Y.; Watanabe, M.; Fukunaga, T.; Ogura, Y.; Hayashi, T.; Oshima, K.; Hattori, M.; Ikeuchi, M.; et al. Solar-panel and parasol strategies shape the proteorhodopsin distribution pattern in marine Flavobacteriia. ISME J. 2018, 12, 1329–1343. [Google Scholar] [CrossRef]
- Seo, Y.L.; Jung, J.; Song, C.; Kwon, Y.M.; Jung, H.S.; Eyun, S.; Jeon, C.O. Nonlabens ponticola sp. nov., isolated from seawater and reclassification of Nonlabens sediminis as a later heterotypic synonym of Nonlabens tegetincola. Int. J. Syst. Evol. Microbiol. 2021, 71, 004603. [Google Scholar] [CrossRef]
- Khan, S.T.; Nakagawa, Y.; Harayama, S. Sandarakinotalea sediminis gen. nov., sp. nov., a novel member of the family Flavobacteriaceae. Int. J. Syts. Evol. Microbiol. 2006, 56, 959–963. [Google Scholar]
- Lau, S.C.K.; Tsoi, M.M.Y.; Li, X.; Plakhotnikova, I.; Dobretsov, S.; Wong, P.K.; Pawlik, J.R.; Qian, P.Y. Nonlabens tegetincola gen. nov., sp. nov., a novel member of the family Flavobacteriaceae isolated from amicrobial mat in a subtropical estuary. Int. J. Syts. Evol. Microbiol. 2005, 55, 2279–2283. [Google Scholar] [CrossRef]
- Park, S.; Kang, C.H.; Yoon, J.H. Nonlabens arenilitoris sp. nov., a member of the family Flavobacteriaceae isolated from seashore sand. Antonie Leeuwenhoek 2013, 103, 1125–1132. [Google Scholar] [CrossRef]
- O’Sullivan, L.A.; Rinna, J.; Humphreys, G.; Weightman, A.J.; Fry, J.C. Culturable phylogenetic diversity of the phylum ‘Bacteroidetes’ from river epilithon and coastal water and description of novel members of the family Flavobacteriaceae: Epilithonimonas tenax gen. nov., sp. nov. and Persicivirga xylanidelens gen. nov., sp. nov. Int. J. Syts. Evol. Microbiol. 2006, 56, 169–180. [Google Scholar]
- Yamano, Y.; Masumoto, M.; Takaichi, S.; Wada, A. Total synthesis of myxol and deoxymyxol stereoisomers and their application to determining the absolute configurations of the natural products. Tetrahedron 2018, 74, 1533–1539. [Google Scholar] [CrossRef]
- Misawa, N.; Sato, Y.; Kondo, K.; Yokoyama, A.; Kajiwara, S.; Saito, T.; Ohtani, T.; Miki, W. Structure and functional analysis of a marine bacterial carotenoid biosynthesis gene cluster and astaxanthin biosynthetic pathway proposed at the gene level. J. Bacteriol. 1995, 177, 6575–6584. [Google Scholar] [CrossRef] [PubMed]
- Takemura, M.; Takagi, C.; Aikawa, M.; Araki, K.; Choi, S.K.; Itaya, M.; Shindo, K.; Misawa, N. Heterologous production of novel and rare C30-carotenoids using Planococcus carotenoid biosynthesis genes. Microb. Cell Fact. 2021, 20, 194. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, T.; Hosaka, T.; Hasegawa-Takano, M.; Nishimura, Y.; Tominaga, K.; Mori, K.; Nishino, S.; Takahashi, Y.; Uchikubo-Kamo, T.; Hanada, K.; et al. Carotenoid pigments enhance rhodopsin-mediated phototrophy by light-harvesting and photocycle-accelerating. bioRxiv 2024. preprint. [Google Scholar] [CrossRef]
- Harada, H.; Misawa, N. Novel approaches and achievements in biosynthesis of functional isoprenoids in Escherichia coli. Appl. Microbiol. Biotechnol. 2009, 84, 1021–1031. [Google Scholar] [CrossRef]
- Sun, Z.; Shen, S.; Wang, C.; Wang, H.; Hu, Y.; Jiao, J.; Ma, T.; Tian, B.; Hua, Y. A novel carotenoid 1,2-hydratase (CruF) from two species of the non-photosynthetic bacterium Deinococcus. Microbiology 2009, 155, 2775–2783. [Google Scholar] [CrossRef][Green Version]
- Ouchane, S.; Picaud, M.; Vernotte, C.; Reiss-Husson, F.; Astier, C. Pleiotropic effects of puf interposon mutagenesis on carotenoid biosynthesis in Rubrivivax gelatinosus. J. Biol. Chem. 1997, 272, 1670–1676. [Google Scholar] [CrossRef]
- Kovács, Á.T.; Rákhely, G.; Kovács, K.L. Genes involved in the biosynthesis of photosynthetic pigments in the purple sulfur photosynthetic bacterium Thiocapsa roseopersicina. App. Environ. Microbiol. 2003, 69, 3093–3102. [Google Scholar] [CrossRef]
- Misawa, N.; Maoka, T.; Takemura, M. Carotenoids: Carotenoid and apocarotenoid analysis—Use of E. coli to produce carotenoid standards. Methods Enzymol. 2022, 670, 87–137. [Google Scholar]
- Wang, C.; Zhao, S.; Shao, X.; Park, J.B.; Jeong, S.H.; Park, H.J.; Kwak, W.J.; Wei, G.; Kim, S.W. Challenges and tackles in metabolic engineering for microbial production of carotenoids. Microb. Cell Fact. 2019, 18, 55. [Google Scholar] [CrossRef] [PubMed]
- Kajiwara, S.; Fraser, P.D.; Kondo, K.; Misawa, N. Expression of an exogenous isopentenyl diphosphate isomerase gene enhances isoprenoid biosynthesis in Escherichia coli. Biochem. J. 1997, 324, 421–426. [Google Scholar] [CrossRef]
- Albrecht, M.; Misawa, N.; Sandmann, G. Metabolic engineering of the terpenoid biosynthetic pathway of Escherichia coli for production of the carotenoids β-carotene and zeaxanthin. Biotechnol. Lett. 1999, 21, 791–795. [Google Scholar] [CrossRef]
- Harada, H.; Yu, F.; Okamoto, S.; Kuzuyama, T.; Utsumi, R.; Misawa, N. Efficient synthesis of functional isoprenoids from acetoacetate through metabolic pathway-engineered Escherichia coli. Appl. Microbiol. Biotechnol. 2009, 81, 915–925. [Google Scholar] [CrossRef]
- Chae, H.S.; Kim, K.-H.; Kim, S.C.; Lee, P.C. Strain-dependent carotenoid productions in metabolically engineered Escherichia coli. Appl. Biochem. Biotechnol. 2010, 162, 2333–2344. [Google Scholar] [CrossRef] [PubMed]

| Gene | Enzyme | Query Sequence for BLASTP | Nonlabens spongiae JCM13191T | ||
|---|---|---|---|---|---|
| Accession No. | Organisms | Accession No. | Identity (%) /e-Value | ||
| crtE | Geranylgeranyl pyrophosphate synthase | BAA14124 | Pantoea ananatis [7] | WP_085767001 | 30/3e-22 |
| crtB | Phytoene synthase | BAA14128 | Pantoea ananatis [7] | WP_085765367 | 26/2e-17 |
| crtI | Phytoene desaturase | BAC77668 | Flavobacterium P99-3 [16] | WP_085765368 | 83/0.0 |
| crtY | Lycopene cyclase | BAC77673 | Flavobacterium P99-3 [16] | WP_085765363 | 47/6e-124 |
| cruF | Carotenoid 1,2-hydratase | BCT98105 | Planococcus maritimus [36] | WP_085765364 | 36/1e-17 |
| crtD | Carotenoid 3,4-desaturase | BAC77671 | Flavobacterium P99-3 [18] | WP_085765365 | 73/0.0 |
| crtA-OH | Carotenoid 2-hydroxylase | BAC77674 | Flavobacterium P99-3 [19] | WP_085765362 | 50/2e-81 |
| crtA | Spheroidene monooxygenase | CAA77539 | Rhodobacter capsulatus [6] | WP_085765362 | 31/2e-30 |
| crtZ | β-Carotene hydroxylase | BAC77670 | Flavobacterium P99-3 [17] | WP_085765366 | 80/1e-79 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakazawa, K.; Mineo, D.; Harayama, T.; Yoshizawa, S.; Takaichi, S.; Sugiyama, K. Product Speculation from Carotenogenic Gene Cluster of Nonlabens spongiae Genome, and Identification of Myxol and Functional Analysis of Each Gene. Genes 2025, 16, 202. https://doi.org/10.3390/genes16020202
Nakazawa K, Mineo D, Harayama T, Yoshizawa S, Takaichi S, Sugiyama K. Product Speculation from Carotenogenic Gene Cluster of Nonlabens spongiae Genome, and Identification of Myxol and Functional Analysis of Each Gene. Genes. 2025; 16(2):202. https://doi.org/10.3390/genes16020202
Chicago/Turabian StyleNakazawa, Keisuke, Daiki Mineo, Takuya Harayama, Susumu Yoshizawa, Shinichi Takaichi, and Kenjiro Sugiyama. 2025. "Product Speculation from Carotenogenic Gene Cluster of Nonlabens spongiae Genome, and Identification of Myxol and Functional Analysis of Each Gene" Genes 16, no. 2: 202. https://doi.org/10.3390/genes16020202
APA StyleNakazawa, K., Mineo, D., Harayama, T., Yoshizawa, S., Takaichi, S., & Sugiyama, K. (2025). Product Speculation from Carotenogenic Gene Cluster of Nonlabens spongiae Genome, and Identification of Myxol and Functional Analysis of Each Gene. Genes, 16(2), 202. https://doi.org/10.3390/genes16020202





