Abstract
(1) Background: Litter size is one of the most important economic traits of sheep. The FecB locus has been extensively studied due to its significant impact on litter size in Hu sheep, and BMP15 and GDF9 have also been reported as major genes associated with litter size in sheep. This study aimed to identify variants of BMP15 and GDF9 and perform an association analysis of these variants with litter size in the Hu sheep breed. (2) Methods: In this study, exons of the BMP15 and GDF9 genes were fully sequenced to identify polymorphisms in Hu sheep. Population genetic parameters and haplotype frequencies were estimated, and an association analysis between these polymorphic loci and litter size was performed. Additionally, the protein structures of the wild-type and mutated BMP15 and GDF9 genes were predicted. (3) Results: The polymorphisms of the BMP15 and GDF9 genes were investigated within their exon regions, revealing mutations at four previously reported sites: BMP15 c.31_33CTTdel and GDF9 (G2, G3, and G4) in Hu sheep, with no novel variants were detected. Genetic analysis indicated that the GDF9-G3 and GDF9-G4 loci have low polymorphisms, whereas the BMP15 c.31_33CTTdel and the GDF9-G2 locus are moderately polymorphic. The mutation sites in the BMP15 and GDF9 genes were under Hardy–Weinberg equilibrium. Association analysis revealed that the BMP15 c.31_33CTTdel and GDF9 (G2, G3, and G4) mutations are not associated with litter size in Hu sheep. Protein structure prediction indicated that the mutations in BMP15 and GDF9 resulted in alterations to their tertiary structures. (4) Conclusions: In this study, four reported mutations in the BMP15 and GDF9 genes can also be detected in the Hu sheep breed. In these mutations, the G2 and G3 mutations of GDF9 did not alter the amino acid sequence, while the BMP15 c.31_33CTTdel mutation and the GDF9 G4 mutation resulted in protein structure alteration. Furthermore, the BMP15 c.31_33CTTdel mutation and the GDF9 mutations (G2, G3, G4) were associated with an increased tendency in litter size. However, no significant difference was observed (p > 0.05). This study provides valuable insights for improving the lambing performance of Hu sheep.
1. Introduction
Sheep (Ovis aries) are important agricultural economic animals [1]. The productive efficiency of the sheep industry largely depends on reproductive performance [2,3]. Most Chinese local sheep breeds, including the Tibetan and Mongolian sheep breeds, typically produce single lambs. Only a few sheep breeds, such as the Hu sheep and the Small-tailed Han sheep, can deliver multiple lambs per litter [4]. Hu sheep, a distinctive local breed in China, demonstrate outstanding reproductive traits, including early sexual maturation, high fecundity, and year-round continuous estrous cycles.
In the past decade, the Chinese government has significantly increased the proportion of domestic sheep raised in housed or semi-housed systems to 70% of total production. This intensive approach incorporates industrial practices into traditional farming, alleviating constraints on environmental resources. Within this large-scale farming framework, over 90% of enterprises have chosen the Hu sheep as their primary maternal breed.
The Hu sheep, originally raised in the Taihu Basin, is a descendant of Mongolian sheep. It was introduced to southern China during the Song Dynasty by northern settlers and subsequently adapted to its new environment. The Hu sheep is predominantly white, exhibiting little to no pigmentation. Both rams and ewes are polled. Its physiological traits make it particularly well-suited for intensive housed feeding systems [5]. First, the Hu sheep boasts exceptionally high fecundity, arguably one of the highest among sheep breeds worldwide. The lambing percentage exceeds 200%, and the annual birth rate averages 300%. Additionally, female Hu sheep reach sexual maturity at 4 to 5 months of age, while males mature at 5 to 6 months, allowing for year-round breeding and continuous productivity [6,7]. Currently, the majority of Hu sheep enterprises operate on a large scale, with herds exceeding 10,000 ewes. Breeding efforts within these enterprises primarily focus on enhancing meat production, as the increased economic value of meat further underscores the importance of reproductive traits. The average weight of an adult male Hu sheep ranges from 70 to 88 kg, while adult females typically weigh between 45 and 56 kg. The slaughter age for Hu sheep is generally between 6 and 7 months [8].
Litter size is influenced by multiple factors, including genetics, environmental conditions, and nutritional status, in which genetics serves as the most critical determinant [9]. At present, studies have identified several major genes that influence litter size in sheep by utilizing different sheep populations and breeds. The known major genes whose mutations can increase ovulation rate are BMPR1B, BMP15, and GDF9, all of which belong to the transforming growth factor beta (TGFβ) superfamily [10]. Previous studies identified a mutation locus (A746G) in the Booroola Merino breed, designated as FecB by the International Committee on Sheep and Goat Genetics. This mutation is located in the coding sequence region (CDS) of the BMPR1B on chromosome 6, resulting in an amino acid substitution from glutamine (Glu) to arginine (Arg) at position 249 (Q249R) [11,12]. The FecB in sheep has been demonstrated to play a crucial role in increasing ovulation numbers [13]. The Hu sheep is a native Chinese breed well known for its high fecundity. The frequency of the G allele in the FecB mutation of Hu sheep can reach 95%, which may explain the breed’s elevated reproductive rate. Currently, the FecB locus is widely used as a molecular marker of litter size in Hu sheep [4,14,15,16]. BMP15, located on the X chromosome in sheep, plays a critical role in ovarian folliculogenesis [17]. Numerous BMP15 mutations have been identified in sheep, including B1, B2 (FecXG), B3, B4 (FecXB) [18], FecXI, FecXH [19], FecXL [20], FecXR [21], FecXGr, and FecXO [22]. Previous studies in sheep indicated that mutants of BMP15 can increase ovulation rate in heterozygotes, while homozygous mutants display primary ovarian failure and lead to infertility, such as FecXI, FecXH, FecXG, etc. [10,20,21,22,23,24]. It is noteworthy that the homozygous ewes of FecXR and FecXO mutations also had an increased ovulation rate without becoming sterile. GDF9 is located on chromosome 5 in sheep and plays a crucial role in the growth and differentiation of follicles. More than a dozen GDF9 mutations have been identified in sheep, including G1, G2, G3, G4, G5, G6, G7, G8, FecGG, FecGE, FecGA, etc. [25,26,27]. In most of these mutations, heterozygous ewes carrying mutations in the GDF9 gene exhibit an increased ovulation rate and litter size. In contrast, homozygous ewes become infertile due to ovarian hypoplasia and failure of primary folliculogenesis.
Owing to the low heritability of litter size in sheep, traditional breeding methods have limited efficiency in increasing litter size. However, marker-assisted selection (MAS) has the potential to increase lambing rates within a shorter timeframe. Following the discovery of the FecB, BMP15, and GDF9 genes have emerged as crucial determinants of litter size in sheep [28], being used to explain the increased litter size and/or ovulation rate in a variety of breeds/strains, including Inverdale, Cambridge, Belclare, and Lacaune sheep. To date, numerous studies have been conducted on the BMP15 and GDF9 genes across various sheep breeds. However, only a few mutations in these genes associated with litter size have been identified in domestic sheep breeds [29]. In this study, we identify the polymorphisms of the GDF9 and BMP15 genes in Hu sheep and investigate the association between these polymorphic loci and litter size in the Hu sheep population. Four previously reported mutations, including BMP15 c.31_33CTTdel and GDF9 (G2, G3, and G4), have been assessed for their genetic diversity within the breed, and the association analysis of those mutations with litter size was performed in Hu sheep. This study provides valuable insights for improving the lambing performance of Hu sheep.
2. Materials and Methods
2.1. Ethics Statement
All experimental procedures adhered to the ethical standards set forth by the Animal Advisory Committee at the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences (Approval No. AP2022015-C1).
2.2. Experimental Animals
Hu sheep were produced and raised by Jiangsu Qianbao Animal Husbandry Co., Ltd. (Yancheng, China). Over a period of five years (2020–2024), data were collected from a nucleus flock with 5188 Hu sheep ewes. All breeding data were recorded using the Pedigree and Sheep Management System with electronic tags. This system allows for the viewing or printing of up to a three-generation pedigree, and its powerful search function enables the retrieval of sheep data at any time. Mating, pregnancy, lactation, litter size, weight, and other phenotypes can be kept for each sheep. Full offspring statistics can be viewed for all sheep used for breeding. The number of lambs per litter was documented, including counts for the first, second, and third litters, with a range of 1 to 6 lambs per litter. A total of 322 healthy adult ewes, aged 4–5 years, were sourced from this Hu sheep nucleus flock for use in this study. The ewes were housed in the same sheds, and supplements (silage and peanut seedlings) were provided as required by commercial standards. All ewes underwent artificial insemination for breeding. They were vaccinated against clostridial diseases and received preventative treatments. Lambs were weighed at birth and tagged, typically within 12 h of birth, with continuous measurements taken for six months post-lambing.
Approximately 5 mL of blood was collected from the jugular vein of each ewe using EDTA blood collection tubes and stored at −80 °C for subsequent DNA extraction.
2.3. DNA Extraction
The genomic DNA of these ewes was extracted with a blood genomic DNA extraction kit (Tiangen Biotech Co., Ltd., Beijing, China). The concentration and quality of genomic DNA were assessed using 1% agarose gel electrophoresis. The concentration and purity of the DNA were quantified with an ultramicro spectrophotometer (NanoDrop One, Thermo Scientific, Waltham, MA, USA). The samples were then stored at −20 °C until further use.
2.4. Primer Design and PCR Amplification
The primer design for PCR amplification of BMP15 and GDF9 was conducted using NCBI BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 1 September 2023), and the reference genome version is ARS-UI Ramb v3.0. The target fragment encompassed the exonic regions of both genes. All primers were synthesized by Sangon Biotech (Shanghai) Co., Ltd. (Shanghai, China). Table 1 provides comprehensive details of the BMP15 and GDF9 primers.

Table 1.
Sequences and characteristics of primers used for amplifying the BMP15 and GDF9 genes in Hu sheep.
2.5. Genotyping and DNA Sequencing
The polymerase chain reaction (PCR) was performed in a 25 µL reaction mixture, which included 1 µL of genomic DNA, 1 µL each of forward and reverse primers, 12.5 µL of 2× Taq Master Mix, and 9.5 µL of ddH2O. The thermal cycling protocol consisted of an initial denaturation step at 95 °C for 3 min, followed by 35 cycles consisting of denaturation at 95 °C for 30 s, annealing at 59 °C for 30 s, and extension at 72 °C for 1 min, concluding with a final extension at 72 °C for 10 min. Subsequently, the PCR products were sequenced by Sangon Biotech to confirm the presence of the mutation.
2.6. Prediction of Protein Structure
The protein tertiary structure prediction of wild-type and mutant BMP15 and GDF9 was performed using a homology modeling approach through the Swiss-Model platform.
2.7. Statistical Analysis
Hardy–Weinberg equilibrium of BMP15 and GDF9 in the Hu sheep population was assessed using the chi-square test. We calculated population genetic indices, including expected heterozygosity (He), the number of effective alleles (NE), and polymorphic information content (PIC), as well as genotype and allele frequencies, following the methodology outlined by Nei [30]. The litter size of different parities was tested for normal distribution using the Kolmogorov–Smirnov test and did not satisfy normal distribution (p < 0.05) (Supplementary Material S1 and S2). We employed the Kruskal–Wallis test to investigate the relationship between the BMP15 c.31_33 polymorphisms and the number of lambs per litter in a sample of 322 Hu sheep. Data are shown as mean and median. The results were considered significant when p < 0.05. Additionally, we examined the associations between mutations in the GDF9 gene (G2, G3, and G4) and the number of lambs per litter.
3. Results
3.1. Amplification of the BMP15 and GDF9
Four pairs of primers of the BMP15 gene (Exon 1 and Exon 2) and the GDF9 gene (Exon 1 and Exon 2) were designed according to reference sequences. The amplification sizes for BMP15-Exon 1, BMP15-Exon 2, GDF9-Exon 1, and GDF9-Exon 2 were 1258 bp, 1080 bp, 813 bp, and 1656 bp, respectively. The amplified fragment (Figure 1) was consistent with the target fragment.
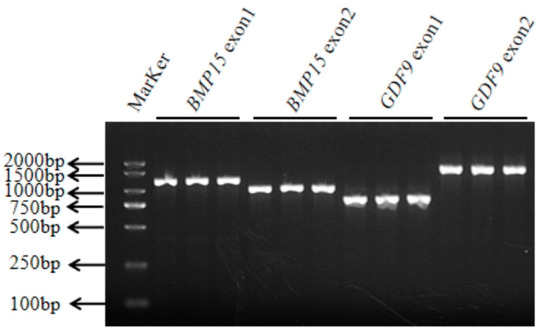
Figure 1.
The specific fragment of the BMP15 and GDF9 genes. The electrophoresis results of the PCR-amplified fragments of the BMP15 and GDF9 genes.
3.2. Identification of Mutations in the BMP15 and GDF9 Genes
The polymorphisms of the BMP15 and GDF9 genes were investigated within their exon regions, revealing mutations at four previously reported sites: BMP15 c.31_33CTTdel and GDF9 (G2, G3, and G4) in Hu sheep, with no novel variants detected. Specifically, a deletion mutation of CTT was identified at positions 31–33 in the coding region of the BMP15 gene, referred to as BMP15 c.31_33CTTdel. In Hu sheep, this mutation resulted in three genotypes: WT, WT/del, and del/del. For the GDF9 gene, the reported mutations included G2, G3, and G4 (see Figure 2). Table 2 provides a comprehensive overview of the polymorphism information for the four mutations in the BMP15 and GDF9 genes in Hu sheep.
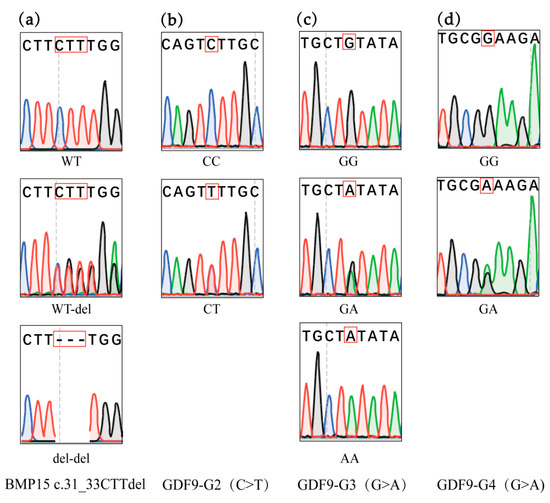
Figure 2.
Mutations of BMP15 and GDF9 by genotyping; (a) genotype of the BMP15 c.31_33CTTdel mutation in Hu sheep; (b) genotypes of the GDF9-G2 site in Hu sheep; (c) genotypes of the GDF9-G3 site in Hu sheep; (d) genotypes of the GDF9-G4 site in Hu sheep. The genotypes of identified mutations were marked by red boxes.

Table 2.
Mutations of GDF9 and BMP15 in Hu sheep.
3.3. Protein Structure Prediction
We conducted predictions of the tertiary protein structures of wild-type and mutant forms of both BMP15 c.31_33CTTdel and GDF9 G4 (G > A), respectively. As shown in Figure 3, despite no significant alterations in the overall spatial configuration of the proteins, the BMP15 protein experienced a loss of leucine at the 11th amino acid position (Figure 3a,b). The mutant GDF9 protein showed the substitution of lysine with glutamate at the 241st amino acid position (Figure 3c,d). These modifications resulted in changes in the tertiary structures of both BMP15 and GDF9.
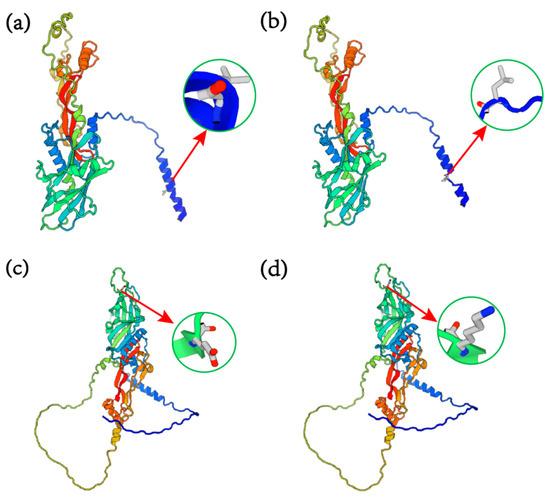
Figure 3.
The tertiary structures of wild-type and mutant BMP15 and GDF9 proteins. (a) The predicted tertiary structures of the wild-type BMP15 protein; (b) the predicted tertiary structures of BMP15 after the BMP15 c.31_33CTTdel mutation; (c) the predicted tertiary structures of wild-type GDF9; (d) the predicted tertiary structures of GDF9 after the mutant GDF9-G4 site.
3.4. Genetic Analysis of Hu Sheep Population
A population genetic analysis was conducted on four mutation sites within the BMP15 and GDF9 genes (Table 3). The results indicated that the GDF9-G2 and GDF9-G4 loci exhibited low polymorphisms (PIC < 0.25), while the BMP15 c.31_33CTTdel and the GDF9-G3 locus demonstrated moderate polymorphisms (0.25 < PIC < 0.5). Furthermore, the chi-square test revealed that the BMP15 c.31_33CTTdel locus and the G2, G3, and G4 loci of GDF9 were under Hardy–Weinberg equilibrium in Hu sheep (p > 0.05).

Table 3.
Genotype, allele frequency, and genetic diversity of the BMP15 Gene (c.31_33 CTTdel) and GDF9 gene (G2, G3, and G4 loci) in Hu sheep.
3.5. Association Analysis of BMP15 and GDF9 Mutations with Litter Size Across Different Parities in Hu Sheep
An evaluation of the association between BMP15 c.31_33CTTdel locus with litter size across different parities was performed in the nucleus flock of Hu sheep (Table 4). The analysis revealed no statistically significant differences in litter size among the various genotypes of the BMP15 c.31_33CTTdel mutation (p > 0.05). In terms of litter size across different parities, ewes with the del/del genotype displayed a numerically higher litter size compared to those with the wild-type (WT) and WT-del genotypes, except in the third parity; however, these differences were not statistically significant (p > 0.05).

Table 4.
The association analysis between BMP15 gene polymorphisms and litter size in Hu sheep.
An evaluation of the association between G2, G3, and G4 loci of GDF9 with litter size across different parities was also performed in the nucleus flock of 195 Hu sheep (Table 5). Analysis of variance revealed no significant association between the polymorphisms at the G2, G3, and G4 loci with litter size in all parities (p > 0.05). Specifically, although the litter sizes for the GDF9-G2, G3, and G4 mutant or heterozygous genotypes were higher than those of the wild type, these differences did not achieve statistical significance.

Table 5.
Association of GDF9 gene polymorphisms and litter size in Hu sheep.
4. Discussion
Litter size is a crucial economic indicator in sheep breeding and has been extensively studied in worldwide sheep breeds. Currently, BMPR1B, BMP15, and GDF9, which are all members of the transforming growth factor β (TGFβ) superfamily, are the reported major genes associated with litter size in sheep. Previous studies have reported that the BMPR1B gene is located in a region under strong selective sweeps in the Hu sheep breed [5,8,15,32]. It also has been shown that Hu sheep carry two major mutations affecting litter size, including FecB [4]. From our findings in the Hu sheep nucleus flock, the allele frequency of the FecB mutant was over 0.85. However, it has been found that the ewes carrying homozygous FecB locus still produced single offspring, and variations in lambing numbers persist in different parities [15]. The molecular mechanisms underlying exceptional reproductive performance in Hu sheep remain extensively studied. BMP15 and GDF9 have been repeatedly identified as major genes influencing the ovulation rate in foreign sheep breeds. The aim of this study was to identify the novel variants of BMP15 and GDF9 in Hu sheep breeds and investigate the association between these polymorphic loci and litter size in the Hu sheep population. This may offer new perspectives for molecular breeding in the Hu sheep breed.
In sheep, BMP15 and GDF9 are exclusively expressed in oocytes and serve as essential growth factors for follicular development. These factors exert a synergistic effect on the number of ovulations in sheep [33]. It has been proposed that the primary functions of BMP15 and GDF9 are to regulate the sensitivity of follicle-stimulating hormone (FSH) in secondary and antral follicles, help prevent premature cell differentiation, and ensure the production of a normal number of preovulatory follicles [34]. During the origination of follicular development, BMP15 regulates granulosa cell proliferation. In the later stages of follicular development, BMP15 is believed to influence the differentiation of granulosa cells by inhibiting the expression of FSH, thereby controlling follicular maturation. Furthermore, BMP15 is involved in regulating cumulus expansion and oocyte composition during the preovulatory period [34,35]. GDF9 is expressed during the type I follicular phase and plays a crucial role in regulating early follicle development through paracrine signaling. It not only promotes the proliferation of granulosa cells but also induces cumulus expansion and cytokine synthesis. Furthermore, GDF9 regulates hormone secretion in ovine cumulus cells, thereby maintaining the stability of the follicular microenvironment [36,37]. BMP15 and GDF9 can form a heterodimer that collaboratively activates the Smad signaling pathway by interacting with bone morphogenetic protein receptor type II (BMPR-II) and protein receptor type I (BMPR1B). This pathway is essential for regulating oocyte maturation and ovulation, which also influences litter size [38,39,40].
BMP15 serves as a crucial signaling molecule for the development of ovarian follicles and plays a significant regulatory role in the growth and differentiation of early oocytes. Initially identified in Romney sheep, this gene was designated as the Inverdale gene (FecXI) [18]. Subsequent research has uncovered other BMP15 mutations, including FecXG, FecXH, FecXB, FecXL, FecXR, and FecXBar. However, these mutations are rarely observed in Chinese local sheep breeds [41]. In this study, a deletion of the CTT sequence at positions 31–33 of exon 1 was identified in the BMP15 gene of Hu sheep, resulting in the loss of a leucine residue in the BMP15 protein. Numerous studies have reported the c.31_33CTTdel mutation in the BMP15 gene across various sheep breeds worldwide. These studies describe leucine-deletion polymorphisms of two leucine codons (CTT) at amino acid positions 10 and 11 in sheep, while another study only reported one amino acid change [18,24,31]. Specifically, the CTT deletion mutation in exon 1 of the BMP15 gene was observed in Chinese Small-tailed Han sheep by analyzing a total of 240 ewes each with a twin litter size, where a significant association was found between litter size and the BMP15 gene deletion (p < 0.05). On average, homozygotes and heterozygotes produced 0.5 and 0.3 more lambs per litter compared to the wild type, respectively [42]. Meanwhile, one study conducted an association analysis examining the relationship between litter size and the BMP15 c.31_33CTTdel mutation in a sample of 325 purebred Ujimqin ewes and found the c.31_33CTTdel mutation in the BMP15 gene was significantly associated with litter size in Ujimqin sheep [43]. The Small-tailed Han sheep, Ujimqin sheep, and Hu sheep are all classified as Mongolian sheep breeds. The c.31_33CTT deletion has been found to be significantly associated with litter size in both the Small-tailed Han and Ujimqin sheep. This may be attributed to the deletion of leucine in the N-terminal signal peptide region, which is hypothesized to influence BMP15 secretion and affect litter size. In this study, three genotypes of the c.31_33CTT deletion were identified, and the association analysis revealed no significant differences in litter size among these genotypes (p > 0.05). The impact of the BMP15 c.31_33CTTdel mutation on litter size varies across different breeds [31]. Furthermore, within this study, the sample size for the del-del genotype is notably limited, and the distribution of the three genotypes is imbalanced, which may influence the association result.
The GDF9 gene, a member of the transforming growth factor superfamily, has been extensively studied in sheep. Research has shown that the timing and pattern of GDF9 gene expression in ruminant oocytes correspond with the initiation and maintenance of folliculogenesis, indicating its critical role in regulating early follicle and oocyte development [44]. In sheep, there are more than a dozen identified mutations of GDF9, including G1, G2, G3, G4, G5, G6, G7, G8, and FecGH. In a domestic study, researchers identified a mutation site g.46547645T > G, in the promoter region of the GDF9 gene in Mongolian sheep, which exhibited a significant correlation with the litter size [45]. They performed whole-coding region sequencing of the GDF9 gene in Shandong Central mutton sheep and discovered two SNPs g.41768501(A > G) and g.41768485(G > A) that were associated with litter size [46]. The GDF9 mutations G1 to G8 were identified in the prolific sheep breeds Belclare and Cambridge, with G1 located on GDF9 exon 1 and the others on exon 2. Among these mutations, G2, G3, and G5 do not result in changes in amino acids. The remaining five mutations—G1, G4, G6, G7, and G8—cause amino acid changes. Specifically, G1, G4, G6, and G7 are all G > A mutations that occur at the furin processing site or before the unprocessed protein, making it unlikely to affect the mature active coding region. However, G8 changes serine to phenylalanine at residue 395 and replaces the uncharged polar group with a nonpolar group at residue 77 in the mature coding region, which may alter the function of ovine GDF9 [17,47]. In Egyptian sheep, the number of litter sizes by GDF9-G4 mutant heterozygous ewes was significantly higher than that of wild-type homozygous ewes (p ≤ 0.05). The polymorphisms of GDF9 may influence the lambing rate in sheep. The G1 and G8 mutations have been identified in Kazakhstan meat-wool sheep, with a distinct differentiation observed between the wild type and G1 mutation carriers, potentially correlating with variations in litter size [48]. The variants GDF9 g.41768501 A > G and g.41768485 G > A were found to be significantly associated with litter size in Luzhong sheep [49]. Several researchers have identified three mutations in the GDF9 gene of Iranian Afshari sheep, designated as G2, G3, and G4. Although the G4 mutation is unlikely to affect the protein activity of GDF9, it may be linked to increased fecundity in Iranian sheep [47]. Several studies have identified the GDF9 G2 mutation in Small-tailed Han sheep, which has no influence on litter size, a finding that aligns with our research outcomes. In this study, G2, G3, and G4 mutations within the GDF9 gene can also be found in the Hu sheep population. It was shown that litter size increased following the mutation, but none of these mutations showed a significant association with the litter size in our nucleus flock of Hu sheep. The analysis of the tertiary structure of the GDF9 protein showed that neither the G2 nor the G3 mutations caused any structural changes. In contrast, the G4 mutation led to a substitution of glutamic acid (Glu) with lysine (Lys) at the 241st amino acid position. However, this alteration did not affect the activity of the mature GDF9 protein.
5. Conclusions
In this study, four known mutation sites were identified in the BMP15 and GDF9 genes in our nucleus flock of Hu sheep population. Among these, the GDF9 G2 and G3 mutations did not affect the amino acid sequence. The BMP15 c.31_33CTTdel mutation led to the deletion of leucine at the eleventh amino acid, thereby altering the protein structure of BMP15. The GDF9-G4 mutation at position 241 replaced glutamic acid with lysine, resulting in a structural change to the protein. Furthermore, the BMP15 c.31_33CTTdel mutation and the GDF9 mutations (G2, G3, and G4) were associated with an increased tendency of litter size, but there was no significant difference.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes16020168/s1, Figure S1: Histogram of normal distribution of BMP15 c.31_33CTTdel locus, Figure S2: Histogram of normal distribution of GDF9 (G2, G3, and G4) locus.
Author Contributions
Conceptualization, Y.Z., H.W., and Q.L.; methodology, T.L.; software, N.Z.; validation, Y.Z., H.W., and Y.W.; formal analysis, J.C.; investigation, H.Y.; resources, Q.L. and R.M.; data curation, Q.L.; writing—original draft preparation, Y.Z.; writing—review and editing, R.M.; visualization, S.P.; supervision, Y.W.; project administration, Q.L. and D.W.; funding acquisition, Q.L.; All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Biological Breeding-National Science and Technology Major Project (2022ZD040130106, 2023ZD0406805, 2023ZD0407106), the Strategic Priority Research Program of Chinese Academy of Sciences (XDA24030205, XDA26040303), the Excellent Young Teachers Training Program of Higher Education Institutions of Anhui Province (YQYB2024039), the Key Project of the Natural Science Foundation of the Anhui Provincial Department of Education (2023AH052002), and the NSFC Incubation Program of Bengbu Medical University (2023byfy009).
Institutional Review Board Statement
This study obtained ethical approval from the Institutional Animal Care and Use Committee at the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences (Approval Code: AP2022015-C1; Approval Date: 11 March 2024), in strict adherence to China’s regulatory framework for experimental animals.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
We thank Zexuan Liu and Yiqiang Zhao from China Agricultural University for their suggestions regarding statistical methods.
Conflicts of Interest
Daxiang Wang is from Jiangsu Qianbao Animal Husbandry Co., Ltd. Other authors declare no conflicts of interest.
References
- Li, T.; Liu, Q.; Li, X.; Wang, H. Research Progress and Applications of Genes Associated with Economic Traits in Sheep. Acta Vet. Et Zootech. Sin. 2022, 53, 2417–2434. [Google Scholar]
- Cao, C.; Zhou, Q.; Kang, Y.; Akhatayeva, Z.; Liu, P.; Bai, Y.; Li, R.; Jiang, Y.; Zhang, Q.; Lan, X.; et al. A Repertoire of Single Nucleotide Polymorphisms (Snps) of Major Fecundity Bmpr1b Gene among 75 Sheep Breeds Worldwide. Theriogenology 2024, 219, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Liu, Y.; Chu, M. Detection of Polymorphisms in Six Genes and Their Association Analysis with Litter Size in Sheep. Anim. Biotechnol. 2024, 35, 3528. [Google Scholar] [CrossRef]
- Wang, W.; La, Y.; Zhou, X.; Zhang, X.; Li, F.; Liu, B. The Genetic Polymorphisms of Tgfβ Superfamily Genes Are Associated with Litter Size in a Chinese Indigenous Sheep Breed (Hu Sheep). Anim. Reprod. Sci. 2018, 189, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Zhang, Y.; Pan, Y.; Xiang, X.; Peng, C.; He, J.; Huang, G.; Wang, Z.; Zhao, P. Genomic Insights into Demographic History, Structural Variation Landscape, and Complex Traits from 514 Hu Sheep Genomes. J. Genet. Genom. 2024, 24, 00330-8. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Shen, W.; Wu, P.; Wang, C.; Li, J.; Wang, D.; Yue, W. How Food Consumption Trends Change the Direction of Sheep Breeding in China. Animals 2024, 14, 3047. [Google Scholar] [CrossRef] [PubMed]
- National Commission on Genetic Resources for Livestock and Poultry. Animal Genetic Resources in China: Sheep and Goats; China Agriculture Press: Beijing, China, 2011. [Google Scholar]
- Zhao, L.; Yuan, L.; Li, F.; Zhang, X.; Tian, H.; Ma, Z.; Zhang, D.; Zhang, Y.; Zhao, Y.; Huang, K.; et al. Whole-Genome Resequencing of Hu Sheep Identifies Candidate Genes Associated with Agronomic Traits. J. Genet. Genom. 2024, 51, 866–876. [Google Scholar] [CrossRef] [PubMed]
- Bian, Z.; Li, K.; Chen, S.; Man, C.; Wang, F.; Li, L. Association between Inha Gene Polymorphisms and Litter Size in Hainan Black Goats. PeerJ 2023, 11, 15381. [Google Scholar] [CrossRef] [PubMed]
- Polley, S.; De, S.; Brahma, B.; Mukherjee, A.; Vinesh, P.V.; Batabyal, S.; Arora, J.S.; Pan, S.; Samanta, A.K.; Datta, T.K.; et al. Polymorphism of Bmpr1b, Bmp15 and Gdf9 Fecundity Genes in Prolific Garole Sheep. Trop. Anim. Health Prod. 2010, 42, 985–993. [Google Scholar] [CrossRef]
- Wilson, T.; Wu, X.Y.; Juengel, J.L.; Ross, I.K.; Lumsden, J.M.; Lord, E.A.; Dodds, K.G.; Walling, G.A.; McEwan, J.C.; O’Connell, A.R.; et al. Highly Prolific Booroola Sheep Have a Mutation in the Intracellular Kinase Domain of Bone Morphogenetic Protein Ib Receptor (Alk-6) That Is Expressed in Both Oocytes and Granulosa Cells. Biol. Reprod. 2001, 64, 1225–1235. [Google Scholar] [CrossRef] [PubMed]
- Souza, C.J.; MacDougall, C.; MacDougall, C.; Campbell, B.K.; McNeilly, A.S.; Baird, D.T. The Booroola (Fecb) Phenotype Is Associated with a Mutation in the Bone Morphogenetic Receptor Type 1 B (Bmpr1b) Gene. J. Endocrinol. 2001, 169, R1–R6. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Hu, W.; Di, R.; Liu, Q.; Wang, X.; Zhang, X.; Zhang, J.; Chu, M. Expression Analysis of the Prolific Candidate Genes, Bmpr1b, Bmp15, and Gdf9 in Small Tail Han Ewes with Three Fecundity (Fecb Gene) Genotypes. Animals 2018, 8, 166. [Google Scholar] [CrossRef] [PubMed]
- Davis, G.H.; Balakrishnan, L.; Ross, I.K.; Wilson, T.; Galloway, S.M.; Lumsden, B.M.; Hanrahan, J.P.; Mullen, M.; Mao, X.Z.; Wang, G.L.; et al. Investigation of the Booroola (Fecb) and Inverdale (Fecx(I)) Mutations in 21 Prolific Breeds and Strains of Sheep Sampled in 13 Countries. Anim. Reprod. Sci. 2006, 92, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Zhong, T.; Hou, D.; Zhao, Q.; Zhan, S.; Wang, L.; Li, L.; Zhang, H.; Zhao, W.; Yang, S.; Niu, L. Comparative Whole-Genome Resequencing to Uncover Selection Signatures Linked to Litter Size in Hu Sheep and Five Other Breeds. BMC Genom. 2024, 25, 480. [Google Scholar] [CrossRef]
- Guan, F.; Liu, S.-R.; Shi, G.-Q.; Yang, L.-G. Polymorphism of Fecb Gene in Nine Sheep Breeds or Strains and Its Effects on Litter Size, Lamb Growth and Development. Anim. Reprod. Sci. 2007, 99, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Davis, G.H.; McEwan, J.C.; Fennessy, P.F.; Dodds, K.G.; Farquhar, P.A. Evidence for the Presence of a Major Gene Influencing Ovulation Rate on the X Chromosome of Sheep. Biol. Reprod. 1991, 44, 620–624. [Google Scholar] [CrossRef] [PubMed]
- Hanrahan, J.P.; Gregan, S.M.; Mulsant, P.; Mullen, M.; Davis, G.H.; Powell, R.; Galloway, S.M. Mutations in the Genes for Oocyte-Derived Growth Factors Gdf9 and Bmp15 Are Associated with Both Increased Ovulation Rate and Sterility in Cambridge and Belclare Sheep (Ovis Aries). Biol. Reprod. 2004, 70, 900–909. [Google Scholar] [CrossRef]
- Davis, G.; Bruce, G.; Dodds, K. Ovulation Rate and Litter Size of Prolific Inverdale (Fecxi) and Hanna (Fecxh) Sheep. Proc. Assoc. Adv. Anim. Breed. Genet. 2001, 14, 74–77. [Google Scholar]
- Bodin, L.; Di Pasquale, E.; Fabre, S.; Bontoux, M.; Monget, P.; Persani, L.; Mulsant, P. A Novel Mutation in the Bone Morphogenetic Protein 15 Gene Causing Defective Protein Secretion Is Associated with Both Increased Ovulation Rate and Sterility in Lacaune Sheep. Endocrinology 2007, 148, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Royo, A.; Dervishi, E.; Alabart, J.L.; Jurado, J.J.; Folch, J.; Calvo, J.H. Freemartinism and Fecxr Allele Determination in Replacement Ewes of the Rasa Aragonesa Sheep Breed by Duplex Pcr. Theriogenology 2009, 72, 1148–1152. [Google Scholar] [CrossRef] [PubMed]
- Demars, J.; Fabre, S.; Sarry, J.; Rossetti, R.; Gilbert, H.; Persani, L.; Tosser-Klopp, G.; Mulsant, P.; Nowak, Z.; Drobik, W.; et al. Genome-Wide Association Studies Identify Two Novel Bmp15 Mutations Responsible for an Atypical Hyperprolificacy Phenotype in Sheep. PLoS Genet. 2013, 9, e1003482. [Google Scholar] [CrossRef]
- Martinez-Royo, A.; Jurado, J.J.; Smulders, J.P.; Marti, J.I.; Alabart, J.L.; Roche, A.; Fantova, E.; Bodin, L.; Mulsant, P.; Serrano, M. A Deletion in the Bone Morphogenetic Protein 15 Gene Causes Sterility and Increased Prolificacy in Rasa Aragonesa Sheep. Anim. Genet. 2008, 39, 294–297. [Google Scholar] [CrossRef]
- Galloway, S.M.; McNatty, K.P.; Cambridge, L.M.; Laitinen, M.P.; Juengel, J.L.; Jokiranta, T.S.; McLaren, R.J.; Luiro, K.; Dodds, K.G.; Montgomery, G.W.; et al. Mutations in an Oocyte-Derived Growth Factor Gene (Bmp15) Cause Increased Ovulation Rate and Infertility in a Dosage-Sensitive Manner. Nat. Genet. 2000, 25, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Bravo, S.; Larama, G.; Paz, E.; Inostroza, K.; Montaldo, H.H.; Sepúlveda, N. Polymorphism of the Gdf9 Gene Associated with Litter Size in Araucana Creole Sheep. Anim. Genet. 2016, 47, 390–391. [Google Scholar] [CrossRef]
- Sae-Foo, P.; Triwutanon, S.; Rukkwamsuk, T. Detection of Booroola Polymorphism of Bone Morphogenetic Protein Receptor 1b and Embrapa Polymorphism of Growth Differentiation Factor 9 in Sheep in Thailand. Animals 2024, 14, 809. [Google Scholar] [CrossRef] [PubMed]
- El Fiky, Z.A.; Hassan, G.M.; Nassar, M.I. Genetic Polymorphism of Growth Differentiation Factor 9 (Gdf9) Gene Related to Fecundity in Two Egyptian Sheep Breeds. J. Assist. Reprod. Genet. 2017, 34, 1683–1690. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Ji, X.; Cang, M.; Wang, J.; Yu, H.; Liu, Y.; Zhang, W.; Wu, Y.; Zhao, S.; Cao, G.; et al. Association Analysis between Novel Variants in Lepr Gene and Litter Size in Mongolia and Ujimqin Sheep Breeds. Theriogenology 2022, 183, 79–89. [Google Scholar] [CrossRef]
- Cuiling, W.; Xinglong, Z.; Zhuo, Z.; Yunhui, Z.; Bo, Z.; Mingxin, Z.; Chunxin, W. Genetic Polymorphism Analysis of Bmp15 and Gdf9 Genes in Six Sheep Breeds. China Anim. Husb. Vet. Med. 2018, 45, 2236–2246. [Google Scholar] [CrossRef]
- Nei, M.; Li, W.H. Mathematical Model for Studying Genetic Variation in Terms of Restriction Endonucleases. Proc. Natl. Acad. Sci. USA 1979, 76, 5269–5273. [Google Scholar] [CrossRef] [PubMed]
- Najafabadi, H.A.; Khansefid, M.; Mahmoud, G.G.; Haruna, I.L.; Zhou, H.; Hickford, J.G.H. Identification of Sequence Variation in the Oocyte-Derived Bone Morphogenetic Protein 15 (Bmp15) Gene (Bmp15) Associated with Litter Size in New Zealand Sheep (Ovis Aries) Breeds. Mol. Biol. Rep. 2021, 48, 6335–6342. [Google Scholar] [CrossRef]
- Zhao, F.; Xie, R.; Fang, L.; Xiang, R.; Yuan, Z.; Liu, Y.; Wang, L. Analysis of 206 Whole-Genome Resequencing Reveals Selection Signatures Associated with Breed-Specific Traits in Hu Sheep. Evol. Appl. 2024, 17, e13697. [Google Scholar] [CrossRef] [PubMed]
- Crawford, J.L.; A Heath, D.; Reader, K.L.; Quirke, L.D.; Hudson, N.L.; Juengel, J.L.; McNatty, K.P. Oocytes in Sheep Homozygous for a Mutation in Bone Morphogenetic Protein Receptor 1b Express Lower Mrna Levels of Bone Morphogenetic Protein 15 but Not Growth Differentiation Factor 9. Reproduction 2011, 142, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.K.; Erickson, G.F.; Shimasaki, S. Are Bmp-15 and Gdf-9 Primary Determinants of Ovulation Quota in Mammals? Trends Endocrinol. Metab. 2004, 15, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Persani, L.; Rossetti, R.; Di Pasquale, E.; Cacciatore, C.; Fabre, S. The Fundamental Role of Bone Morphogenetic Protein 15 in Ovarian Function and Its Involvement in Female Fertility Disorders. Hum. Reprod. Updat. 2014, 20, 869–883. [Google Scholar] [CrossRef] [PubMed]
- Elvin, J.A.; Clark, A.T.; Wang, P.; Wolfman, N.M.; Matzuk, M.M. Paracrine Actions of Growth Differentiation Factor-9 in the Mammalian Ovary. Mol. Endocrinol. 1999, 13, 1035–1048. [Google Scholar] [CrossRef] [PubMed]
- Vitt, U.; Hayashi, M.; Klein, C.; Hsueh, A. Growth Differentiation Factor-9 Stimulates Proliferation but Suppresses the Follicle-Stimulating Hormone-Induced Differentiation of Cultured Granulosa Cells from Small Antral and Preovulatory Rat Follicles. Biol. Reprod. 2000, 62, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Mottershead, D.G.; Sugimura, S.; Al-Musawi, S.L.; Li, J.-J.; Richani, D.; White, M.A.; Martin, G.A.; Trotta, A.P.; Ritter, L.J.; Shi, J.; et al. Cumulin, an Oocyte-Secreted Heterodimer of the Transforming Growth Factor-Β Family, Is a Potent Activator of Granulosa Cells and Improves Oocyte Quality. J. Biol. Chem. 2015, 290, 24007–24020. [Google Scholar] [CrossRef]
- Xu, Y.; Li, E.; Han, Y.; Chen, L.; Xie, Z. Differential Expression of Mrnas Encoding Bmp/Smad Pathway Molecules in Antral Follicles of High- and Low-Fecundity Hu Sheep. Anim. Reprod. Sci. 2010, 120, 47–55. [Google Scholar] [CrossRef]
- Monestier, O.; Servin, B.; Auclair, S.; Bourquard, T.; Poupon, A.; Pascal, G.; Fabre, S. Evolutionary Origin of Bone Morphogenetic Protein 15 and Growth and Differentiation Factor 9 and Differential Selective Pressure between Mono- and Polyovulating Species. Biol. Reprod. 2014, 91, 83. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-F.; Jiang, Y.-L.; Du, L.-X. Studies of Bmpr-Ib and Bmp15 as Candidate Genes for Fecundity in Little Tailed Han Sheep. Yi Chuan Xue Bao 2003, 30, 755–760. [Google Scholar] [PubMed]
- Guo, W.; Chu, M.X.; Deng, X.M.; Feng, J.D.; Li, N.; Wu, C. Association of a Single Codon Deletion in Bone Morphogenetic Protein 15 Gene with Prolificacy in Small Tail Han Sheep. Asian-Australas. J. Anim. Sci. 2004, 17, 1491–1495. [Google Scholar] [CrossRef]
- Ji, X.; Cao, Z.; Hao, Q.; He, M.; Cang, M.; Yu, H.; Ma, Q.; Li, X.; Bao, S.; Wang, J.; et al. Effects of New Mutations in Bmprib, Gdf9, Bmp15, Lepr, and B4galnt2 Genes on Litter Size in Sheep. Vet. Sci. 2023, 10, 258. [Google Scholar] [CrossRef] [PubMed]
- Bodensteiner, K.; Clay, C.; Moeller, C.; Sawyer, H. Molecular Cloning of the Ovine Growth/Differentiation Factor-9 Gene and Expression of Growth/Differentiation Factor-9 in Ovine and Bovine Ovaries. Biol. Reprod. 1999, 60, 381–386. [Google Scholar] [CrossRef]
- Cheng, Z.; Liu, J.; Li, Y.; Zhang, X.; Chao, L.; Cang, M.; Wang, J.; Yu, H.; Li, G.; Tong, B. Effect of G. 46547645t>G Locus of Gdf9 Gene on Promoter Activity and Litter Size of Mongolia Sheep (Ovis Aries). J. Agric. Biotechnol. 2021, 29, 540–549. [Google Scholar]
- Wang, F.; Chu, M.; Pan, L.; Wang, X.; He, X.; Zhang, R.; Tao, L.; La, Y.; Ma, L.; Di, R. Polymorphism Detection of Gdf9 Gene and Its Association with Litter Size in Luzhong Mutton Sheep (Ovis Aries). Animals 2021, 11, 571. [Google Scholar] [CrossRef] [PubMed]
- Eghbalsaied, S.; Ghaedi, K.; Shahmoradi, S.; Pirestani, A.; Amini, H.; Saiedi, T.; Nicol, L.; McNeilly, A. Presence of Snps in Gdf9 Mrna of Iranian Afshari Sheep. Int. J. Fertil. Steril. 2012, 5, 225–230. [Google Scholar]
- Amandykova, M.; Orazymbetova, Z.; Kapassuly, T.; Kozhakhmet, A.; Khamzina, S.; Iskakov, K.; Dossybayev, K. Detection of Genetic Variations in the Gdf9 and Bmp15 Genes in Kazakh Meat-Wool Sheep. Arch. Anim. Breed. 2023, 66, 401–409. [Google Scholar] [CrossRef]
- Di, R.; Wang, F.; Yu, P.; Wang, X.; He, X.; Mwacharo, J.M.; Pan, L.; Chu, M. Detection of Novel Variations Related to Litter Size in Bmp15 Gene of Luzhong Mutton Sheep (Ovis Aries). Animals 2021, 11, 3528. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).