Vacuolar (H+)-ATPase Genes Are Essential for Cuticle and Wing Development in Locusta migratoria
Abstract
1. Introduction
2. Materials and Methods
2.1. Insects
2.2. Identification and Bioinformatics Analysis of LmV-ATPase Genes
2.3. Total RNA Isolation and First-Strand cDNA Synthesis
2.4. RT-qPCR
2.5. RNA Interference (RNAi)
2.6. Hematoxylin–Eosin (H&E) Staining
2.7. Transmission Electron Microscopy (TEM)
3. Results
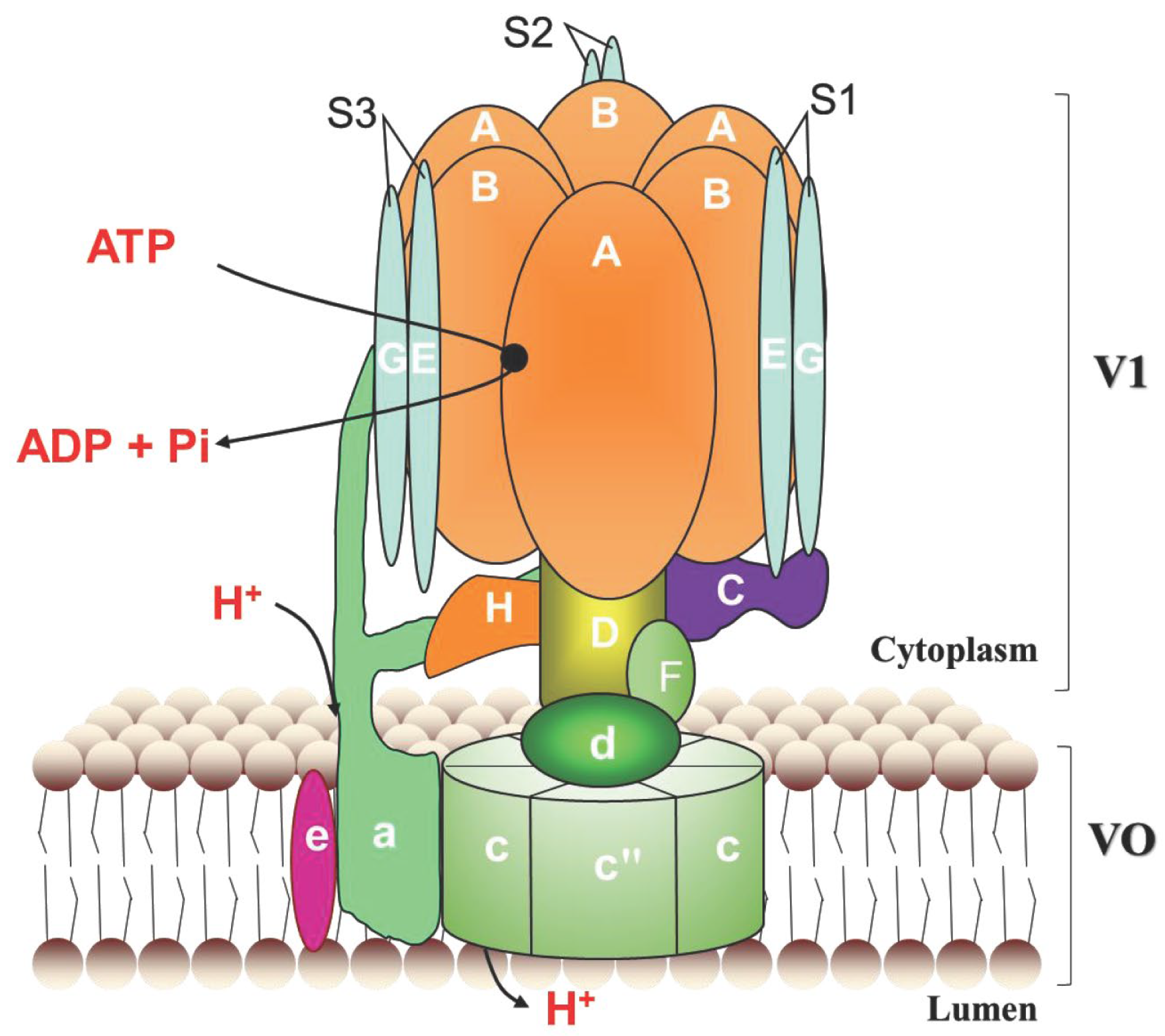
3.1. Identification and Characterization of the LmV-ATPase Genes
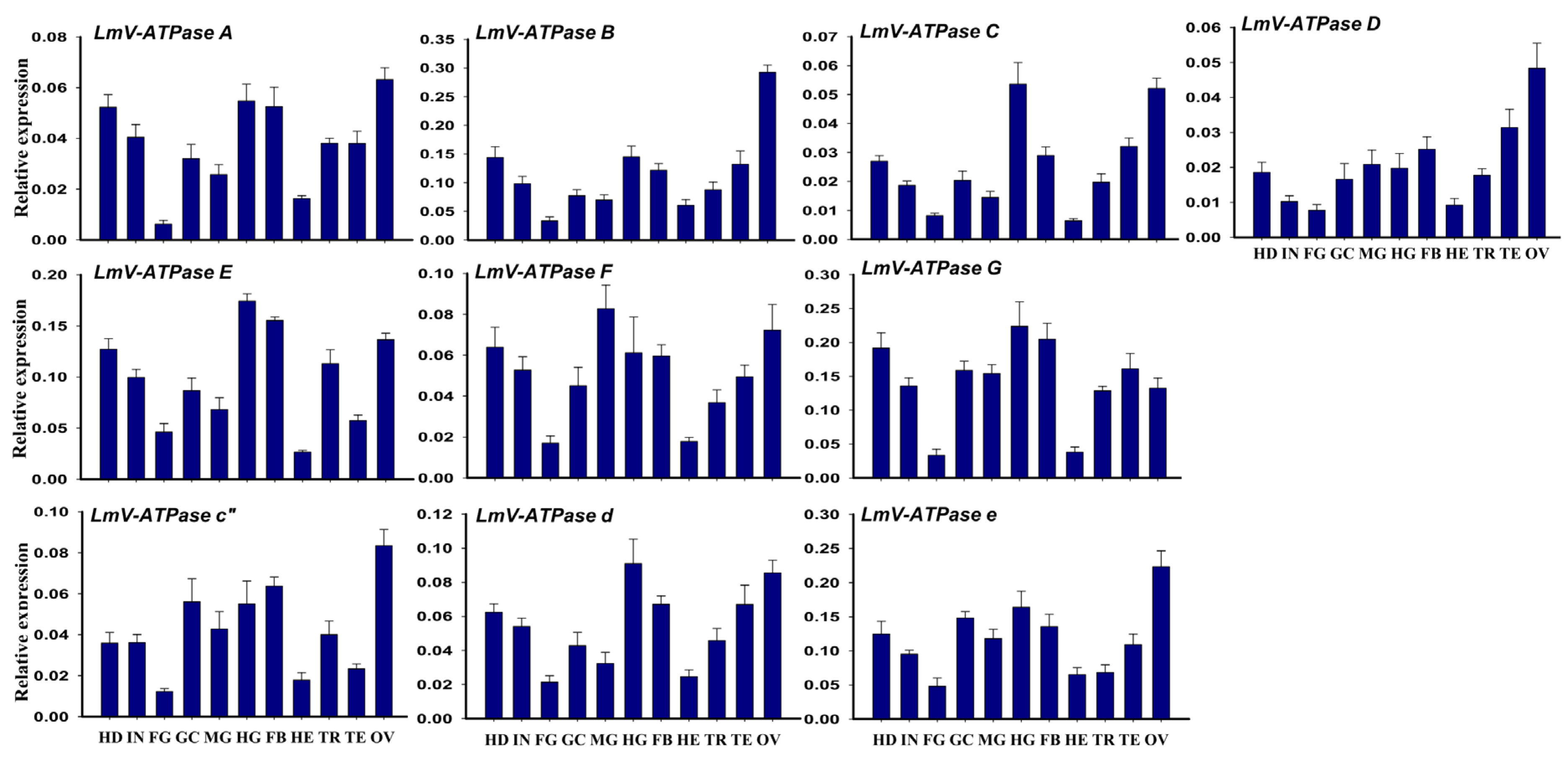
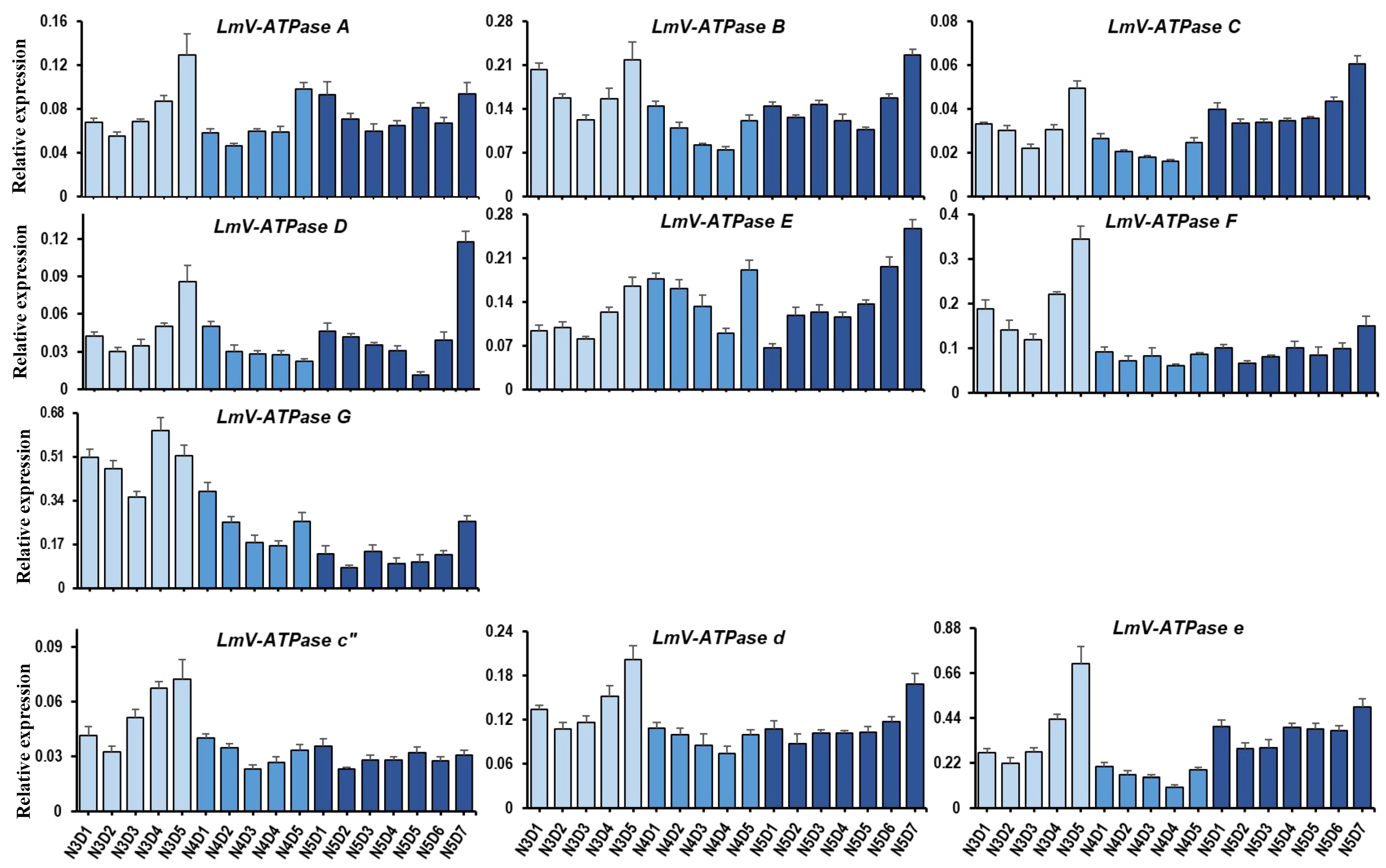
3.2. Expression Patterns of LmV-ATPase Genes
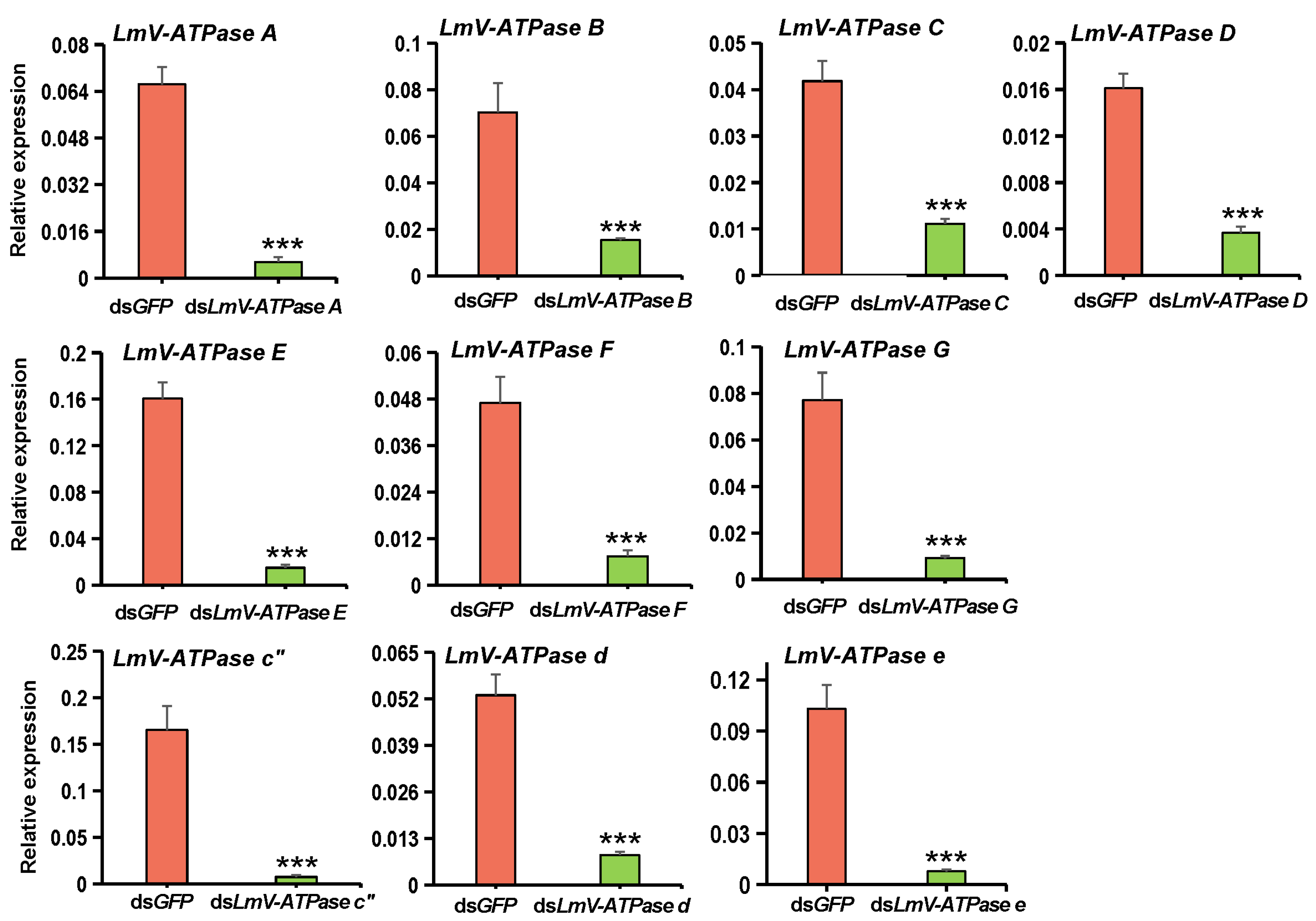
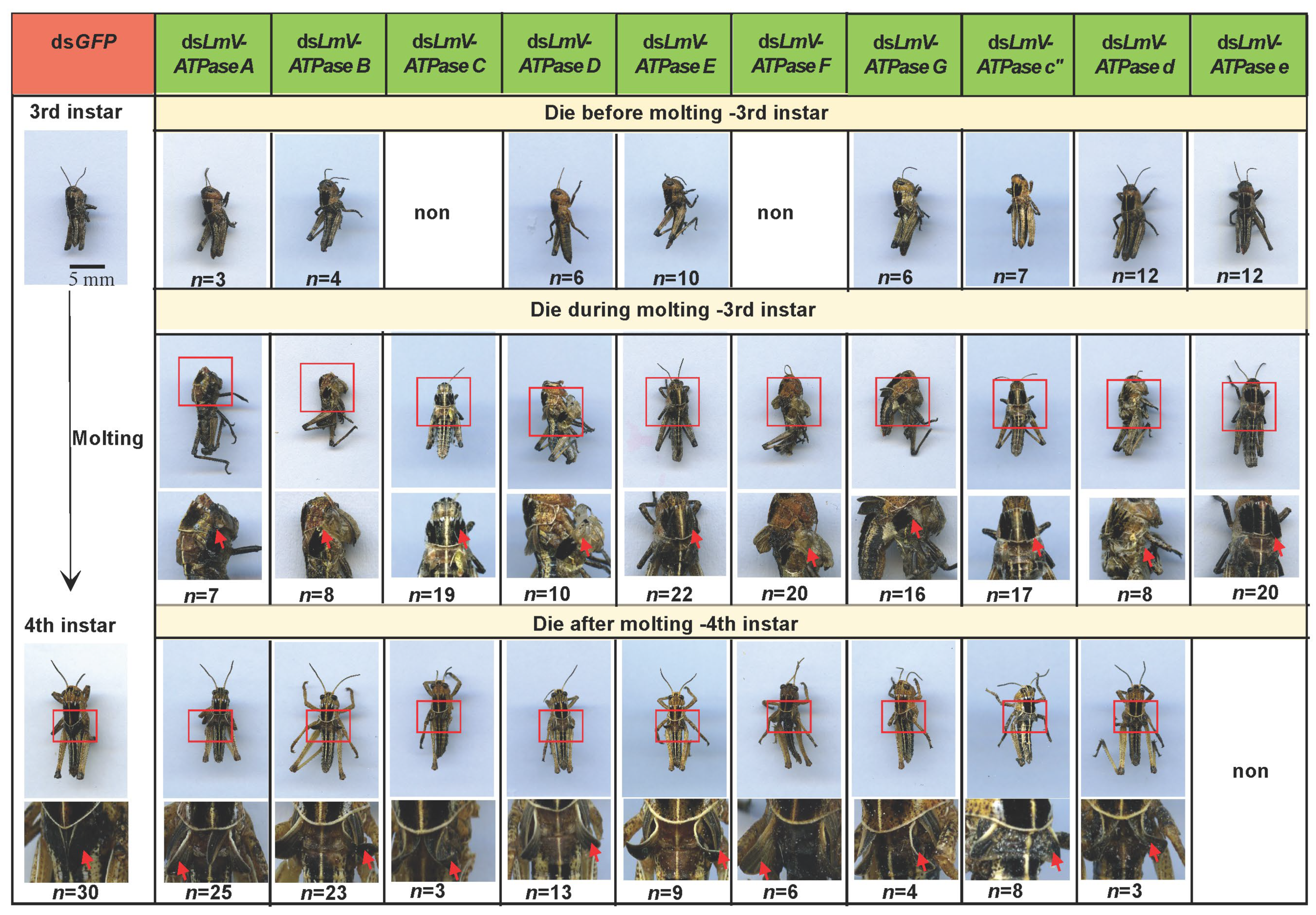
3.3. Effect on Locust Survival After LmV-ATPase Genes Knockdown
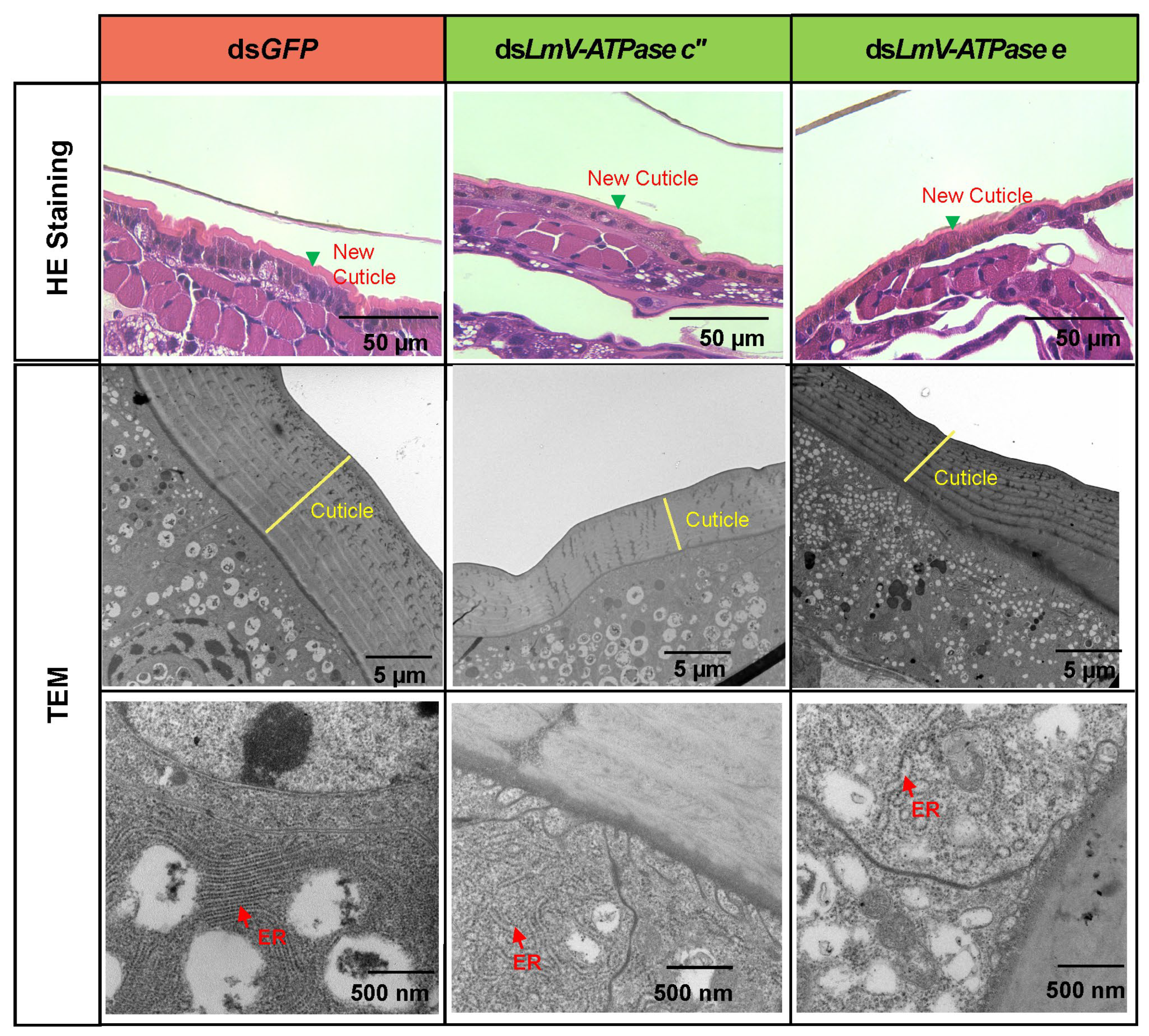
3.4. Effect on the Locust Cuticle After Knockdown LmV-ATPase c″ and LmV-ATPase e
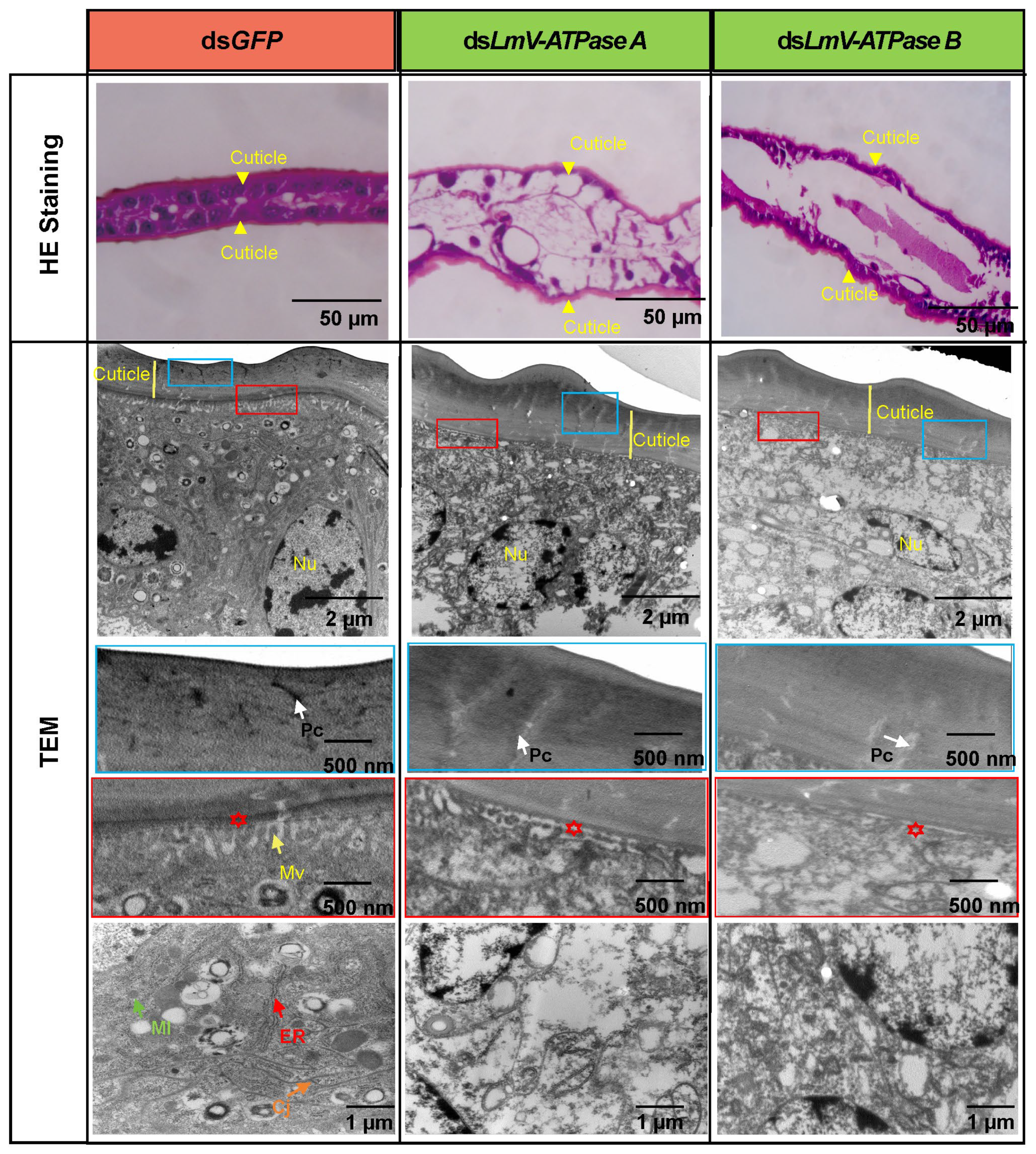
3.5. Effect on the Locust Wing After Knockdown LmV-ATPase A and LmV-ATPase B
4. Discussion
4.1. Identification and Molecular Characterization of the Locust V-ATPase Genes
4.2. LmV-ATPases Are Essential for the Survival of Locusts
4.3. LmV-ATPases Are Involved in the Cuticle and Wing Development
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kakinuma, Y.; Ohsumi, Y.; Anraku, Y. Properties of H+-translocating adenosine triphosphatase in vacuolar membranes of Saccharomyces cerevisiae. J. Biol. Chem. 1981, 256, 10859–10863. [Google Scholar] [CrossRef] [PubMed]
- Maxson, M.E.; Grinstein, S. The vacuolar-type H+-ATPase at a glance—More than a proton pump. J. Cell Sci. 2014, 127, 4987–4993. [Google Scholar] [CrossRef] [PubMed]
- Cotter, K.; Stransky, L.; McGuire, C.; Forgac, M. Recent insights into the structure, regulation, and function of the V-ATPases. Trends Biochem. Sci. 2015, 40, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Oot, R.A.; Couoh-Cardel, S.; Sharma, S.; Stam, N.J.; Wilkens, S. Breaking up and making up: The secret life of the vacuolar H+-ATPase. Protein Sci. 2017, 26, 896–909. [Google Scholar] [CrossRef] [PubMed]
- Abbas, Y.M.; Wu, D.; Bueler, S.A.; Robinson, C.V.; Rubinstein, J.L. Structure of V-ATPase from the mammalian brain. Science 2020, 367, 1240–1246. [Google Scholar] [CrossRef]
- Toei, M.; Saum, R.; Forgac, M. Regulation and isoform function of the V-ATPases. Biochemistry 2010, 49, 4715–4723. [Google Scholar] [CrossRef]
- Julian, A.T.D. The multifunctional Drosophila melanogaster V-ATPase is encoded by a multigene family. J. Bioenerg. Biomembr. 1999, 31, 75–83. [Google Scholar]
- Allan, A.K.; Juan, D.; Davies, S.A.; Julian, A.T.D. Genome-wide survey of V-ATPase genes in Drosophila reveals a conserved renal phenotype for lethal alleles. Physiol. Genom. 2005, 22, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Denef, N.; Schucpbach, T. The vacuolar proton pump, V-ATPase, is required for notch signaling and endosomal trafficking in Drosophila. Dev. Cell 2009, 17, 387–402. [Google Scholar] [CrossRef]
- Buechling, T.; Bartscherer, K.; Ohkawara, B.; Chaudhary, V.; Spirohn, K.; Niehrs, C.; Boutros, M. Wnt/Frizzled signaling requires dPRR, the Drosophila homolog of the prorenin receptor. Curr. Biol. 2010, 20, 1263–1268. [Google Scholar] [CrossRef] [PubMed]
- Hermle, T.; Saltukoglu, D.; Grucnewald, J.; Walz, G.; Simons, M. Regulation of Frizzled-dependent planar polarity signaling by a V-ATPase subunit. Curr. Biol. 2010, 20, 1269–1276. [Google Scholar] [CrossRef]
- Chang, Y.W.; Wang, Y.C.; Zhang, X.X.; Iqbal, J.; Du, Y.Z. RNA interference of genes encoding the Vacuolar-ATPase in Liriomyza trifolii. Insects 2021, 12, 41. [Google Scholar] [CrossRef]
- Jin, S.; Singh, N.D.; Li, L.; Zhang, X. Engineered chloroplast dsRNA silences cytochrome p450 monooxygenase, V-ATPase and chitin synthase genes in the insect gut and disrupts Helicoverpa armigera larval development and pupation. Plant Biotechnol. J. 2015, 13, 435–446. [Google Scholar] [CrossRef]
- Mao, J.J.; Zhang, P.Z.; Liu, C.Y.; Zeng, F.R. Co–silence of the coatomer β and vATPase A genes by siRNA feeding reduces larval survival rate and weight gain of cotton bollworm, Helicoverpa armigera. Pestic. Biochem. Physiol. 2015, 118, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.J.; Zhao, D.; Wang, Q.; Liu, Y.N.; Lu, X.J.; Guo, W. Identification and functional analysis of V-ATPaseA and C genes in Hyphantria cunea. Insects 2024, 15, 515. [Google Scholar] [CrossRef] [PubMed]
- Basnet, S.; Kamble, S.T. RNAi-mediated knockdown of vATPase subunits affects survival and reproduction of bed bugs (Hemiptera: Cimicidae). J. Med. Entomol. 2018, 55, 540–546. [Google Scholar] [CrossRef]
- Upadhyay, S.K.; Chandrashekar, K.; Thakur, N.; Verma, P.C.; Borgio, J.F.; KSingh, P.; Tuli, R.; Biosci, J. RNA interference for the control of whiteflies (Bemisia tabaci) by oral route. J. Biosci. 2011, 36, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.M.; Ashfaq, M.; Khan, A.A.; Naseem, M.T.; Mansoor, S. Evaluation of potential RNA-interference-target genes to control cotton mealybug, Phenacoccus solenopsis (Hemiptera: Pseudococcuidae). Insect Sci. 2018, 25, 778–786. [Google Scholar] [CrossRef] [PubMed]
- Rebijith, K.B.; Asokan, R.; Ranjitha, H.H.; Rajendra, B.S.; Krishna, V.; Krishna Kumar, N.K. Diet-Delivered dsRNAs for Juvenile Hormone-Binding Protein and Vacuolar ATPase-H Implied Their Potential in the Management of the Melon Aphid (Hemiptera: Aphididae). Environ. Entomol. 2016, 45, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.Z.; Yang, B.; Zhang, A.H.; Ding, D.R.; Wang, G.R. Plant-Mediated RNAi for Controlling Apolygus lucorum. Front. Plant Sci. 2019, 10, 64. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Rotenberg, D.; Afsharifar, A.; Barandoc-Alviar, K.; Whitfield, A.E. Development of RNAi methods for Peregrinus maidis, the corn planthopper. PLoS ONE 2013, 8, e70243. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.M.; Ashfaq, M.; Kiss, Z.; Khan, A.A.; Mansoor, S.; Falk, B.W. Use of recombinant tobacco mosaic virus to achieve RNA interference in plants against the citrus mealybug, Planococcus citri (Hemiptera: Pseudococcidae). PLoS ONE 2013, 8, e73657. [Google Scholar] [CrossRef]
- Sato, K.; Miyata, K.; Ozawa, S.; Hasegawa, K. Systemic RNAi of V-ATPase subunit B causes molting defect and developmental abnormalities in Periplaneta fuliginosa. Insect Sci. 2019, 26, 721–731. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.D.; Chen, Y.H.; Zhang, Y.X.; Lin, J.W.; Gao, S.J.; Tang, B.Z.; Hou, Y.M. Establishment of RNAi-Mediated Pest Control Method for Red Imported Fire Ant, Solenopsis invicta. J. Agric. Food Chem. 2024, 72, 10936–10943. [Google Scholar] [CrossRef] [PubMed]
- Baum, J.A.; Bogaert, T.; Clinton, W.; Heck, G.R.; Feldmann, P.; Ilagan, O.; Johnson, S.; Plaetinck, G.; Munyikwa, T.; Pleau, M.; et al. Control of coleopteran insect pests through RNA interference. Nature Biotechnol. 2007, 25, 1322–1326. [Google Scholar] [CrossRef]
- Pereira, A.E.; Vélez, A.M.; Meinke, L.J.; Siegfried, B.D. Sublethal effects of vATPase-A and Snf7 dsRNAs on biology of southern corn rootworm, Diabrotica undecimpunctata howardi barber. J. Econ. Entomol. 2017, 110, 2545–2553. [Google Scholar] [CrossRef]
- Whyard, S.; Singh, A.D.; Wong, S. Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem. Mol. Biol. 2009, 39, 824–832. [Google Scholar] [CrossRef]
- Cao, M.; Gatehouse, J.A.; Fitches, E.C. A systematic study of RNAi effects and dsRNA stability in Tribolium castaneum and Acyrthosiphon pisum, following injection and ingestion of analogous dsRNAs. Int. J. Mol. Sci. 2018, 19, 1079. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Xu, J.; Palli, R.; Ferguson, J.; Palli, S.R. Ingested RNA interference for managing the populations of the Colorado potato beetle, Leptinotarsa decemlineata. Pest. Manag. Sci. 2011, 67, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Fu, K.Y.; Guo, W.C.; Lü, F.G.; Liu, X.P.; Li, G.Q. Response of the vacuolar ATPase subunit E to RNA interference and four chemical pesticides in Leptinotarsa decemlineata (say). Pestic. Biochem. Physiol. 2014, 114, 16–23. [Google Scholar] [CrossRef]
- Powell, M.E.; Bradish, H.M.; Gatehouse, J.A.; Fitches, E.C. Systemic RNAi in the small hive beetle Aethina tumida Murray (Coleoptera: Nitidulidae), a serious pest of the European honey bee Apis mellifera. Pest. Manag. Sci. 2017, 73, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Lü, J.; Guo, M.; Chen, S.; Noland, J.E.; Guo, W.; Sang, W.; Qi, Y.; Qiu, B.; Zhang, Y.; Yang, C.; et al. Double-stranded RNA targeting vATPase B reveals a potential target for pest management of Henosepilachna vigintioctopunctata. Pestic. Biochem. Physiol. 2020, 165, 104555. [Google Scholar]
- Zeng, J.; Kang, W.N.; Jin, L.; Anjum, A.A.; Li, G.Q. Vacuolar ATPase subunit F is critical for larval survival in Henosepilachna vigintioctopunctata. Insect Mol. Biol. 2022, 31, 177–189. [Google Scholar] [CrossRef]
- Zeng, J.; Kang, W.N.; Jin, L.; Anjum, A.A.; Li, G.Q. Knockdown of Vacuolar ATPase Subunit G Gene Affects Larval Survival and Impaired Pupation and Adult Emergence in Henosepilachna vigintioctopunctata. Insects 2021, 12, 935. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Mu, L.L.; Jin, L.; Anjum, A.; Li, G.Q. RNAi of vacuolar-type H(+)-ATPase genes causes growth delay and molting defect in Henosepilachna vigintioctopunctata. Bull. Entomol. Res. 2021, 11, 705–714. [Google Scholar] [CrossRef]
- Guo, W.; Guo, M.; Yang, C.; Liu, Z.; Chen, S.; Lü, J.; Qiu, B.; Zhang, Y.; Zhou, X.; Pan, H. RNA interference-mediated silencing of vATPase subunits A and E affect survival and development of the 28-spotted ladybeetle, Henosepilachna vigintioctopunctata. Insect Sci. 2021, 28, 1664–1676. [Google Scholar] [CrossRef] [PubMed]
- Badillo-Vargas, I.E.; Rotenberg, D.; Schneweis, B.A.; Whitfield, A.E. RNA interference tools for the western flower thrips, Frankliniella occidentalis. J. Insect Phys. 2015, 76, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Xia, Y.X. Vacuolar ATPase subunit H is essential for the survival and moulting of Locusta migratoria manilensis. Insect Mol. Biol. 2012, 21, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.J.; Liang, X.Y.; Guo, J.; Shi, X.K.; Hans, M.; Zhu, K.Y.; Zhang, J.Z. V-ATPase subunit a is required for survival and midgut development of Locusta migratoria. Insect Mol. Biol. 2022, 31, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.K.; Liu, X.J.; Cooper, A.M.; Silver, K.; Hans, M.; Zhu, K.Y.; Zhang, J.Z. Vacuolar (H+)-ATPase subunit c is essential for the survival and systemic RNA interference response in Locusta migratoria. Pest. Manag. Sci. 2022, 78, 1555–1566. [Google Scholar] [CrossRef]
- Zhao, Y.G.; Li, P.; Yao, X.; Li, Y.P.; Tian, Y.; Xie, G.Y.; Deng, Z.Y.; Xu, S.X.; Wei, J.Z.; Li, X.C.; et al. V-ATPase E mediates Cry2Ab binding and toxicity in Helicoverpa armigera. Pestic. Biochem. Physiol. 2024, 198, 105744. [Google Scholar]
- Xie, C.; Xiong, L.; Ye, M.; Shen, L.L.; Li, J.G.; Zhang, Z.; You, M.S.; You, S.J. Genome-wide analysis of V-ATPase genes in Plutella xylostella (L) and the potential role of PxVHA-G1 in resistance to Bacillus thuringiensis Cry1Ac toxin. Int. J. Biol. Macromol. 2022, 194, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.M.; Gou, X.; Qin, Z.Y.; Li, D.Q.; Wang, Y.; Ma, E.B.; Li, S.; Zhang, J.Z. Identification and expression of cuticular protein genes based on Locusta migratoria transcriptome. Sci. Rep. 2017, 7, 45462. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.J.; Li, F.; Li, D.Q.; Zhang, W.Q.; Ma, E.B.; Zhu, K.Y.; Zhang, J.Z. Molecular and Functional Analysis of UDP- N-Acetylglucosamine Pyrophosphorylases from the Migratory Locust, Locusta migratoria. PLoS ONE 2013, 8, e71970. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.M.; Xie, Y.P.; Xue, J.L.; Gao, Y.; Zhang, Y.F.; Zhang, X.M.; Tan, J.S. Histopathological changes of Ceroplastes japonicus infected by Lecanicillium lecanii. J. Invertebr. Pathol. 2009, 101, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Fang, X.; Yang, P.; Jiang, X.; Jiang, F.; Zhao, D.; Li, B.; Cui, F.; Wei, J.; Ma, C.; et al. The locust genome provides insight into swarm formation and long-distance flight. Nature Commun. 2014, 5, 2957. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Mu, L.L.; Jin, L.; Anjum, A.A.; Li, G.Q. Evaluation of three vacuolar ATPase genes as potential RNAi target in Henosepilachn vigintioctopunctata. J. Asia Pac. Entomol. 2021, 24, 55–63. [Google Scholar] [CrossRef]
- Liu, X.J.; Li, Y.; Gao, Y.; Wakil, A.E.; Moussian, B.; Zhang, J.Z. RNA interference-mediated silencing of coat protein II (COPII) genes affects the gut homeostasis and cuticle development in Locusta migratoria. Int. J. Biol. Macromol. 2024, 266, 131137. [Google Scholar] [CrossRef]
- Norum, M.; Tång, E.; Chavoshi, T.; Schwarz, H.; Linke, D.; Uv, A.; Moussian, B. Trafficking through COPII stabilises cell polarity and drives secretion during Drosophila epidermal differentiation. PLoS ONE. 2010, 5, e10802. [Google Scholar] [CrossRef]
- Hughes, H.; Stephens, D.J. Assembly, organization, and function of the COPII coat. Cell Biol. 2008, 129, 129–151. [Google Scholar] [CrossRef]
- Liégeois, S.; Benedetto, A.; Michaux, G.; Belliard, G.; Labouesse, M. Genes required for osmoregulation and apical secretion in Caenorhabditis elegans. Genetics 2007, 175, 709–724. [Google Scholar] [CrossRef] [PubMed]
- Mo, D.; Chen, Y.; Jiang, N.; Shen, J.; Zhang, J. Investigation of isoform specific functions of the V-ATPase a subunit during Drosophila wing development. Front. Genet. 2020, 11, 723. [Google Scholar] [CrossRef]

| Name | CDS (bp) | Amino Acids | Mw (KDa) | pI | TM (Position; aa) |
|---|---|---|---|---|---|
| LmV-ATPase A | 1848 | 615 | 68.2 | 5.11 | - |
| LmV-ATPase B | 1503 | 500 | 55.7 | 5.33 | - |
| LmV-ATPase C | 1158 | 385 | 44.4 | 8.16 | - |
| LmV-ATPase D | 753 | 250 | 28.2 | 9.63 | - |
| LmV-ATPase E | 684 | 227 | 26.2 | 8.51 | - |
| LmV-ATPase F | 372 | 123 | 13.8 | 5.92 | - |
| LmV-ATPase G | 357 | 118 | 14.0 | 9.66 | - |
| LmV-ATPase H | 1590 | 529 | 60.8 | 5.54 | - |
| LmV-ATPase c″ | 627 | 208 | 21.7 | 8.87 | 5–27; 47–69; 89–111; 137–159; 172–194 |
| LmV-ATPase d | 1047 | 348 | 39.6 | 4.93 | - |
| LmV-ATPase e | 255 | 84 | 9.2 | 9.35 | 4–26; 33–51 |
| Gene Name | Gene Silencing Efficiency | Nymphs (n) | Die Before Molting | Die During Molting | Die After Molting | The Accumulative Mortality |
|---|---|---|---|---|---|---|
| LmV-ATPase A | 90.2% | 38 | 7.8% | 18.4% | 65.8% | 92.0% |
| LmV-ATPase B | 78.1% | 39 | 10.3% | 20.5% | 58.9% | 89.7% |
| LmV-ATPase C | 73.3% | 32 | non | 59.3% | 9.4% | 76.7% |
| LmV-ATPase D | 77.2% | 31 | 19.3% | 32.3% | 41.9% | 93.5% |
| LmV-ATPase E | 90.6% | 43 | 23.2% | 51.2% | 20.9% | 95.3% |
| LmV-ATPase F | 84.2% | 31 | non | 64.5% | 19.4% | 83.9% |
| LmV-ATPase G | 88.2% | 36 | 16.7% | 44.4% | 11.1% | 72.2% |
| LmV-ATPase c” | 95.5% | 33 | 21.2% | 51.5% | 24.3% | 97.0% |
| LmV-ATPase d | 84.2% | 29 | 44.8% | 27.6% | 10.3% | 82.7% |
| LmV-ATPase e | 92.5% | 35 | 34.3% | 57.1% | non | 91.4% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Liang, X.; Shi, X.; Zhang, J. Vacuolar (H+)-ATPase Genes Are Essential for Cuticle and Wing Development in Locusta migratoria. Genes 2025, 16, 145. https://doi.org/10.3390/genes16020145
Liu X, Liang X, Shi X, Zhang J. Vacuolar (H+)-ATPase Genes Are Essential for Cuticle and Wing Development in Locusta migratoria. Genes. 2025; 16(2):145. https://doi.org/10.3390/genes16020145
Chicago/Turabian StyleLiu, Xiaojian, Xiaoyu Liang, Xuekai Shi, and Jianzhen Zhang. 2025. "Vacuolar (H+)-ATPase Genes Are Essential for Cuticle and Wing Development in Locusta migratoria" Genes 16, no. 2: 145. https://doi.org/10.3390/genes16020145
APA StyleLiu, X., Liang, X., Shi, X., & Zhang, J. (2025). Vacuolar (H+)-ATPase Genes Are Essential for Cuticle and Wing Development in Locusta migratoria. Genes, 16(2), 145. https://doi.org/10.3390/genes16020145






