Exploring miR-21 Knock-Out Using CRISPR/Cas as a Treatment for Lung Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
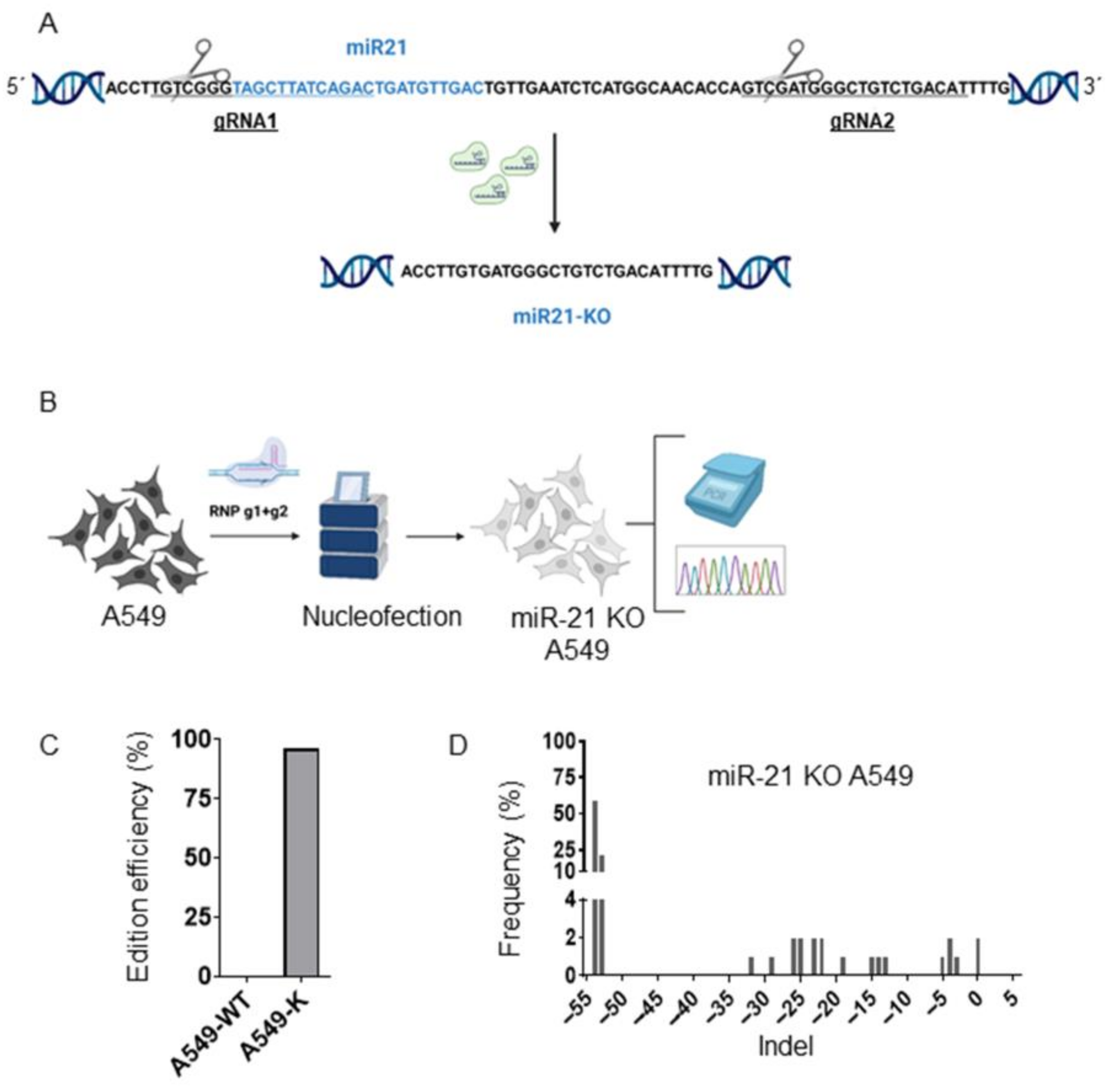
2.2. Generation of miR-21 KO A549 Cells by Cas9 RNP Nucleofection
2.3. ICE (Inference of CRISPR Edits)
2.4. Proliferation Assay (Sulforhodamine B, SBR)
2.5. Wound-Healing Assay
2.6. Colony Formation
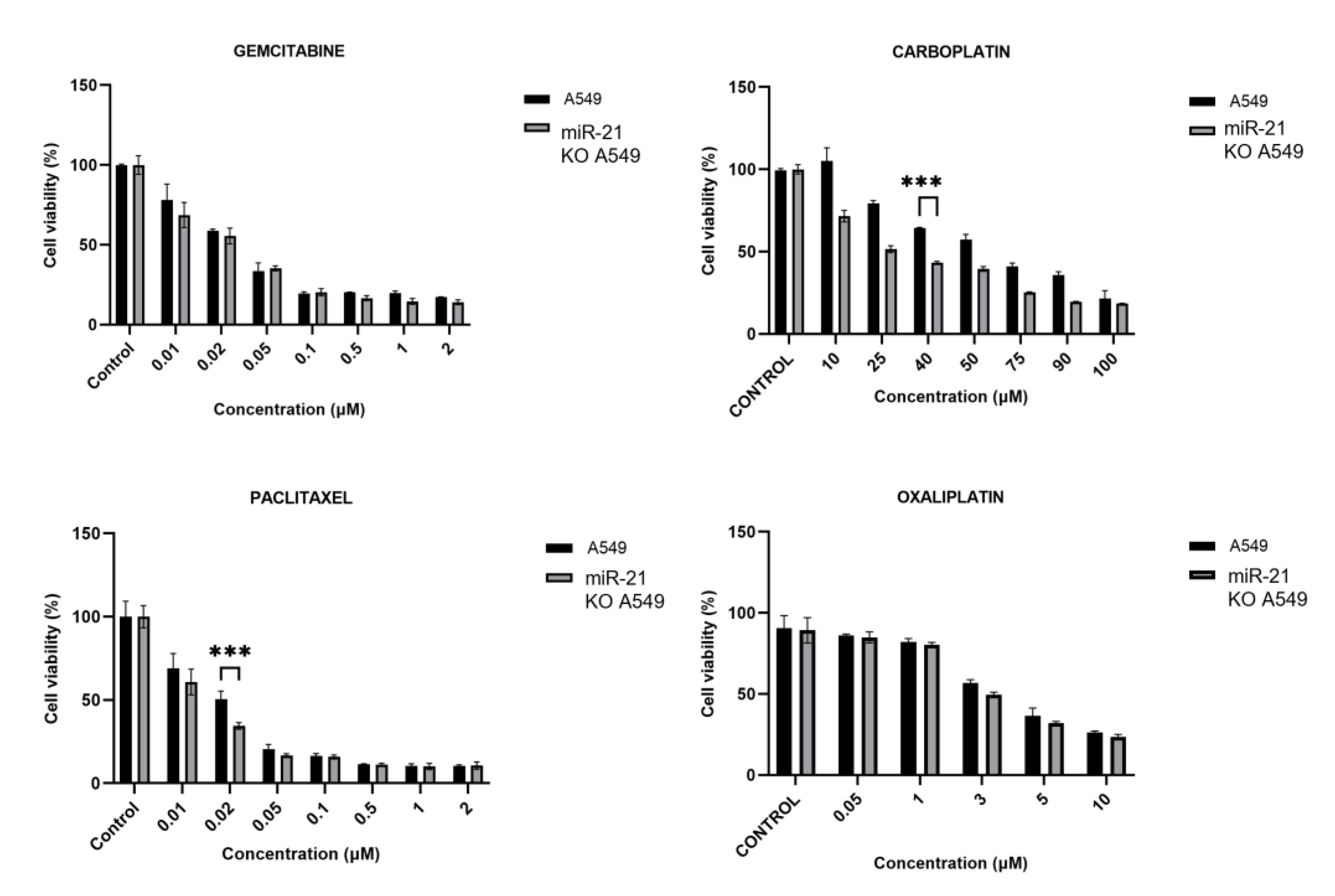
2.7. Determination of Half-Maximal Inhibitory Concentration (IC50)
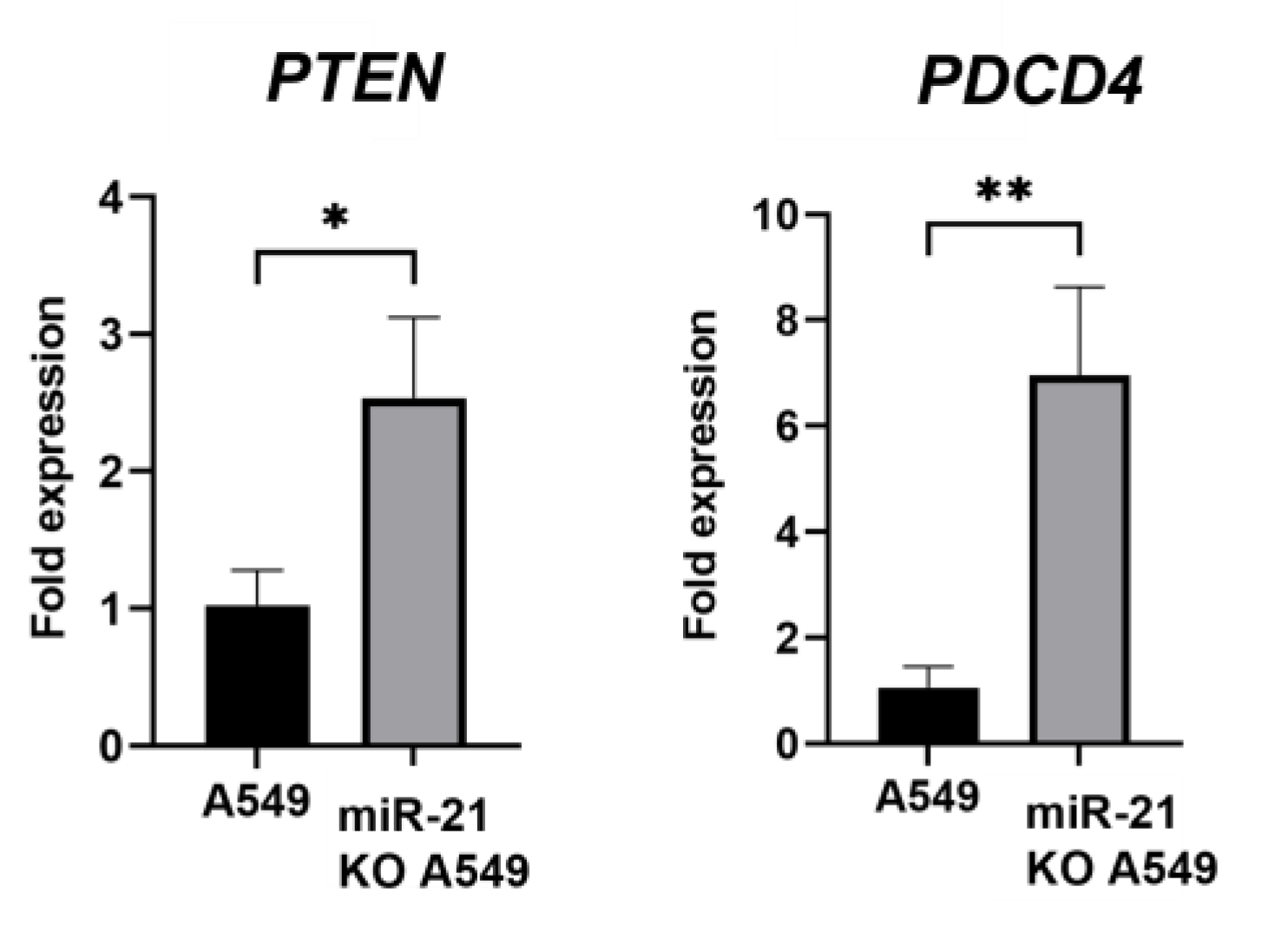
2.8. RNA Extraction and Quantitative Real-Time PCR
2.9. Statistical Analysis
3. Results
3.1. Generation of A549 miR-21 Knock-Out Cell Models by CRISPR/Cas9
3.2. MiR-21 Knock-Out Decreases Proliferation, Migration, and Colony Formation
3.3. MiR-21 Knock-Out Increases Carboplatin and Paclitaxel Activity
3.4. Gene Expression Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, I.V.; Schwartz, D.A. Epigenetic Control of Gene Expression in the Lung. Am. J. Respir. Crit. Care Med. 2011, 183, 1295–1301. [Google Scholar] [CrossRef]
- Li, H.; Zhao, J.; Jia, X.; Zhang, Y.; Du, Y.; Li, H.; Ma, L.; Huang, J. MiR-21 Promotes Growth, Invasion and Migration of Lung Cancer Cells by AKT/P-AKT/Cleaved-Caspase 3/MMP-2/MMP-9 Signaling Pathway. Int. J. Clin. Exp. Pathol. 2020, 13, 692–700. [Google Scholar] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Pelosi, G.; Sonzogni, A.; Viale, G. The Classification of Lung Carcinoma: Time to Change the Morphology-Based Approach? Int. J. Surg. Pathol. 2010, 18, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Menon, A.; Abd-Aziz, N.; Khalid, K.; Poh, C.L.; Naidu, R. MiRNA: A Promising Therapeutic Target in Cancer. Int. J. Mol. Sci. 2022, 23, 11502. [Google Scholar] [CrossRef]
- He, B.; Zhao, Z.; Cai, Q.; Zhang, Y.; Zhang, P.; Shi, S.; Xie, H.; Peng, X.; Yin, W.; Tao, Y.; et al. MiRNA-Based Biomarkers, Therapies, and Resistance in Cancer. Int. J. Biol. Sci. 2020, 16, 2628–2647. [Google Scholar] [CrossRef] [PubMed]
- Abdelaal, A.M.; Sohal, I.S.; Iyer, S.; Sudarshan, K.; Kothandaraman, H.; Lanman, N.A.; Low, P.S.; Kasinski, A.L. A First-in-Class Fully Modified Version of MiR-34a with Outstanding Stability, Activity, and Anti-Tumor Efficacy. Oncogene 2023, 42, 2985–2999. [Google Scholar] [CrossRef]
- Abdelaal, A.M.; Sohal, I.S.; Iyer, S.G.; Sudarshan, K.; Orellana, E.A.; Ozcan, K.E.; dos Santos, A.P.; Low, P.S.; Kasinski, A.L. Selective Targeting of Chemically Modified MiR-34a to Prostate Cancer Using a Small Molecule Ligand and an Endosomal Escape Agent. Mol. Ther. Nucleic Acids 2024, 35, 102193. [Google Scholar] [CrossRef]
- Shi, Y.P.; Liu, G.L.; Li, S.; Liu, X.L. MiR-17-5p Knockdown Inhibits Proliferation, Autophagy and Promotes Apoptosis in Thyroid Cancer via Targeting PTEN. Neoplasma 2020, 67, 249–258. [Google Scholar] [CrossRef]
- Pfeffer, S.R.; Yang, C.H.; Pfeffer, L.M. The Role of MiR-21 in Cancer. Drug Dev. Res. 2015, 76, 270–277. [Google Scholar] [CrossRef]
- Zhao, W.; Zhao, J.J.; Zhang, L.; Xu, Q.F.; Zhao, Y.M.; Shi, X.Y.; Xu, A.G. Serum MiR-21 Level: A Potential Diagnostic and Prognostic Biomarker for Non-Small Cell Lung Cancer. Int. J. Clin. Exp. Med. 2015, 8, 14759. [Google Scholar] [PubMed]
- Gao, W.; Lu, X.; Liu, L.; Xu, J.; Feng, D.; Shu, Y. MiRNA-21: A Biomarker Predictive for Platinum-Based Adjuvant Chemotherapy Response in Patients with Non-Small Cell Lung Cancer. Cancer Biol. Ther. 2012, 13, 330–340. [Google Scholar] [CrossRef]
- Ribas, J.; Ni, X.; Castanares, M.; Liu, M.M.; Esopi, D.; Yegnasubramanian, S.; Rodriguez, R.; Mendell, J.T.; Lupold, S.E. A Novel Source for MiR-21 Expression through the Alternative Polyadenylation of VMP1 Gene Transcripts. Nucleic Acids Res. 2012, 40, 6821–6833. [Google Scholar] [CrossRef]
- Zhou, C.; Wu, L.; Fan, Y.; Wang, Z.; Liu, L.; Chen, G.; Zhang, L.; Huang, D.; Cang, S.; Yang, Z.; et al. Sintilimab Plus Platinum and Gemcitabine as First-Line Treatment for Advanced or Metastatic Squamous NSCLC: Results From a Randomized, Double-Blind, Phase 3 Trial (ORIENT-12). J. Thorac. Oncol. 2021, 16, 1501–1511. [Google Scholar] [CrossRef]
- Vasconcellos, V.F.; Marta, G.N.; da Silva, E.M.; Gois, A.F.; de Castria, T.B.; Riera, R. Cisplatin versus Carboplatin in Combination with Third-Generation Drugs for Advanced Non-Small Cell Lung Cancer. Cochrane Database Syst. Rev. 2020, 2020, CD009256. [Google Scholar] [CrossRef]
- Bi, Y.; Li, F.; Ren, J.; Han, X. The Safety and Efficacy of Oxaliplatin-Loaded Drug-Eluting Beads Transarterial Chemoembolization for the Treatment of Unresectable or Advanced Lung Cancer. Front. Pharmacol. 2022, 13, 1079707. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Gu, A.; Tu, H.; Huang, C.; Wang, H.; Yu, Z.; Wang, X.; Cao, L.; Shu, Y.; Yang, R.; et al. Comparing Nanoparticle Polymeric Micellar Paclitaxel and Solvent-Based Paclitaxel as First-Line Treatment of Advanced Non-Small-Cell Lung Cancer: An Open-Label, Randomized, Multicenter, Phase III Trial. Ann. Oncol. 2021, 32, 85–96. [Google Scholar] [CrossRef]
- Min, H.Y.; Lee, H.Y. Mechanisms of Resistance to Chemotherapy in Non-Small Cell Lung Cancer. Arch. Pharmacal Res. 2021, 44, 146–164. [Google Scholar] [CrossRef]
- Sun, L.H.; Tian, D.; Yang, Z.C.; Li, J.L. Exosomal MiR-21 Promotes Proliferation, Invasion and Therapy Resistance of Colon Adenocarcinoma Cells through Its Target PDCD4. Sci. Rep. 2020, 10, 8271. [Google Scholar] [CrossRef]
- Chen, Z.; Yuan, Y.C.; Wang, Y.; Liu, Z.; Chan, H.J.; Chen, S. Down-Regulation of Programmed Cell Death 4 (PDCD4) Is Associated with Aromatase Inhibitor Resistance and a Poor Prognosis in Estrogen Receptor-Positive Breast Cancer. Breast Cancer Res. Treat. 2015, 152, 29–39. [Google Scholar] [CrossRef]
- Wei, Z.T.; Zhang, X.; Wang, X.Y.; Gao, F.; Zhou, C.J.; Zhu, F.L.; Wang, Q.; Gao, Q.; Ma, C.H.; Sun, W.S.; et al. PDCD4 Inhibits the Malignant Phenotype of Ovarian Cancer Cells. Cancer Sci. 2009, 100, 1408–1413. [Google Scholar] [CrossRef]
- De Marco, C.; Laudanna, C.; Rinaldo, N.; Oliveira, D.M.; Ravo, M.; Weisz, A.; Ceccarelli, M.; Caira, E.; Rizzuto, A.; Zoppoli, P.; et al. Specific Gene Expression Signatures Induced by the Multiple Oncogenic Alterations That Occur within the PTEN/PI3K/AKT Pathway in Lung Cancer. PLoS ONE 2017, 12, e0178865. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; He, Y.; Wang, X.; Peng, D.; Chen, X.; Li, X.; Wang, Q. Overexpression of MiR-21 in Stem Cells Improves Ovarian Structure and Function in Rats with Chemotherapy-Induced Ovarian Damage by Targeting PDCD4 and PTEN to Inhibit Granulosa Cell Apoptosis. Stem Cell Res. Ther. 2017, 8, 187. [Google Scholar] [CrossRef] [PubMed]
- Tirpe, A.; Gulei, D.; Razvan Tirpe, G.; Nutu, A.; Irimie, A.; Campomenosi, P.; Ancutapop, L.; Berindan-Neagoe, I. Beyond Conventional: The New Horizon of Anti-Angiogenic MicroRNAs in Non-Small Cell Lung Cancer Therapy. Int. J. Mol. Sci. 2020, 21, 8002. [Google Scholar] [CrossRef]
- Toscano, M.G.; Anderson, P.; Muñoz, P.; Lucena, G.; Cobo, M.; Benabdellah, K.; Gregory, P.D.; Holmes, M.C.; Martin, F. Use of Zinc-Finger Nucleases to Knock out the WAS Gene in K562 Cells: A Human Cellular Model for Wiskott-Aldrich Syndrome. DMM Dis. Models Mech. 2013, 6, 544–554. [Google Scholar]
- Toscano, M.G.; Muñoz, P.; Sánchez-Gilabert, A.; Cobo, M.; Benabdellah, K.; Anderson, P.; Ramos-Mejía, V.; Real, P.J.; Neth, O.; Molinos-Quintana, A.; et al. Absence of WASp Enhances Hematopoietic and Megakaryocytic Differentiation in a Human Embryonic Stem Cell Model. Mol. Ther. 2016, 24, 342–353. [Google Scholar] [CrossRef]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA Targeting Specificity of RNA-Guided Cas9 Nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome Engineering Using the CRISPR-Cas9 System. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Maldonado-Pérez, N.; Tristán-Manzano, M.; Justicia-Lirio, P.; Martínez-Planes, E.; Muñoz, P.; Pavlovic, K.; Cortijo-Gutiérrez, M.; Blanco-Benítez, C.; Castella, M.; Juan, M.; et al. Efficacy and Safety of Universal (TCRKO) ARI-0001 CAR-T Cells for the Treatment of B-Cell Lymphoma. Front. Immunol. 2022, 13, 1011858. [Google Scholar] [CrossRef]
- Gutierrez-Guerrero, A.; Sanchez-Hernandez, S.; Galvani, G.; Pinedo-Gomez, J.; Martin-Guerra, R.; Sanchez-Gilabert, A.; Aguilar-González, A.; Cobo, M.; Gregory, P.; Holmes, M.; et al. Comparison of Zinc Finger Nucleases Versus CRISPR-Specific Nucleases for Genome Editing of the Wiskott-Aldrich Syndrome Locus. Hum. Gene Ther. 2018, 29, 366–380. [Google Scholar] [CrossRef]
- Sánchez-Hernández, S.; Aguilar-González, A.; Guijarro-Albaladejo, B.; Maldonado-Pérez, N.; Ramos-Hernández, I.; Cortijo-Gutiérrez, M.; Sánchez Martín, R.M.; Benabdellah, K.; Martin, F. Development of Cellular Models to Study Efficiency and Safety of Gene Edition by Homologous Directed Recombination Using the CRISPR/Cas9 System. Cells 2020, 9, 1492. [Google Scholar] [CrossRef] [PubMed]
- Benabdellah, K.; Sánchez-Hernández, S.; Aguilar-González, A.; Maldonado-Pérez, N.; Gutierrez-Guerrero, A.; Cortijo-Gutierrez, M.; Ramos-Hernández, I.; Tristán-Manzano, M.; Galindo-Moreno, P.; Herrera, C.; et al. Genome-Edited Adult Stem Cells: Next-Generation Advanced Therapy Medicinal Products. Stem Cells Transl. Med. 2020, 9, 674–685. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Rivera, F.J.; Jacks, T. Applications of the CRISPR–Cas9 System in Cancer Biology. Nat. Rev. Cancer 2015, 15, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Tzelepis, K.; Koike-Yusa, H.; De Braekeleer, E.; Li, Y.; Metzakopian, E.; Dovey, O.M.; Mupo, A.; Grinkevich, V.; Li, M.; Mazan, M.; et al. A CRISPR Dropout Screen Identifies Genetic Vulnerabilities and Therapeutic Targets in Acute Myeloid Leukemia. Cell Rep. 2016, 17, 1193. [Google Scholar] [CrossRef]
- Sorrentino, C.; D’Antonio, L.; Ciummo, S.L.; Fieni, C.; Landuzzi, L.; Ruzzi, F.; Vespa, S.; Lanuti, P.; Lotti, L.V.; Lollini, P.L.; et al. CRISPR/Cas9-Mediated Deletion of Interleukin-30 Suppresses IGF1 and CXCL5 and Boosts SOCS3 Reducing Prostate Cancer Growth and Mortality. J. Hematol. Oncol. 2022, 15, 145. [Google Scholar] [CrossRef]
- D’Antonio, L.; Fieni, C.; Ciummo, S.L.; Vespa, S.; Lotti, L.; Sorrentino, C.; Di Carlo, E. Inactivation of Interleukin-30 in Colon Cancer Stem Cells via CRISPR/Cas9 Genome Editing Inhibits Their Oncogenicity and Improves Host Survival. J. Immunother. Cancer 2023, 11, e006056. [Google Scholar] [CrossRef]
- McAndrews, K.M.; Xiao, F.; Chronopoulos, A.; LeBleu, V.S.; Kugeratski, F.G.; Kalluri, R. Exosome-Mediated Delivery of CRISPR/Cas9 for Targeting of Oncogenic KrasG12D in Pancreatic Cancer. Life Sci. Alliance 2021, 4, e202000875. [Google Scholar] [CrossRef]
- Soucek, L.; Whitfield, J.; Martins, C.P.; Finch, A.J.; Murphy, D.J.; Sodir, N.M.; Karnezis, A.N.; Swigart, L.B.; Nasi, S.; Evan, G.I. Modelling Myc Inhibition as a Cancer Therapy. Nature 2008, 455, 679–683. [Google Scholar] [CrossRef]
- Menegatti, J.; Nakel, J.; Stepanov, Y.K.; Caban, K.M.; Ludwig, N.; Nord, R.; Pfitzner, T.; Yazdani, M.; Vilimova, M.; Kehl, T.; et al. Changes of Protein Expression after CRISPR/Cas9 Knockout of MiRNA-142 in Cell Lines Derived from Diffuse Large B-Cell Lymphoma. Cancers 2022, 14, 5031. [Google Scholar] [CrossRef]
- Lin, S.C.; Wu, H.L.; Yeh, L.Y.; Yang, C.C.; Kao, S.Y.; Chang, K.W. Activation of the MiR-371/372/373 MiRNA Cluster Enhances Oncogenicity and Drug Resistance in Oral Carcinoma Cells. Int. J. Mol. Sci. 2020, 21, 9442. [Google Scholar] [CrossRef]
- Caramori, G.; Casolari, P.; Cavallesco, G.N.; Giuffr, S.; Adcock, I.; Papi, A. Mechanisms Involved in Lung Cancer Development in COPD. Int. J. Biochem. Cell Biol. 2011, 43, 1030–1044. [Google Scholar] [CrossRef] [PubMed]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A MicroRNA Expression Signature of Human Solid Tumors Defines Cancer Gene Targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, H.; Sun, L.; Yang, M.; Pan, C.; Chen, W.; Wu, D.; Lin, Z.; Zeng, C.; Yao, Y.; et al. MiR-21 Indicates Poor Prognosis in Tongue Squamous Cell Carcinomas as an Apoptosis Inhibitor. Clin. Cancer Res. 2009, 15, 3998–4008. [Google Scholar] [CrossRef]
- Dai, L.; Chen, F.; Zheng, Y.; Zhang, D.; Qian, B.; Ji, H.; Long, F.; Cretoiu, D. MiR-21 Regulates Growth and EMT in Lung Cancer Cells via PTEN/Akt/GSK3β Signaling. Front. Biosci. 2019, 24, 1426–1439. [Google Scholar]
- Rama, A.R.; Quiñonero, F.; Mesas, C.; Melguizo, C.; Prados, J. Synthetic Circular MiR-21 Sponge as Tool for Lung Cancer Treatment. Int. J. Mol. Sci. 2022, 23, 2963. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zeng, X.; Ma, R.; Wang, L. MicroRNA-21 Promotes the Proliferation, Migration and Invasion of Non-Small Cell Lung Cancer A549 Cells by Regulating Autophagy Activity via AMPK/ULK1 Signaling Pathway. Exp. Ther. Med. 2018, 16, 2038. [Google Scholar] [CrossRef]
- Huo, W.; Zhao, G.; Yin, J.; Ouyang, X.; Wang, Y.; Yang, C.; Wang, B.; Dong, P.; Wang, Z.; Watari, H.; et al. Lentiviral CRISPR/Cas9 Vector Mediated MiR-21 Gene Editing Inhibits the Epithelial to Mesenchymal Transition in Ovarian Cancer Cells. J. Cancer 2017, 8, 57–64. [Google Scholar] [CrossRef]
- Nieland, L.; van Solinge, T.S.; Cheah, P.S.; Morsett, L.M.; El Khoury, J.; Rissman, J.I.; Kleinstiver, B.P.; Broekman, M.L.D.; Breakefield, X.O.; Abels, E.R. CRISPR-Cas Knockout of MiR21 Reduces Glioma Growth. Mol. Ther. Oncolytics 2022, 25, 121–136. [Google Scholar] [CrossRef]
- Liu, H.; Liu, T.; Zhou, Y.; Song, X.; Wei, R. Overexpression of Long Non-Coding RNA Cancer Susceptibility 11 Is Involved in the Development of Chemoresistance to Carboplatin in Hepatocellular Carcinoma. Oncol. Lett. 2020, 19, 1993–1998. [Google Scholar] [CrossRef]
- Alharbi, M.; Sharma, S.; Guanzon, D.; Lai, A.; Zuñiga, F.; Shiddiky, M.J.A.; Yamauchi, Y.; Salas-Burgos, A.; He, Y.; Pejovic, T.; et al. MiRNa Signature in Small Extracellular Vesicles and Their Association with Platinum Resistance and Cancer Recurrence in Ovarian Cancer. Nanomedicine 2020, 28, 102207. [Google Scholar] [CrossRef]
- Gamal-Eldeen, A.M.; Alrehaili, A.A.; Alharthi, A.; Raafat, B.M. Perftoran Inhibits Hypoxia-Associated Resistance in Lung Cancer Cells to Carboplatin. Front. Pharmacol. 2022, 13, 860898. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Cheng, X.; Li, Y.; Han, Y.; Song, X.; Yu, D.; Cao, X.; Liu, Z. MiR-21 Improves Invasion and Migration of Drug-resistant Lung Adenocarcinoma Cancer Cell and Transformation of EMT through Targeting HBP1. Cancer Med. 2018, 7, 2485–2503. [Google Scholar] [CrossRef] [PubMed]
- Du, G.; Cao, D.; Meng, L. MiR-21 Inhibitor Suppresses Cell Proliferation and Colony Formation through Regulating the PTEN/AKT Pathway and Improves Paclitaxel Sensitivity in Cervical Cancer Cells. Mol. Med. Rep. 2017, 15, 2713–2719. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; Wu, Y.Q.; Zhang, S.P. MiR-21-5p Enhances the Progression and Paclitaxel Resistance in Drug-Resistant Breast Cancer Cell Lines by Targeting PDCD4. Neoplasma 2019, 66, 746–755. [Google Scholar] [CrossRef]
- Farasati Far, B.; Vakili, K.; Fathi, M.; Yaghoobpoor, S.; Bhia, M.; Naimi- Jamal, M.R. The Role of MicroRNA-21 (MiR-21) in Pathogenesis, Diagnosis, and Prognosis of Gastrointestinal Cancers: A Review. Life Sci. 2023, 316, 121340. [Google Scholar] [CrossRef]
- Li, L.; Zhang, H.; Wang, X.; Wang, J.; Wei, H. Long Non-Coding RNA CASC2 Enhanced Cisplatin-Induced Viability Inhibition of Non-Small Cell Lung Cancer Cells by Regulating the PTEN/PI3K/Akt Pathway through down-Regulation of MiR-18a and MiR-21. RSC Adv. 2018, 8, 15923. [Google Scholar] [CrossRef]
- Zheng, X.; Dong, L.; Zhao, S.; Li, Q.; Liu, D.; Zhu, X.; Ge, X.; Li, R.; Wang, G. Propofol Affects Non–Small-Cell Lung Cancer Cell Biology By Regulating the MiR-21/PTEN/AKT Pathway In Vitro and In Vivo. Anesth. Analg. 2020, 131, 1270–1280. [Google Scholar] [CrossRef]
- Ding, Y.; Hou, Y.; Liu, Y.; Xie, X.; Cui, Y.; Nie, H. Prospects for MiR-21 as a Target in the Treatment of Lung Diseases. Curr. Pharm. Des. 2021, 27, 415–422. [Google Scholar] [CrossRef]
- Li, B.; Ren, S.; Li, X.; Wang, Y.; Garfield, D.; Zhou, S.; Chen, X.; Su, C.; Chen, M.; Kuang, P.; et al. MiR-21 Overexpression Is Associated with Acquired Resistance of EGFR-TKI in Non-Small Cell Lung Cancer. Lung Cancer 2014, 83, 146–153. [Google Scholar] [CrossRef]
- Yang, Y.; Meng, H.; Peng, Q.; Yang, X.; Gan, R.; Zhao, L.; Chen, Z.; Lu, J.; Meng, Q.H. Downregulation of MicroRNA-21 Expression Restrains Non-Small Cell Lung Cancer Cell Proliferation and Migration through Upregulation of Programmed Cell Death 4. Cancer Gene Therapy 2014, 22, 23–29. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lara, P.; Aguilar-González, A.; Martín, F.; Mesas, C.; Moreno, J.; Rama, A.R. Exploring miR-21 Knock-Out Using CRISPR/Cas as a Treatment for Lung Cancer. Genes 2025, 16, 133. https://doi.org/10.3390/genes16020133
Lara P, Aguilar-González A, Martín F, Mesas C, Moreno J, Rama AR. Exploring miR-21 Knock-Out Using CRISPR/Cas as a Treatment for Lung Cancer. Genes. 2025; 16(2):133. https://doi.org/10.3390/genes16020133
Chicago/Turabian StyleLara, Patricia, Araceli Aguilar-González, Francisco Martín, Cristina Mesas, Javier Moreno, and Ana R. Rama. 2025. "Exploring miR-21 Knock-Out Using CRISPR/Cas as a Treatment for Lung Cancer" Genes 16, no. 2: 133. https://doi.org/10.3390/genes16020133
APA StyleLara, P., Aguilar-González, A., Martín, F., Mesas, C., Moreno, J., & Rama, A. R. (2025). Exploring miR-21 Knock-Out Using CRISPR/Cas as a Treatment for Lung Cancer. Genes, 16(2), 133. https://doi.org/10.3390/genes16020133





