Structural Analysis of Missense Mutations on the Stability of APOE3 and APOE4
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequence and Structural Alignment
2.2. Computational Saturated Mutagenesis
2.3. Energy Calculation
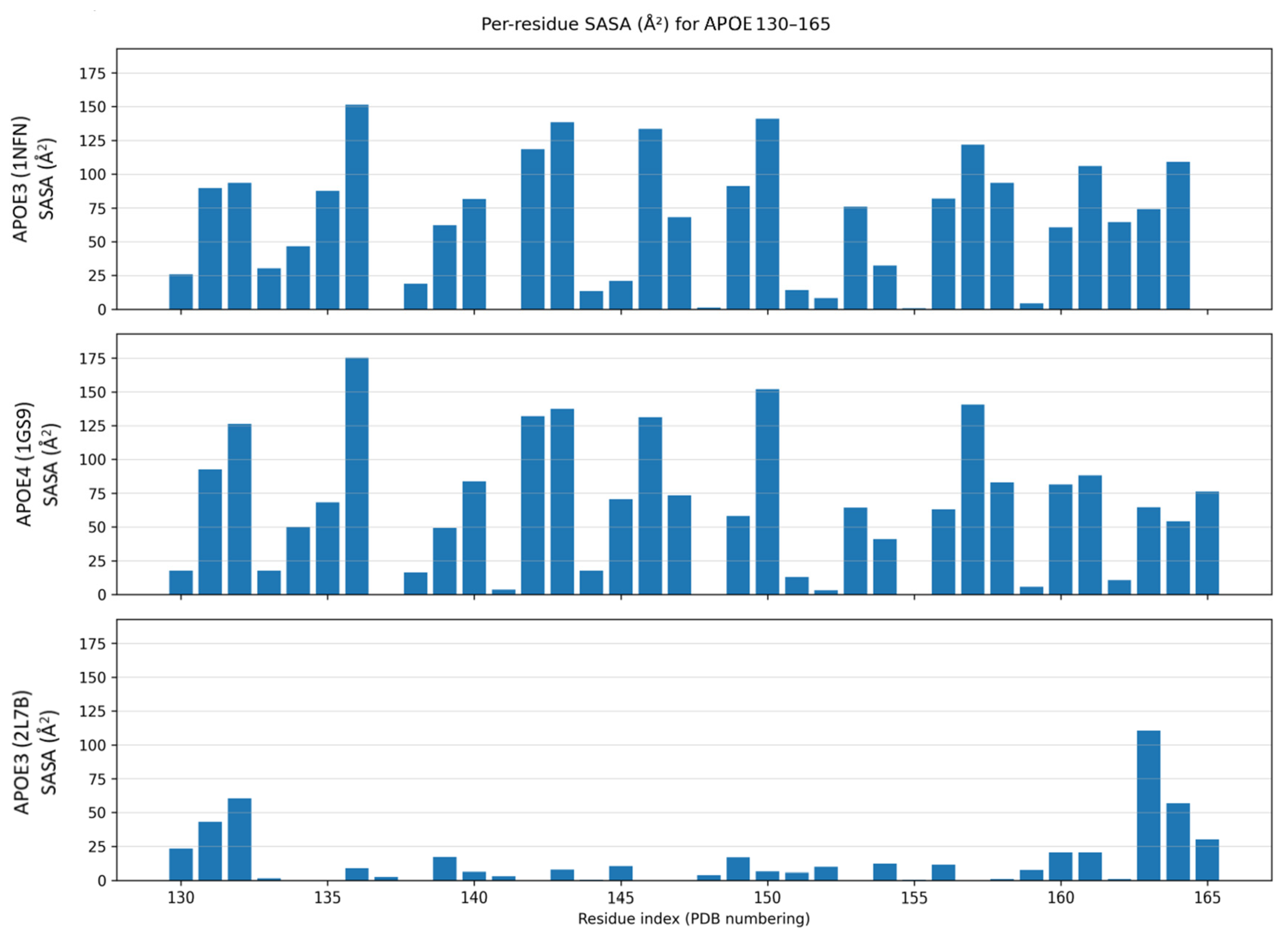
2.4. Solvent-Accessible Surface Area (SASA) Analysis
2.5. Supplementary Stability Predictions via DynaMut2 and DUET
2.6. Molecular Dynamics Simulation
3. Results
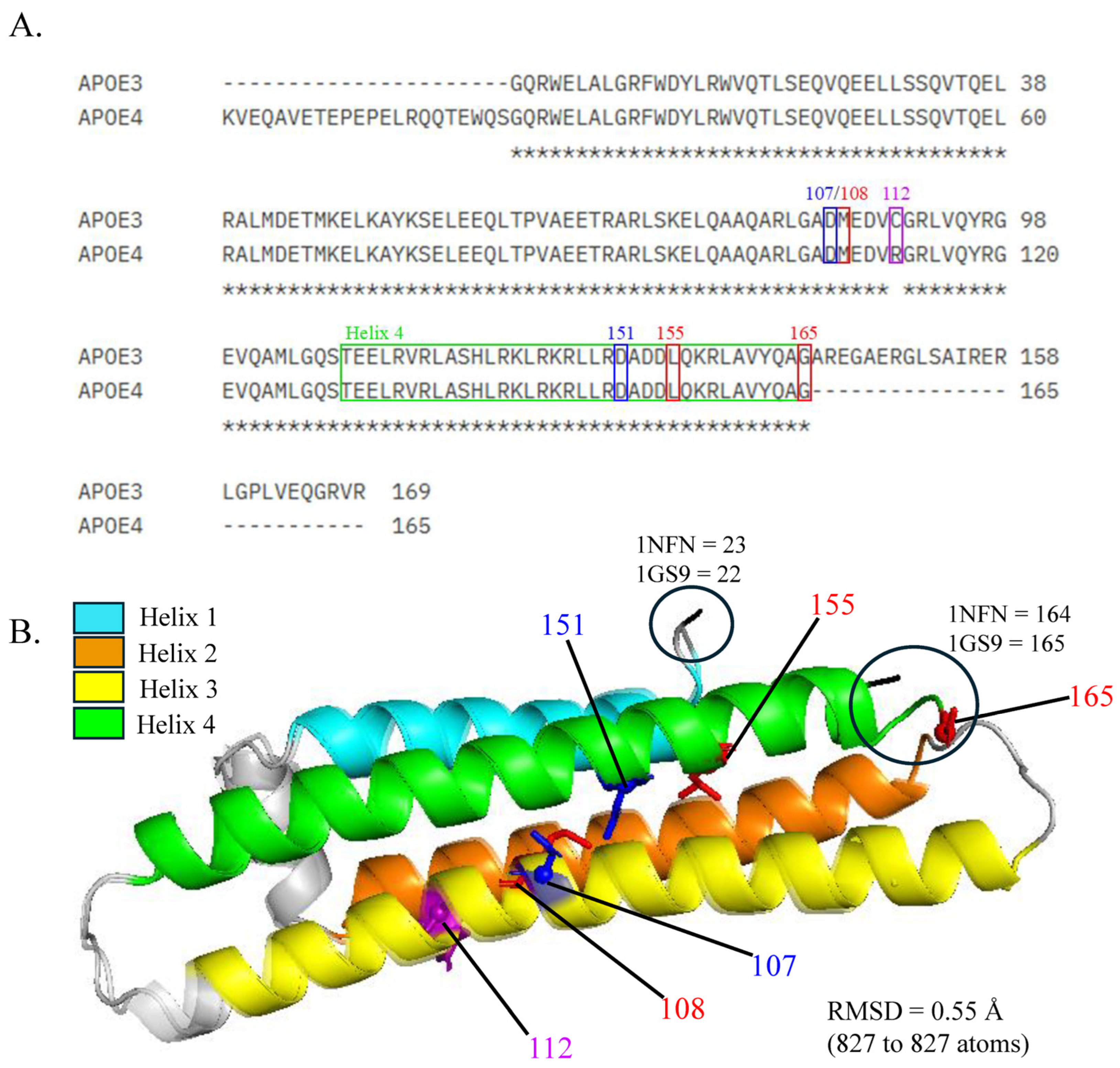
3.1. Sequence and Structural Alignment of APOE3 and APOE4
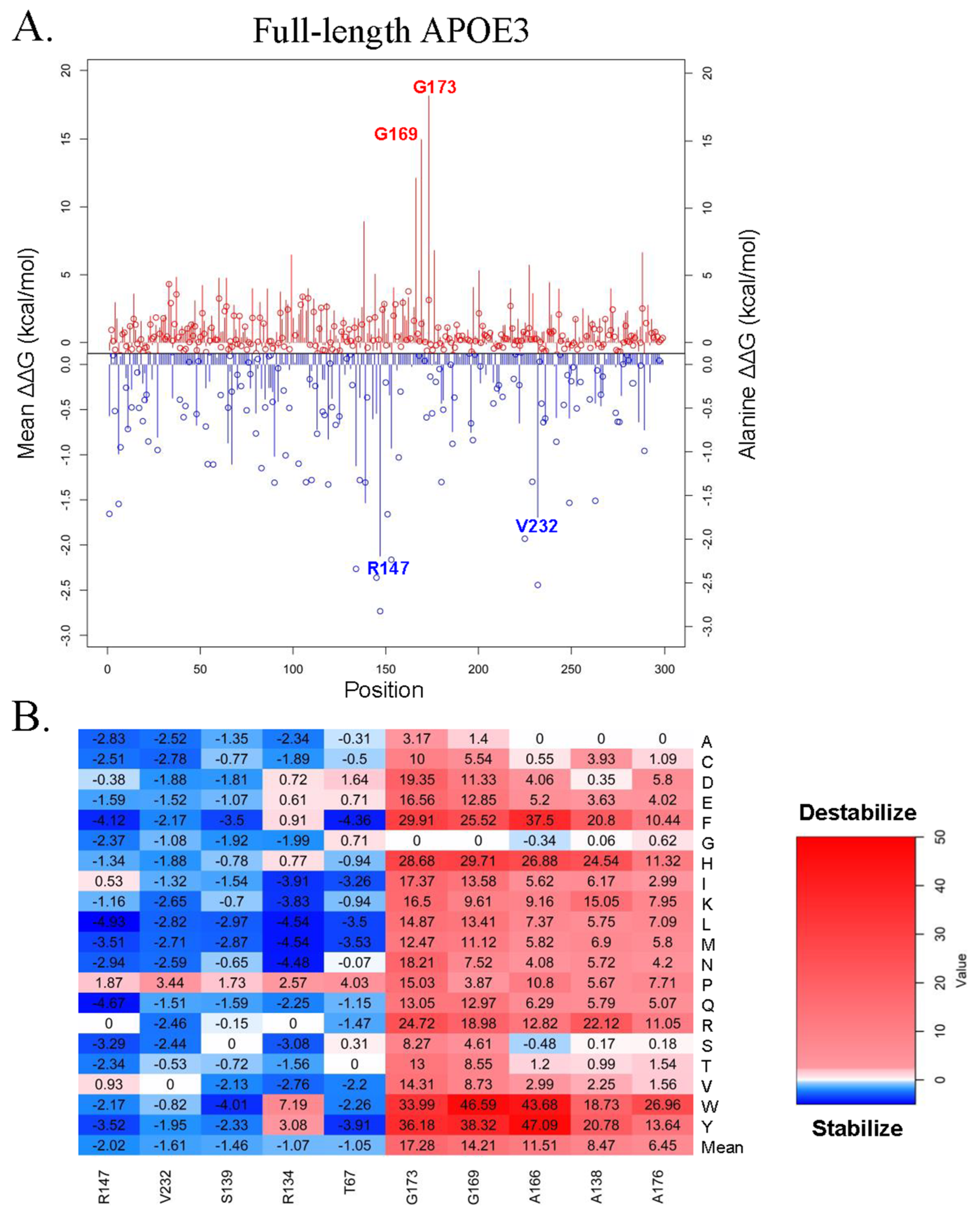
3.2. Computational Analysis of the Effect of Missense Mutations on APOE3 and APOE4
3.3. Molecular Dynamics Simulations of APOE3 and APOE4 Missense Variants
4. Discussion
Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s Disease |
| APOE | Apolipoprotein E |
| CTD | C-terminal Domain |
| DSSP | Dictionary of Secondary Structure Proteins |
| fs | Femtosecond |
| nm | Nanometer |
| NTD | N-terminal Domain |
| Rg | Radius of Gyration |
| RMSD | Root-Mean-Square Deviation |
| RMSF | Root-Mean-Square Fluctuation |
| SASA | Solvent-Accessible Surface Area |
| TREM2 | Triggering Receptor Expressed on Myeloid Cells 2 |
| WT | Wild Type |
References
- Alzheimer’s Association. 2025 Alzheimer’s Disease Facts and Figures (PDF). Available online: http://www.alz.org/getmedia/ef8f48f9-ad36-48ea-87f9-b74034635c1e/alzheimers-facts-and-figures.pdf (accessed on 9 October 2025).
- Serrano-Pozo, A.; Frosch, M.P.; Masliah, E.; Hyman, B.T. Neuropathological alterations in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2011, 1, a006189. [Google Scholar] [CrossRef]
- Holtzman, D.M.; Herz, J.; Bu, G. Apolipoprotein E and apolipoprotein E receptors: Normal biology and roles in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2012, 2, a006312. [Google Scholar] [CrossRef]
- Khalil, Y.A.; Rabès, J.-P.; Boileau, C.; Varret, M. APOE gene variants in primary dyslipidemia. Atherosclerosis 2021, 328, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Corder, E.H.; Saunders, A.M.; Strittmatter, W.J.; Schmechel, D.E.; Gaskell, P.C.; Small, G.W.; Roses, A.D.; Haines, J.L.; Pericak-Vance, M.A. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 1993, 261, 921–923. [Google Scholar] [CrossRef] [PubMed]
- Farrer, L.A.; Cupples, L.A.; Haines, J.L.; Hyman, B.; Kukull, W.A.; Mayeux, R.; Myers, R.H.; Pericak-Vance, M.A.; Risch, N.; Van Duijn, C.M. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA 1997, 278, 1349–1356. [Google Scholar] [CrossRef]
- Huang, Y.; Mahley, R.W. Apolipoprotein E: Structure and function in lipid metabolism, neurobiology, and Alzheimer’s diseases. Neurobiol. Dis. 2014, 72 Pt A, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Michaelson, D.M. APOE ε4: The most prevalent yet understudied risk factor for Alzheimer’s disease. Alzheimer’s Dement. 2014, 10, 861–868. [Google Scholar] [CrossRef]
- Liu, C.C.; Kanekiyo, T.; Xu, H.; Bu, G. Apolipoprotein E and Alzheimer disease: Risk, mechanisms and therapy. Nat. Rev. Neurol. 2013, 9, 106–118. [Google Scholar] [CrossRef]
- Colonna, M.; Wang, Y. TREM2 variants: New keys to decipher Alzheimer disease pathogenesis. Nat. Rev. Neurosci. 2016, 17, 201–207. [Google Scholar] [CrossRef]
- Atagi, Y.; Liu, C.-C.; Painter, M.M.; Chen, X.-F.; Verbeeck, C.; Zheng, H.; Li, X.; Rademakers, R.; Kang, S.S.; Xu, H.; et al. Apolipoprotein E Is a Ligand for Triggering Receptor Expressed on Myeloid Cells 2 (TREM2). J. Biol. Chem. 2015, 290, 26043–26050. [Google Scholar] [CrossRef]
- Yeh, F.L.; Wang, Y.; Tom, I.; Gonzalez, L.C.; Sheng, M. TREM2 Binds to Apolipoproteins, Including APOE and CLU/APOJ, and Thereby Facilitates Uptake of Amyloid-Beta by Microglia. Neuron 2016, 91, 328–340. [Google Scholar] [CrossRef]
- Guerreiro, R.; Wojtas, A.; Bras, J.; Carrasquillo, M.; Rogaeva, E.; Majounie, E.; Cruchaga, C.; Sassi, C.; Kauwe, J.S.; Younkin, S.; et al. TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 2013, 368, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Mucke, L. Alzheimer mechanisms and therapeutic strategies. Cell 2012, 148, 1204–1222. [Google Scholar] [CrossRef] [PubMed]
- Allen, P.; Smith, A.C.; Benedicto, V.; Abdulhasan, A.; Narayanaswami, V.; Tapavicza, E. Molecular dynamics simulation of apolipoprotein E3 lipid nanodiscs. Biochim. Biophys. Acta (BBA)-Biomembr. 2024, 1866, 184230. [Google Scholar] [CrossRef] [PubMed]
- Strickland, M.R.; Rau, M.J.; Summers, B.; Basore, K.; Wulf, J.; Jiang, H.; Chen, Y.; Ulrich, J.D.; Randolph, G.J.; Zhang, R.; et al. Apolipoprotein E secreted by astrocytes forms antiparallel dimers in discoidal lipoproteins. Neuron 2024, 112, 1100–1109.e5. [Google Scholar] [CrossRef]
- Pourmousa, M.; Pastor, R.W. Molecular dynamics simulations of lipid nanodiscs. Biochim. Biophys. Acta (BBA)-Biomembr. 2018, 1860, 2094–2107. [Google Scholar] [CrossRef]
- Mamchur, A.; Ivanov, M.; Matkava, L.; Yudin, V.; Keskinov, A.; Yudin, S.; Kashtanova, D. Tackling APOE’s structural challenges via in silico modeling in the era of neural networks: Can AlphaFold II help circumvent the problem of lacking full-length protein structure? Arch. Biochem. Biophys. 2024, 761, 110185. [Google Scholar] [CrossRef]
- Lewkowicz, E.; Nakamura, M.N.; Rynkiewicz, M.J.; Gursky, O. Molecular modeling of apoE in complexes with Alzheimer’s amyloid-β fibrils from human brain suggests a structural basis for apolipoprotein co-deposition with amyloids. Cell. Mol. Life Sci. 2023, 80, 376. [Google Scholar] [CrossRef]
- Dong, L.-M.; Parkin, S.; Trakhanov, S.D.; Rupp, B.; Simmons, T.; Arnold, K.S.; Newhouse, Y.M.; Innerarity, T.L.; Weisgraber, K.H. Novel mechanism for defective receptor binding of apolipoprotein E2 in type III hyperlipoproteinemia. Nat. Struct. Mol. Biol. 1996, 3, 718–722. [Google Scholar] [CrossRef]
- RCSB Protein Data Bank—Entry 1GS9. Available online: https://www.rcsb.org/structure/1GS9 (accessed on 13 October 2025).
- Chen, J.; Li, Q.; Wang, J. Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions. Proc. Natl. Acad. Sci. USA 2011, 108, 14813–14818. [Google Scholar] [CrossRef]
- FoldX. Protein Design Algorithm. Available online: http://foldxsuite.crg.eu/ (accessed on 9 October 2025).
- PyMOL Molecular Graphics System. Available online: https://pymol.org/2/ (accessed on 13 October 2025).
- Rodrigues, C.H.; Pires, D.E.; Ascher, D.B. DynaMut2: Assessing changes in stability and flexibility upon single and multiple point missense mutations. Protein Sci. 2021, 30, 60–69. [Google Scholar] [CrossRef]
- Pires, D.E.V.; Ascher, D.B.; Blundell, T.L. DUET: A server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res. 2014, 42, W314–W319. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Hauser, P.S.; Narayanaswami, V.; Ryan, R.O. Apolipoprotein E: From lipid transport to neurobiology. Prog. Lipid Res. 2011, 50, 62–74. [Google Scholar] [CrossRef] [PubMed]
- Mahley, R.W.; Weisgraber, K.H.; Huang, Y. Apolipoprotein E: Structure determines function, from atherosclerosis to Alzheimer’s disease to AIDS. J. Lipid Res. 2009, 50, S183–S188. [Google Scholar] [CrossRef] [PubMed]
- Lessard, C.B.; Malnik, S.L.; Zhou, Y.; Ladd, T.B.; Cruz, P.E.; Ran, Y.; Mahan, T.E.; Chakrabaty, P.; Holtzman, D.M.; Ulrich, J.D.; et al. High-affinity interactions and signal transduction between Aβ oligomers and TREM 2. EMBO Mol. Med. 2018, 10, e201809027. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Yang, Y.; Hu, Z. The Novel Apolipoprotein E Mutation ApoE Chengdu (c.518T>C, p.L173P) in a Chinese Patient with Lipoprotein Glomerulopathy. J. Atheroscler. Thromb. 2018, 25, 733–740. [Google Scholar] [CrossRef]




| Mutation | APOE Variant | FoldX ΔΔG (kcal/mol) | DynaMut2 ΔΔG (kcal/mol) | DUET ΔΔG (kcal/mol) |
|---|---|---|---|---|
| G165W | APOE4 | 25.07 D | −0.82 D | −0.824 D |
| L155W | APOE4 | 17.93 D | −1.97 D | −2.273 D |
| L155W | APOE3 | 15.08 D | −2.11 D | −2.232 D |
| M108W | APOE4 | 16.22 D | −1.91 D | −1.595 D |
| M108W | APOE3 | 11.03 D | 1.71 S | −1.603 D |
| D151I | APOE4 | −4.42 S | 1.04 S | 0.742 S |
| D151F | APOE3 | −3.81 S | −1.31 D | −0.905 D |
| D107M | APOE4 | −4.49 S | 1.13 S | 0.598 S |
| D107L | APOE3 | −4.25 S | 0.32 S | 0.73 S |
| C112R | APOE3 | −0.53 S | −0.98 D | −0.912 D |
| R158C | APOE3 | 1.05 D | −1.23 D | −1.614 D |
| G165W | APOE3 (2L7B) | 3.71 D | −1.3 D | −1.312 D |
| L155W | APOE3 (2L7B) | 1.42 D | −2.01 D | −2.258 D |
| M108W | APOE3 (2L7B) | 7.96 D | −1.82 D | −1.609 D |
| D151I | APOE3 (2L7B) | −0.32 S | 0.2 S | 0.758 S |
| D151F | APOE3 (2L7B) | 6.31 D | −1.26 D | −1.144 D |
| D107M | APOE3 (2L7B) | 3.08 D | −0.89 D | −0.938 D |
| D107L | APOE3 (2L7B) | −1.49 S | −1.13 D | −0.865 D |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anthony, M.; Xie, Y.; O’Neil, J.N.; Teng, S. Structural Analysis of Missense Mutations on the Stability of APOE3 and APOE4. Genes 2025, 16, 1509. https://doi.org/10.3390/genes16121509
Anthony M, Xie Y, O’Neil JN, Teng S. Structural Analysis of Missense Mutations on the Stability of APOE3 and APOE4. Genes. 2025; 16(12):1509. https://doi.org/10.3390/genes16121509
Chicago/Turabian StyleAnthony, Malcolm, Yixin Xie, Jahn N. O’Neil, and Shaolei Teng. 2025. "Structural Analysis of Missense Mutations on the Stability of APOE3 and APOE4" Genes 16, no. 12: 1509. https://doi.org/10.3390/genes16121509
APA StyleAnthony, M., Xie, Y., O’Neil, J. N., & Teng, S. (2025). Structural Analysis of Missense Mutations on the Stability of APOE3 and APOE4. Genes, 16(12), 1509. https://doi.org/10.3390/genes16121509






