Functional Interpretation of a Novel Homozygous METTL5 Variant Associated with ADHD and Neurodevelopmental Abnormalities: A Case Report and Literature Review
Abstract
1. Introduction and Clinical Significance
2. Case Presentation
3. Methods and Results
3.1. DNA Extraction and Sequencing
3.2. Variant Annotation
3.3. Structural and Domain Analysis
3.4. Evolutionary Conservation
3.5. Subcellular Localization and Disorder Prediction
3.6. Post-Translational Modification Analysis
3.7. Protein Interaction Network
4. Discussion
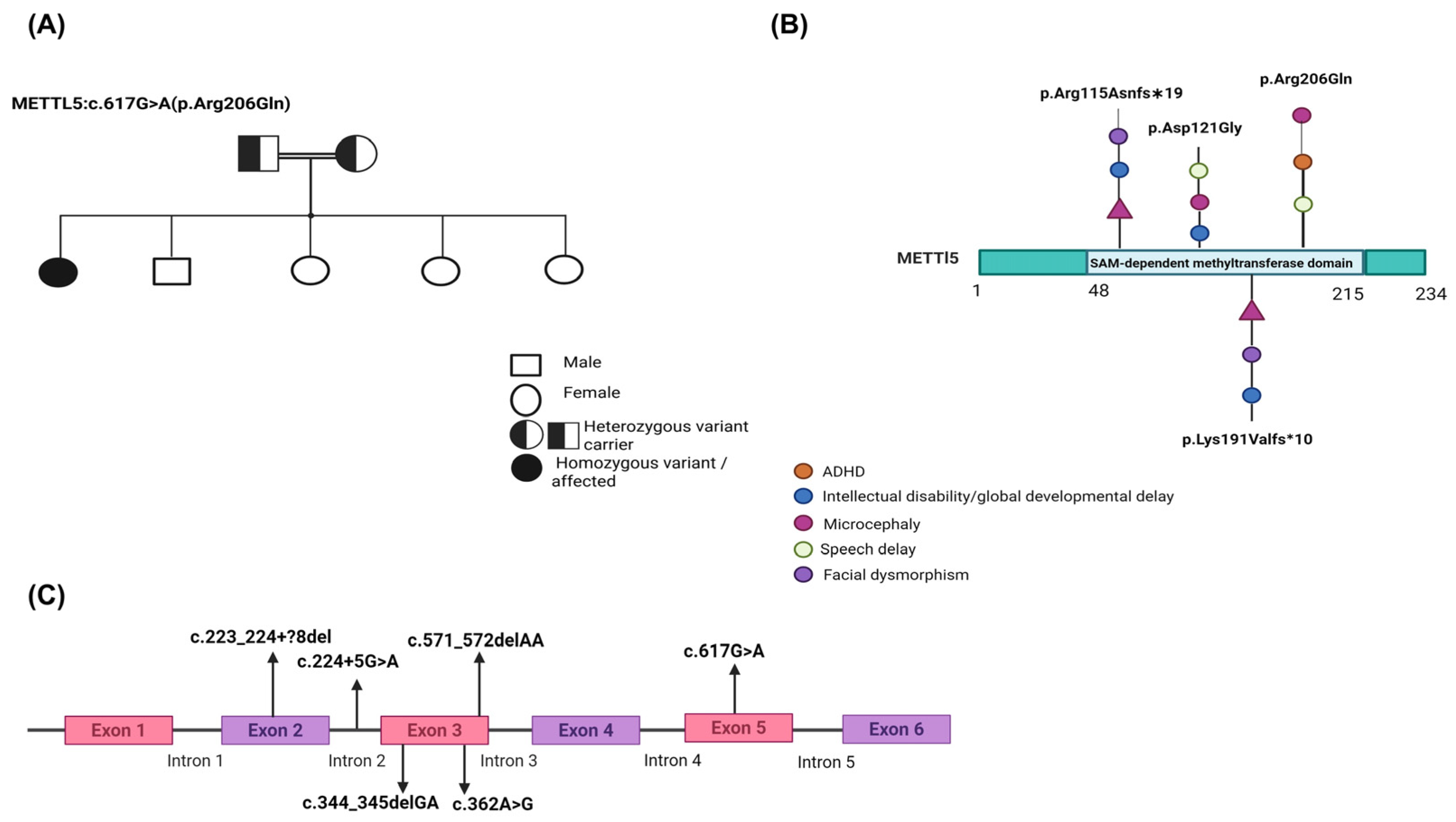
| Report | Individual/Case | Variant (HGVS) | Zygosity | Inheritance | Clinical Features | Reference |
|---|---|---|---|---|---|---|
| Current case: Sidra Medicine | Individual | c.617G > A (p.Arg206Gln) | Homozygous | Autosomal Recessive | ADHD, speech delay, learning challenges, microcephaly and family history of learning difficulties | |
| Iranian Family (3 siblings) | c.223_224 + ?8del (splice-site exon 2) | Homozygous Deletion | Autosomal Recessive | Primary microcephaly, ID, ADHD, speech delay, short stature, GDD | [7] | |
| Consanguineous Pakistani family | c.344_345delGA (p.Arg115Asnfs∗19) | Homozygous Frameshift | Autosomal Recessive | Moderate–severe ID, microcephaly, facial dysmorphism (e.g., large ears, dental anomalies) | [4] | |
| Consanguineous Yemenite family | c.571_572delAA (p.Lys191Valfs∗10) | Homozygous Frameshift | Autosomal Recessive | Moderate–severe ID, microcephaly, facial dysmorphism (large nose) | [4] | |
| Chinese family | c.224 + 5G > A (splice donor site, intron 2) | Homozygous | Autosomal Recessive | Microcephaly-related GDD | [5] | |
| Afghan Family (3 siblings) | c.362A > G (p.Asp121Gly) | Homozygous | Autosomal Recessive | Intellectual disability, microcephaly, poor/absent speech, delayed walking, aggressive behavior, large/posteriorly rotated ears, broad nasal base and short stature aggression, short stature, self-mutilation | [6] |
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| WGS | Whole-genome sequencing |
| METTL5 | Methyltransferase-like 5 |
| ADHD | Attention-deficit/hyperactivity disorder |
References
- Patel, D.R.; Cabral, M.D.; Ho, A.; Merrick, J. A clinical primer on intellectual disability. Transl. Pediatr. 2020, 9, S23–S35. [Google Scholar] [CrossRef]
- Sepich-Poore, C.; Zheng, Z.; Schmitt, E.; Wen, K.; Zhang, Z.S.; Cui, X.L.; Dai, Q.; Zhu, A.C.; Zhang, L.; Sanchez Castillo, A.; et al. The METTL5-TRMT112 N6-methyladenosine methyltransferase complex regulates mRNA translation via 18S rRNA methylation. J. Biol. Chem. 2022, 298, 101590. [Google Scholar] [CrossRef] [PubMed]
- (OMIM) OMIiM. OMIM Entry #618665—Intellectual Developmental Disorder, Autosomal Recessive 72; MRT722025-05-13. Available online: https://www.omim.org/entry/618665 (accessed on 16 May 2024).
- Richard, E.M.; Polla, D.L.; Assir, M.Z.; Contreras, M.; Shahzad, M.; Khan, A.A.; Razzaq, A.; Akram, J.; Tarar, M.N.; Blanpied, T.A.; et al. Bi-allelic Variants in METTL5 Cause Autosomal-Recessive Intellectual Disability and Microcephaly. Am. J. Hum. Genet. 2019, 105, 869–878. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Teng, C.; Zhao, W.; Yang, W.; Yang, Y.; Chen, Q.; He, M.; Zhang, J. Microcephaly-related global developmental delay caused by a pathogenic METTL5 splicing mutation in a Chinese family. J. Hum. Genet. 2025, 70, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Torun, D.; Arslan, M.; Cavdarli, B.; Akar, H.; Cram, D.S. Three Afghani siblings with a novel homozygous variant and further delineation of the clinical features of METTL5 related intellectual disability syndrome. Turk. J. Pediatr. 2022, 64, 956–963. [Google Scholar] [CrossRef]
- Shakarami, F.; Nouri, Z.; Khanahmad, H.; Ghazavi, M.; Amin Tabatabaiefar, M. A novel METTL5 variant disrupting a donor splice site leads to primary microcephaly-related intellectual disability in an Iranian family: Clinical features and literature review. J. Genet. 2023, 102, 45. [Google Scholar] [CrossRef]
- Turkalj, E.M.; Vissers, C. The emerging importance of METTL5-mediated ribosomal RNA methylation. Exp. Mol. Med. 2022, 54, 1617–1625. [Google Scholar] [CrossRef]
- Wang, L.; Liang, Y.; Lin, R.; Xiong, Q.; Yu, P.; Ma, J.; Cheng, M.; Han, H.; Wang, X.; Wang, G.; et al. Mettl5 mediated 18S rRNA N6-methyladenosine (m6A) modification controls stem cell fate determination and neural function. Genes Dis. 2022, 9, 268–274. [Google Scholar] [CrossRef]
- van Tran, N.; Ernst, F.G.M.; Hawley, B.R.; Zorbas, C.; Ulryck, N.; Hackert, P.; Bohnsack, K.E.; Bohnsack, M.T.; Jaffrey, S.R.; Graille, M.; et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019, 47, 7719–7733. [Google Scholar] [CrossRef]
- Fernandez-Castillo, N.; Cabana-Dominguez, J.; Kappel, D.B.; Torrico, B.; Weber, H.; Lesch, K.P.; Lao, O.; Reif, A.; Cormand, B. Exploring the Contribution to ADHD of Genes Involved in Mendelian Disorders Presenting with Hyperactivity and/or Inattention. Genes 2021, 13, 93. [Google Scholar] [CrossRef]
- Jung, Y.; Goldman, D. Role of RNA modifications in brain and behavior. Genes Brain Behav. 2018, 17, e12444. [Google Scholar] [CrossRef]
- Kuehner, J.N.; Bruggeman, E.C.; Wen, Z.; Yao, B. Epigenetic Regulations in Neuropsychiatric Disorders. Front. Genet. 2019, 10, 268. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.H.; Pires, D.E.; Ascher, D.B. DynaMut: Predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018, 46, W350–W355. [Google Scholar] [CrossRef] [PubMed]
- Ashkenazy, H.; Abadi, S.; Martz, E.; Chay, O.; Mayrose, I.; Pupko, T.; Ben-Tal, N. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016, 44, W344–W350. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2018, 47, D607–D613. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Bener, A.; Alali, K.A. Consanguineous marriage in a newly developed country: The Qatari population. J. Biosoc. Sci. 2006, 38, 239–246. [Google Scholar] [CrossRef]
- Abdu, Y.; Ahmed, K.; Ibrahim, M.I.M.; Abdou, M.; Ali, A.; Alsiddig, H.; Selim, N.A.; Yassin, M.A. Perception of consanguineous marriage among the qatari population. Front. Public Health 2023, 11, 1228010. [Google Scholar] [CrossRef]
- Bittles, A.H.; Black, M.L. Evolution in health and medicine Sackler colloquium: Consanguinity, human evolution, and complex diseases. Proc. Natl. Acad. Sci. USA 2010, 107, 1779–1786. [Google Scholar] [CrossRef]
- Tadmouri, G.O.; Nair, P.; Obeid, T.; Al Ali, M.T.; Al Khaja, N.; Hamamy, H.A. Consanguinity and reproductive health among Arabs. Reprod. Health 2009, 6, 17. [Google Scholar] [CrossRef]


| Genomic Position | chr14:16661234 (GRCh38) |
|---|---|
| AnnotationMethod | CADD v1.6 |
| Database | Genome Aggregation Database (gnomAD) |
| Variant Classification | Missense (Arginine → Glutamine) |
| Key Findings | |
| CADD Phred Score | 23.7 (deleterious) |
| Percentile Rank | >99% (top 1% of deleterious variants) |
| Functional Impact | Non-conservative amino acid change in the catalytic domain |
| Structural Implications | |
| Effect 1 | Disrupts hydrogen bonding with the SAM cofactor |
| Effect 2 | Loss of positive charge at the substrate recognition site |
| Effect 3 | Predicted stability change (ΔΔG = +1.8 kcal/mol) |
| Conclusion This variant is classified as likely pathogenic based on the following evidence: | |
| Criterion 1 | High CADD score (23.7 > 20 deleterious threshold) |
| Criterion 2 | Location in evolutionarily conserved residue (PhyloP = 5.1) |
| Criterion 3 | Critical position in the methyltransferase domain |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hashem, S.; Elhag, S.F.; Bhat, A.A.; Aamer, W.; Al-Maraghi, A.; Alhaboub, H.; Abuthaher, D.; Akil, A.S.A.-S.; Haris, M.; Fakhro, K.; et al. Functional Interpretation of a Novel Homozygous METTL5 Variant Associated with ADHD and Neurodevelopmental Abnormalities: A Case Report and Literature Review. Genes 2025, 16, 1502. https://doi.org/10.3390/genes16121502
Hashem S, Elhag SF, Bhat AA, Aamer W, Al-Maraghi A, Alhaboub H, Abuthaher D, Akil ASA-S, Haris M, Fakhro K, et al. Functional Interpretation of a Novel Homozygous METTL5 Variant Associated with ADHD and Neurodevelopmental Abnormalities: A Case Report and Literature Review. Genes. 2025; 16(12):1502. https://doi.org/10.3390/genes16121502
Chicago/Turabian StyleHashem, Sheema, Saba F. Elhag, Ajaz A. Bhat, Waleed Aamer, Aljazi Al-Maraghi, Hala Alhaboub, Dalya Abuthaher, Ammira S. Al-Shabeeb Akil, Mohammad Haris, Khalid Fakhro, and et al. 2025. "Functional Interpretation of a Novel Homozygous METTL5 Variant Associated with ADHD and Neurodevelopmental Abnormalities: A Case Report and Literature Review" Genes 16, no. 12: 1502. https://doi.org/10.3390/genes16121502
APA StyleHashem, S., Elhag, S. F., Bhat, A. A., Aamer, W., Al-Maraghi, A., Alhaboub, H., Abuthaher, D., Akil, A. S. A.-S., Haris, M., Fakhro, K., Nemer, G., & Kamal, M. (2025). Functional Interpretation of a Novel Homozygous METTL5 Variant Associated with ADHD and Neurodevelopmental Abnormalities: A Case Report and Literature Review. Genes, 16(12), 1502. https://doi.org/10.3390/genes16121502






