G2H: A Precise Block-Scanning Strategy for Genetic Background Assessment in Maize Backcross Breeding
Abstract
1. Introduction
2. Materials and Methods
2.1. Test Materials
2.2. Genotyping and Loci Filtering
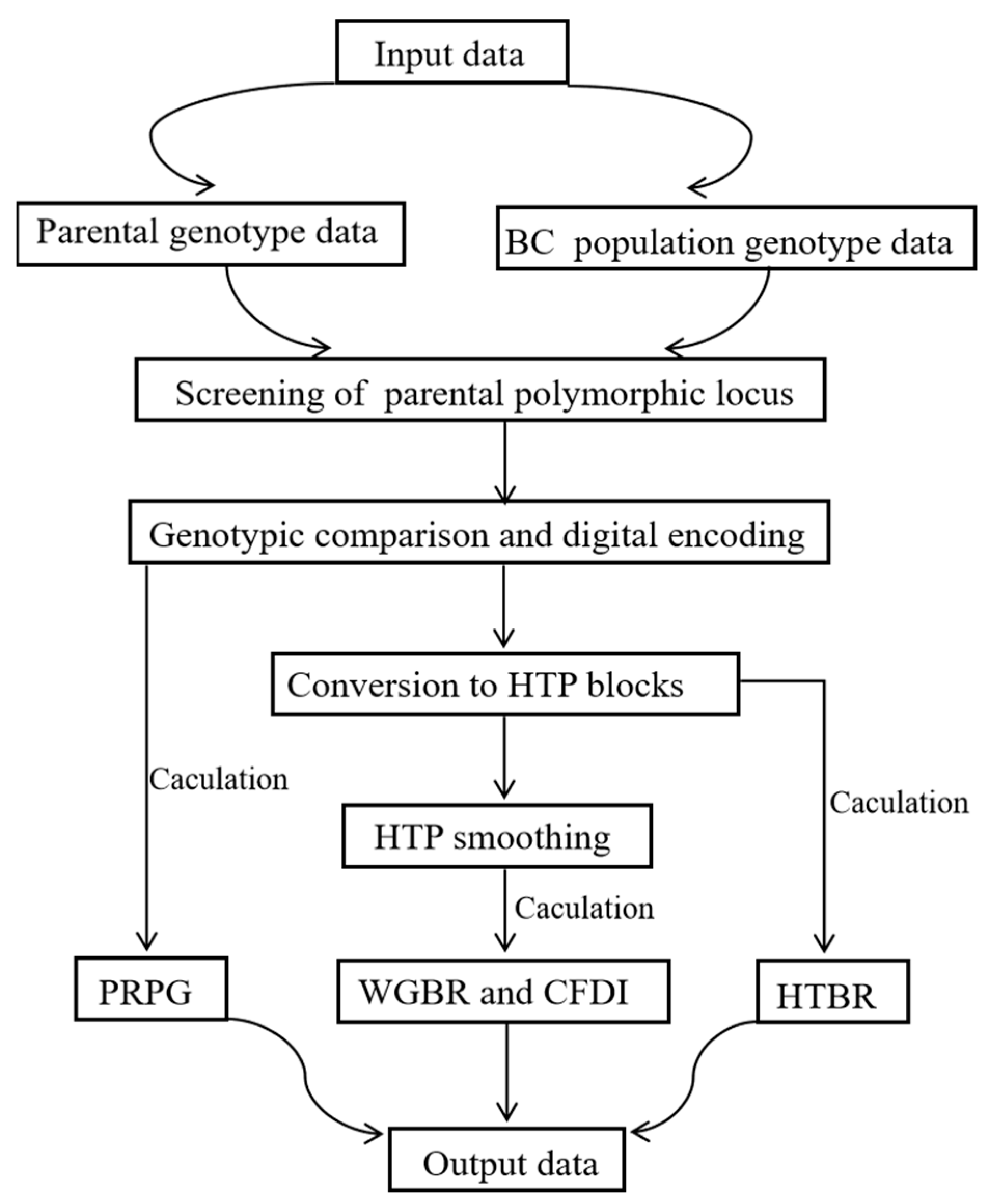
2.3. Construction and Application of the G2H System
2.3.1. Parental Polymorphic Loci
2.3.2. Genotype Comparison and Digital Coding
2.3.3. Conversion of Digitally Encoded Genotypes to HTP Blocks
2.3.4. HTP Smoothing
2.3.5. Calculation of WGBR and CFDI
3. Results
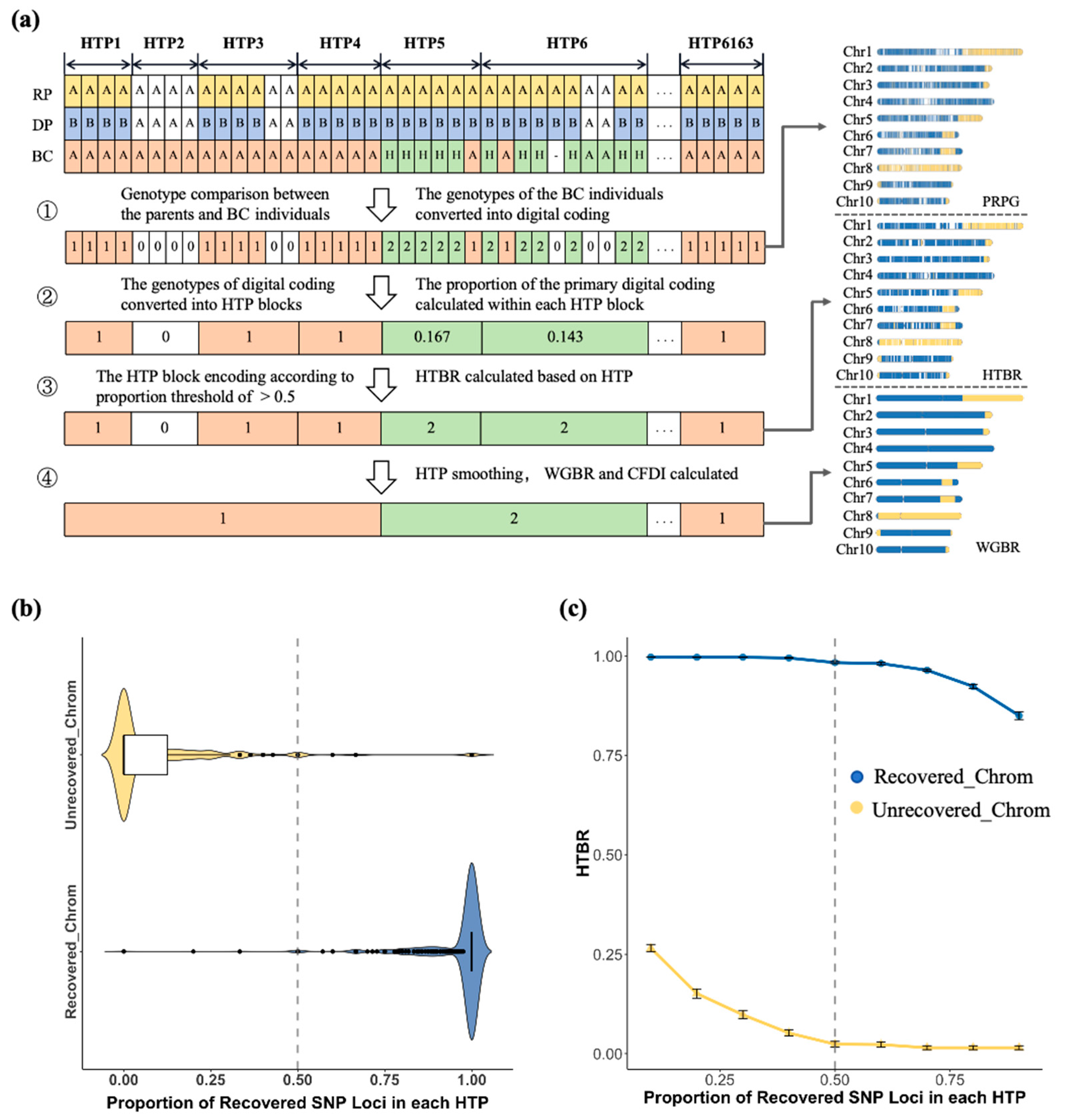
3.1. Design of the G2H Block-Based Genetic Background Screening Strategy
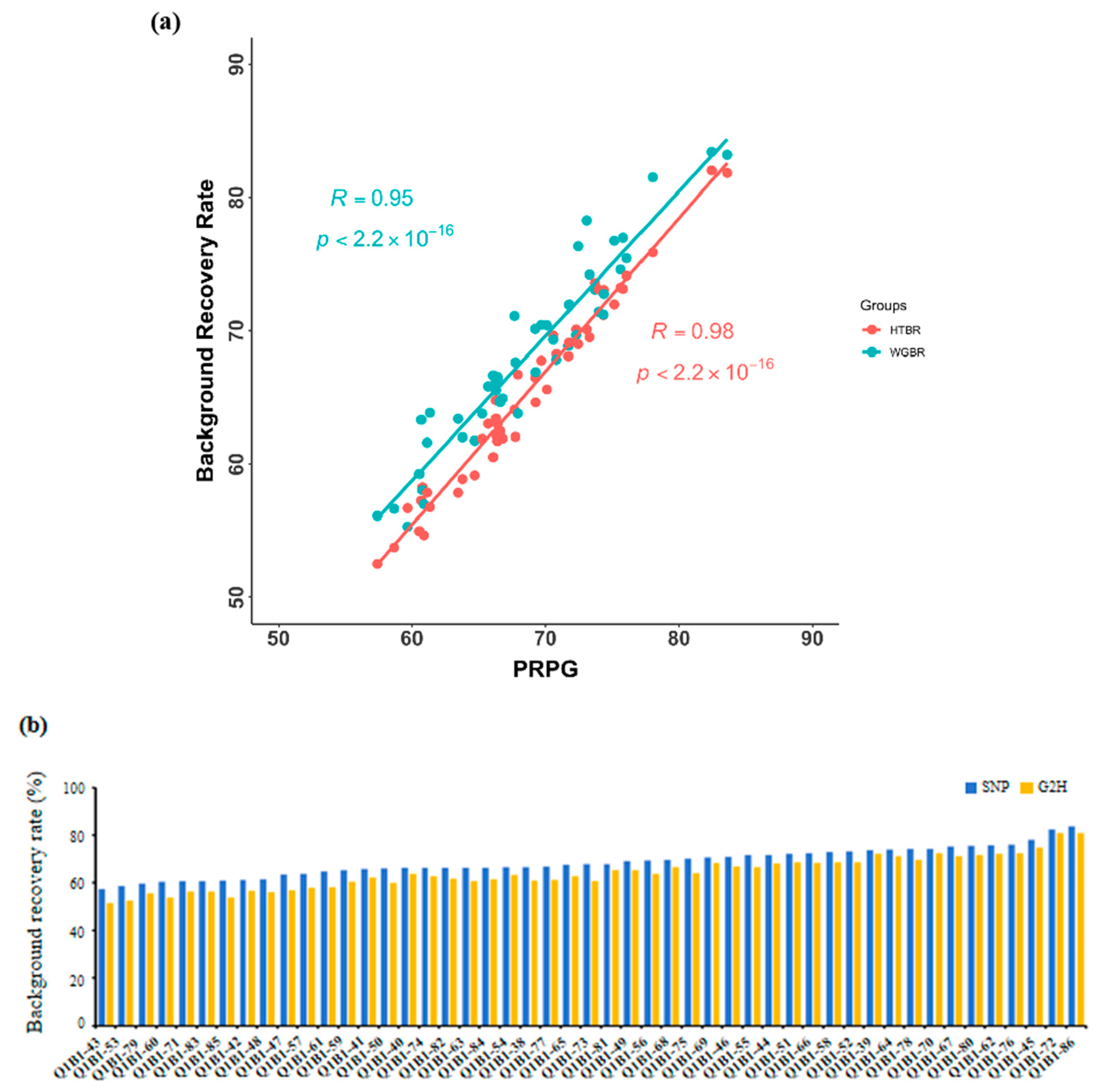
3.2. Comparative Analysis of G2H Strategy and Traditional Point Marking Methods
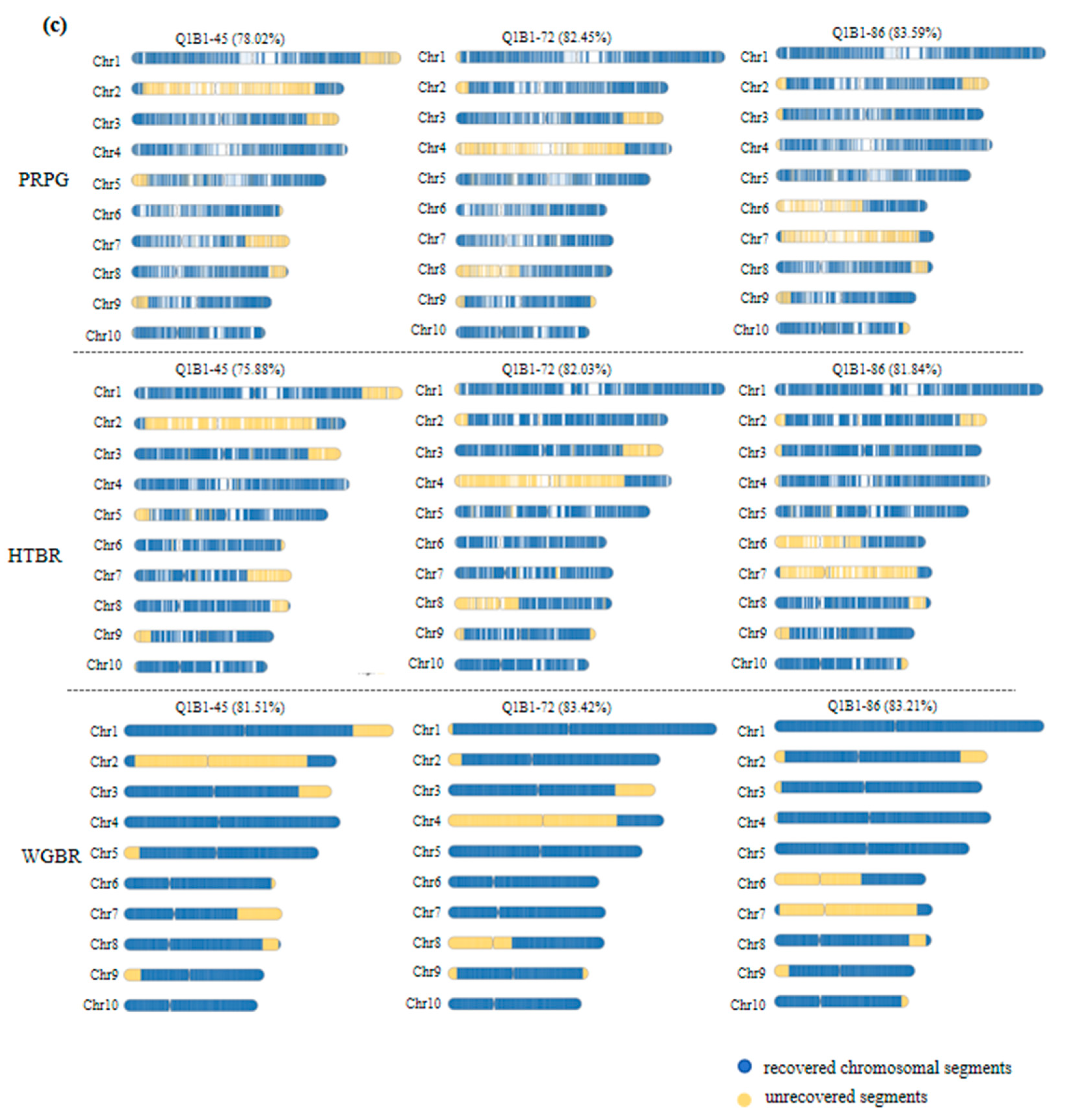
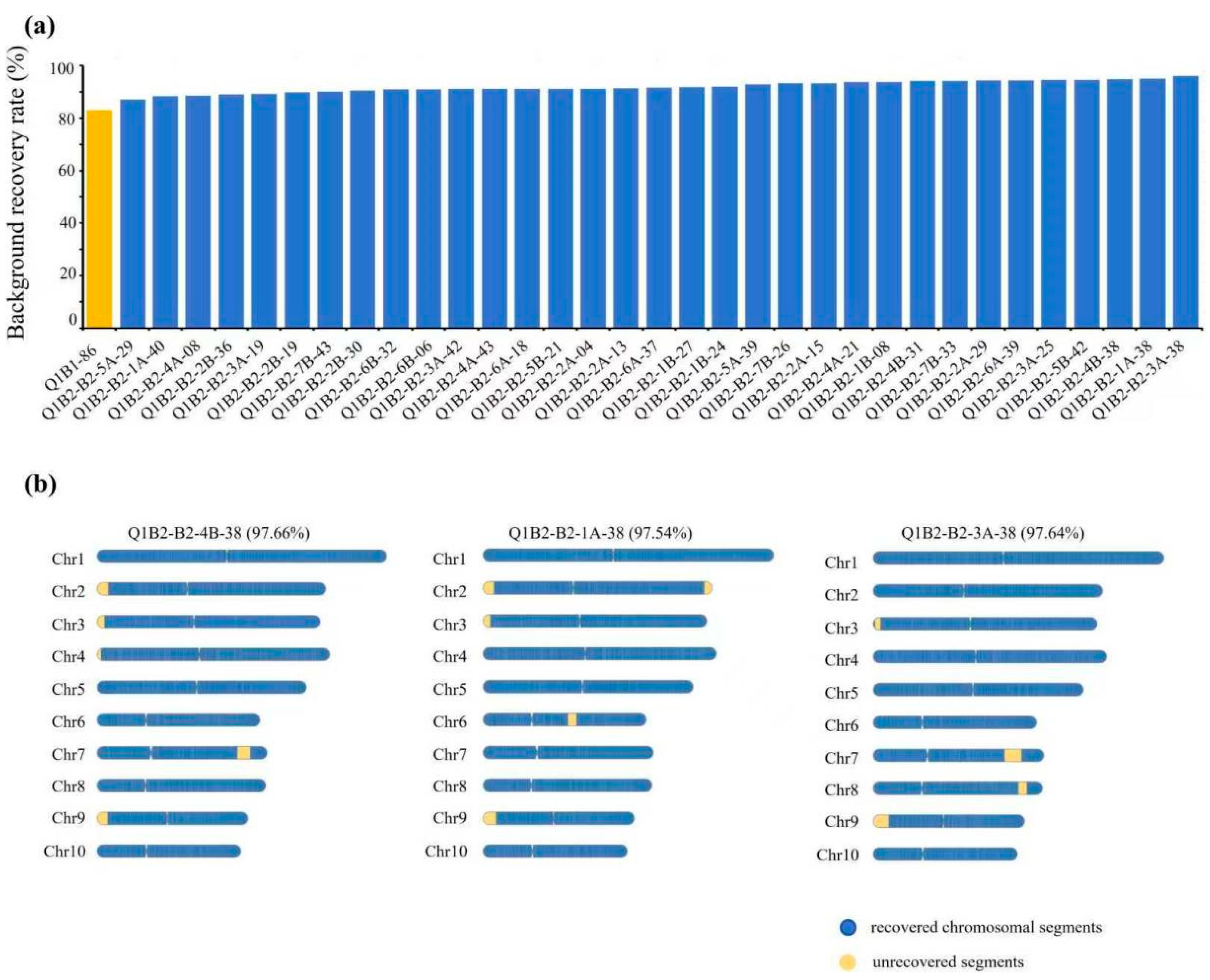
3.3. Evaluation of the High-Generation Backcross Population Background in G2H Strategy
4. Discussion
4.1. A Paradigm Shift from Point-Based to Block-Based Genetic Background Assessment
4.2. The CFDI: An Innovative Metric for Addressing Small Fragment Residuals
4.3. G2H Technology Advantages and Application Prospects
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BC | Backcross |
| G2H | Genotype-to-Haplotype |
| CFDI | Chromosomal Fragment Distribution Index |
| DP | Donor Parent |
| RP | Recurrent Parent |
| MAS | Marker-Assisted Selection |
| SSR | Simple Sequence Repeat |
| SNP | Single-Nucleotide Polymorphism |
| BRR | Background Recovery Rate |
| HTP | Haplotype Tag Polymorphism |
| PIC | Polymorphism Information Content |
| RIL | Recombinant Inbred Line |
| PRPG | Proportion of Recurrent Parent Genome |
| HTBR | HTP-based Recurrent Parental Background Recovery Rate |
| WGBR | Whole-Genome Background Recovery Rate |
References
- Kumar, S.; Thakur, A.; Sharma, M.; Thakur, N.; Kumar, S.; Singh, A. Anther-Mediated Haploid Induction and Genetic Stability of Phytophthora-Resistant Backcross Lines of Capsicum annuum L. J. Genet. Eng. Biotechnol. 2025, 23, 100498. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Bie, X.; Qiu, Y.; Wang, K.; Yang, Z.; Jia, Y.; Xu, Z.; Yu, M.; Du, L.; Lin, Z.; et al. Development of Powdery Mildew Resistant Derivatives of Wheat Variety Fielder for Use in Genetic Transformation. Crop J. 2023, 11, 573–583. [Google Scholar] [CrossRef]
- Herzog, E.; Frisch, M. Selection Strategies for Marker-Assisted Backcrossing with High-Throughput Marker Systems. Theor. Appl. Genet. 2011, 123, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Ni, Z.; Moeinizade, S.; Kusmec, A.; Hu, G.; Wang, L.; Schnable, P.S. New Insights into Trait Introgression with the Look-Ahead Intercrossing Strategy. G3 Genes Genomes Genet. 2023, 13, jkad042. [Google Scholar] [CrossRef] [PubMed]
- Hasan, N.; Choudhary, S.; Naaz, N.; Sharma, N.; Laskar, R.A. Recent Advancements in Molecular Marker-Assisted Selection and Applications in Plant Breeding Programmes. J. Genet. Eng. Biotechnol. 2021, 19, 128. [Google Scholar] [CrossRef] [PubMed]
- Davey, J.W.; Hohenlohe, P.A.; Etter, P.D.; Boone, J.Q.; Catchen, J.M.; Blaxter, M.L. Genome-Wide Genetic Marker Discovery and Genotyping Using next-Generation Sequencing. Nat. Rev. Genet. 2011, 12, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Rafalski, A. Applications of Single Nucleotide Polymorphisms in Crop Genetics. Curr. Opin. Plant Biol. 2002, 5, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Bhat, J.A.; Ali, S.; Salgotra, R.K.; Mir, Z.A.; Dutta, S.; Jadon, V.; Tyagi, A.; Mushtaq, M.; Jain, N.; Singh, P.K.; et al. Genomic Selection in the Era of Next Generation Sequencing for Complex Traits in Plant Breeding. Front. Genet. 2016, 7, 221. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, P.; Yang, Z.; Xu, C. Genetic Mapping of Quantitative Trait Loci in Crops. Crop J. 2017, 5, 175–184. [Google Scholar] [CrossRef]
- Varshney, R.K.; Graner, A.; Sorrells, M.E. Genic Microsatellite Markers in Plants: Features and Applications. Trends Biotechnol. 2005, 23, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Tian, H.; Li, C.; Yi, H.; Zhang, Y.; Li, X.; Zhao, H.; Huo, Y.; Wang, R.; Kang, D.; et al. HTPdb and HTPtools: Exploiting Maize Haplotype-Tag Polymorphisms for Germplasm Resource Analyses and Genomics-Informed Breeding. Plant Commun. 2022, 3, 100331. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Yang, Y.; Yi, H.; Xu, L.; He, H.; Fan, Y.; Wang, L.; Ge, J.; Liu, Y.; Wang, F.; et al. New Resources for Genetic Studies in Maize (Zea mays L.): A Genome-wide Maize6H-60K Single Nucleotide Polymorphism Array and Its Application. Plant J. 2021, 105, 1113–1122. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.A. Marker-Assisted Selection for Fusarium Head Blight Resistance in Wheat. Int. J. Food Microbiol. 2007, 119, 51–53. [Google Scholar] [CrossRef] [PubMed]
- Olshen, A.B.; Venkatraman, E.S.; Lucito, R.; Wigler, M. Circular Binary Segmentation for the Analysis of Array-Based DNA Copy Number Data. Biostatistics 2004, 5, 557–572. [Google Scholar] [CrossRef] [PubMed]
- Venkatraman, E.S.; Olshen, A.B. A Faster Circular Binary Segmentation Algorithm for the Analysis of Array CGH Data. Bioinformatics 2007, 23, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Hao, Z.; Lv, D.; Ge, Y.; Shi, J.; Weijers, D.; Yu, G.; Chen, J. RIdeogram: Drawing SVG Graphics to Visualize and Map Genome-Wide Data on the Idiograms. PeerJ Comput. Sci. 2020, 6, e251. [Google Scholar] [CrossRef] [PubMed]
- Hameed, A.; Poznanski, P.; Nadolska-Orczyk, A.; Orczyk, W. Graph Pangenomes Track Genetic Variants for Crop Improvement. Int. J. Mol. Sci. 2022, 23, 13420. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; An, L.; Yang, W.; Yang, L.; Wei, T.; Shi, J.; Wang, J.; Doonan, J.H.; Xie, K.; Fernie, A.R.; et al. Integrated Biotechnological and AI Innovations for Crop Improvement. Nature 2025, 643, 925–937. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qing, X.; Wang, W.; Xu, L.; Zhang, Y.; Zhao, Y.; Ge, J.; Shen, X.; Wang, R.; Xue, Y.; Wang, F. G2H: A Precise Block-Scanning Strategy for Genetic Background Assessment in Maize Backcross Breeding. Genes 2025, 16, 1480. https://doi.org/10.3390/genes16121480
Qing X, Wang W, Xu L, Zhang Y, Zhao Y, Ge J, Shen X, Wang R, Xue Y, Wang F. G2H: A Precise Block-Scanning Strategy for Genetic Background Assessment in Maize Backcross Breeding. Genes. 2025; 16(12):1480. https://doi.org/10.3390/genes16121480
Chicago/Turabian StyleQing, Xiangyu, Weiwei Wang, Liwen Xu, Yunlong Zhang, Yikun Zhao, Jianrong Ge, Xuelei Shen, Rui Wang, Yingjie Xue, and Fengge Wang. 2025. "G2H: A Precise Block-Scanning Strategy for Genetic Background Assessment in Maize Backcross Breeding" Genes 16, no. 12: 1480. https://doi.org/10.3390/genes16121480
APA StyleQing, X., Wang, W., Xu, L., Zhang, Y., Zhao, Y., Ge, J., Shen, X., Wang, R., Xue, Y., & Wang, F. (2025). G2H: A Precise Block-Scanning Strategy for Genetic Background Assessment in Maize Backcross Breeding. Genes, 16(12), 1480. https://doi.org/10.3390/genes16121480




