Comparative Transcriptome Analysis Reveals the Regulation of Growth Activity in Poria cocos Under Different Light Duration Stimulations
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. RNA-Seq Sequencing and Transcriptome Assembly
2.3. Analysis of Differential Expressed Genes
2.4. WGCNA Constructs Co-Expression Modules and Regulatory Networks
2.5. Real-Time Fluorescence Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR) Analysis
2.6. Detection of Physiological Indicators of Poria cocos
3. Results
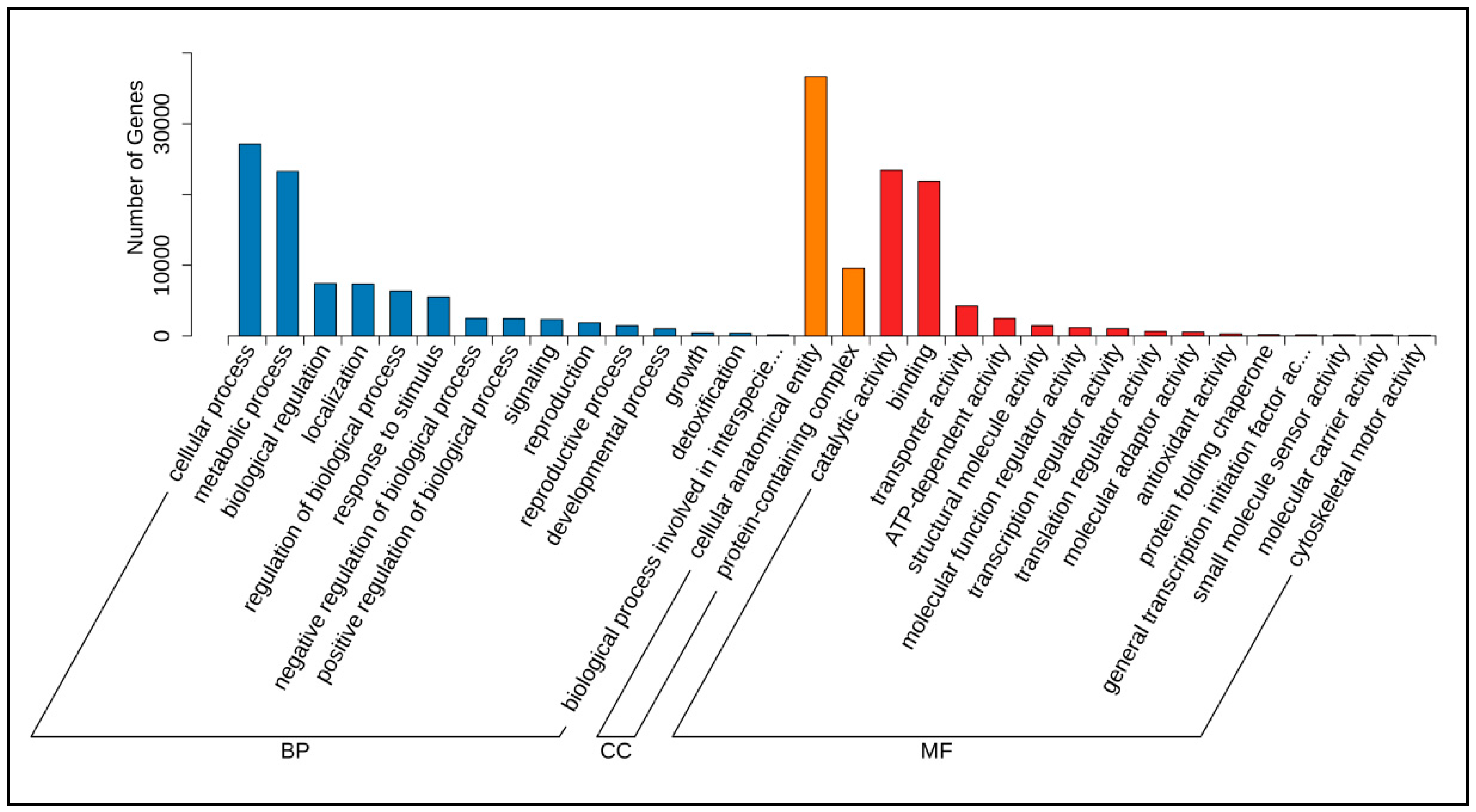
3.1. Identification and Functional Annotation of Poria cocos Transcriptome Unigene Under Different Light Exposure Times
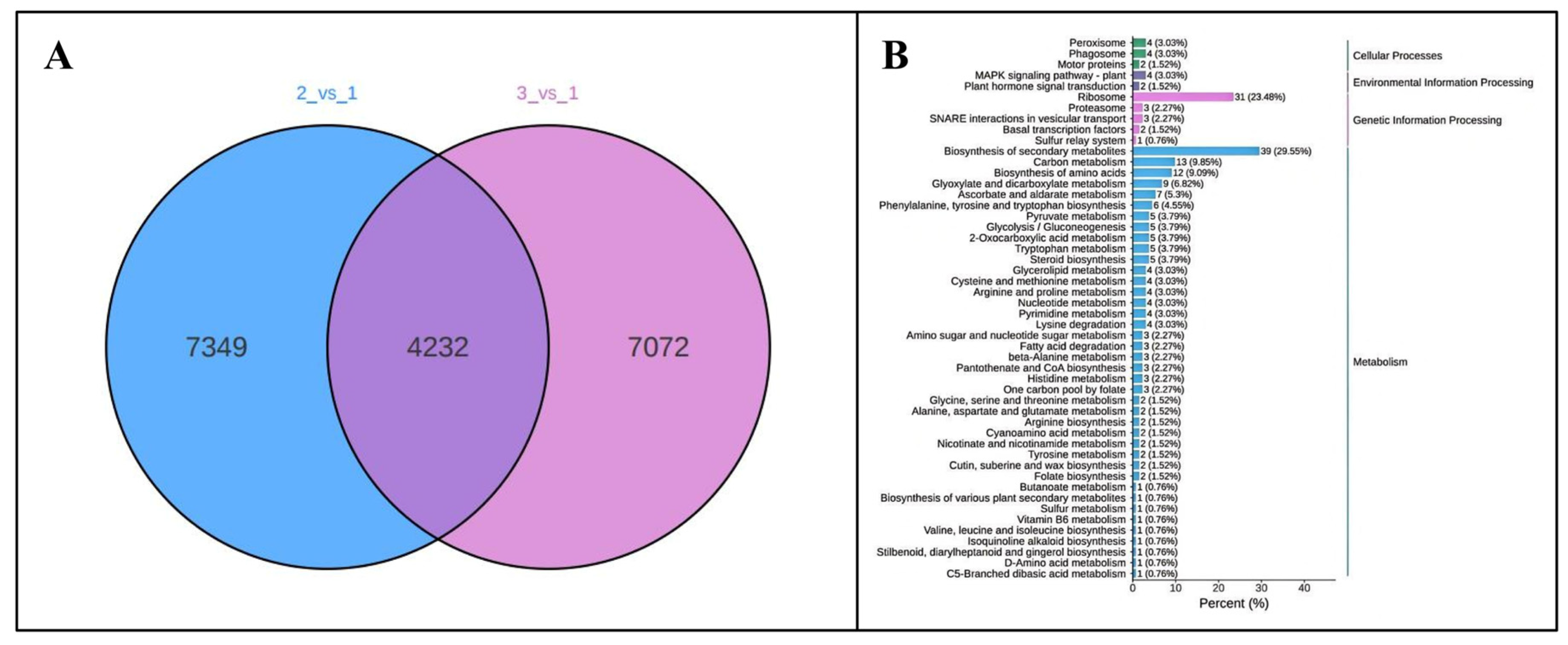
3.2. Identification and Characterization of Total Differentially Expressed Genes (DEGs) in Poria cocos Under Different Light Exposure Times
3.3. Differentially Expressed Genes (DEGs) in Response to Photoperiodic Stimulation in Poria cocos
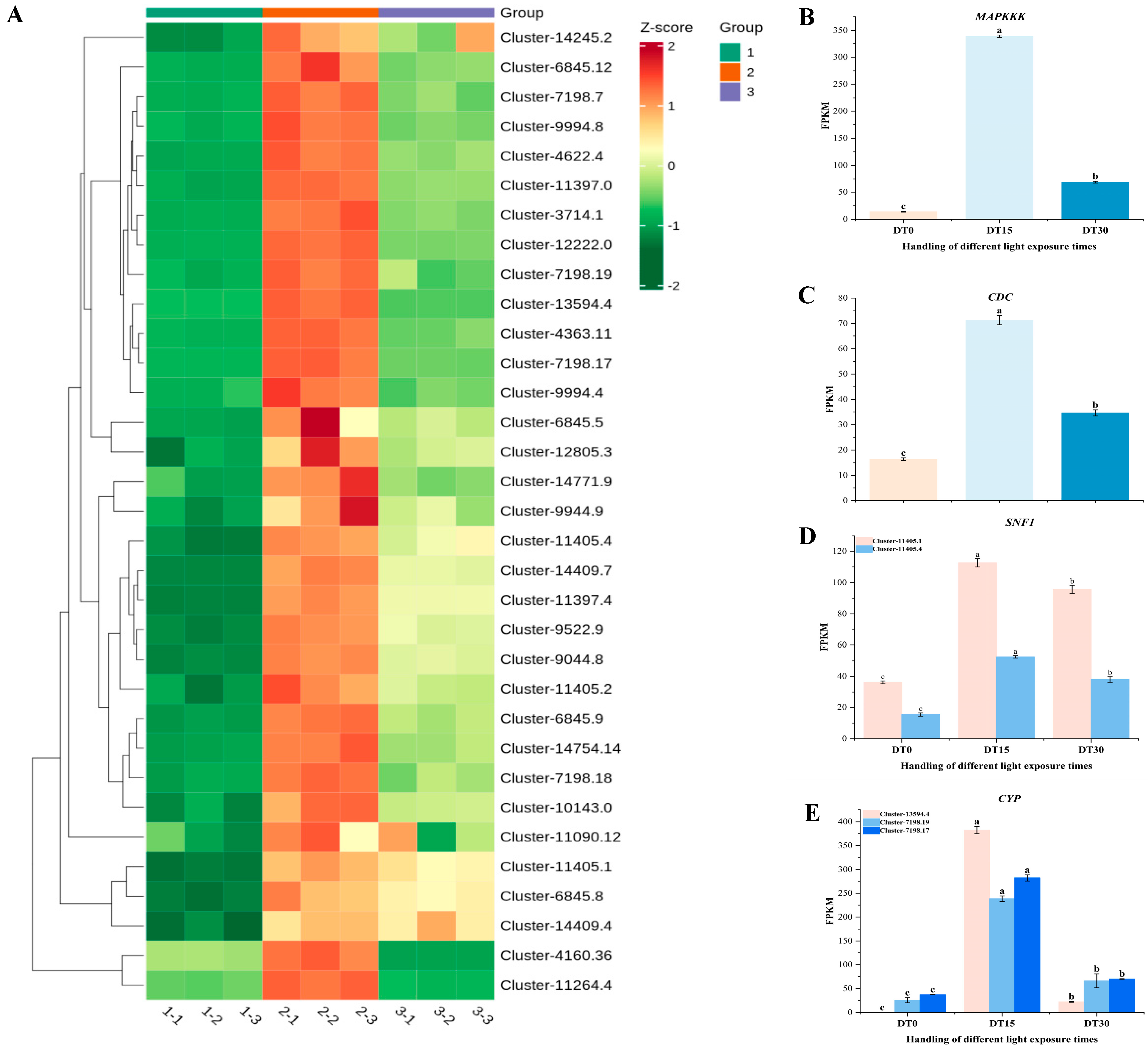
3.4. Regulation of Differential Genes in the Growth and Metabolic Pathways of Poria cocos by Light Duration
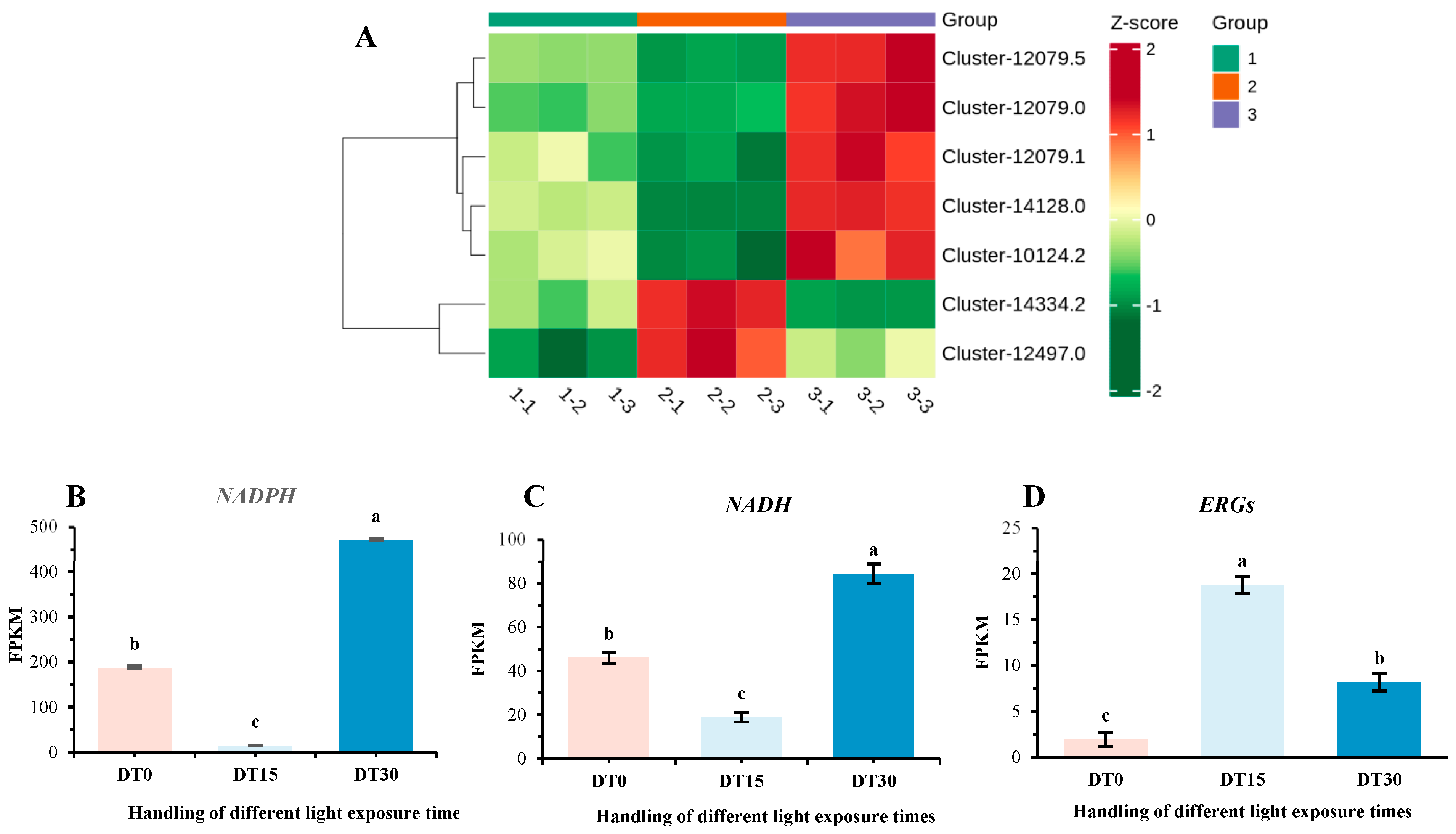
3.5. DEG Response of Cell Damage in Poria cocos Under Different Light Time Stimulation
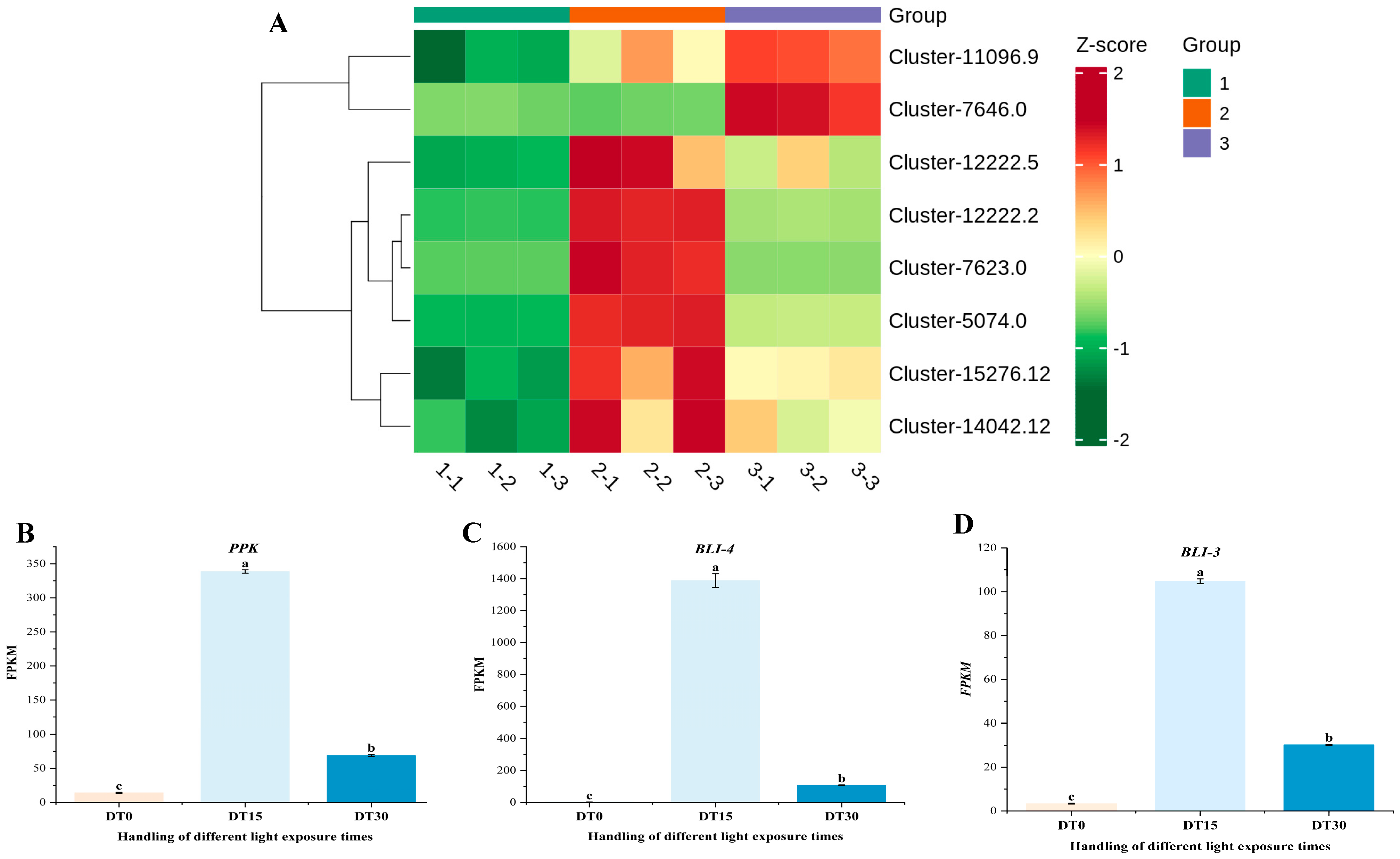
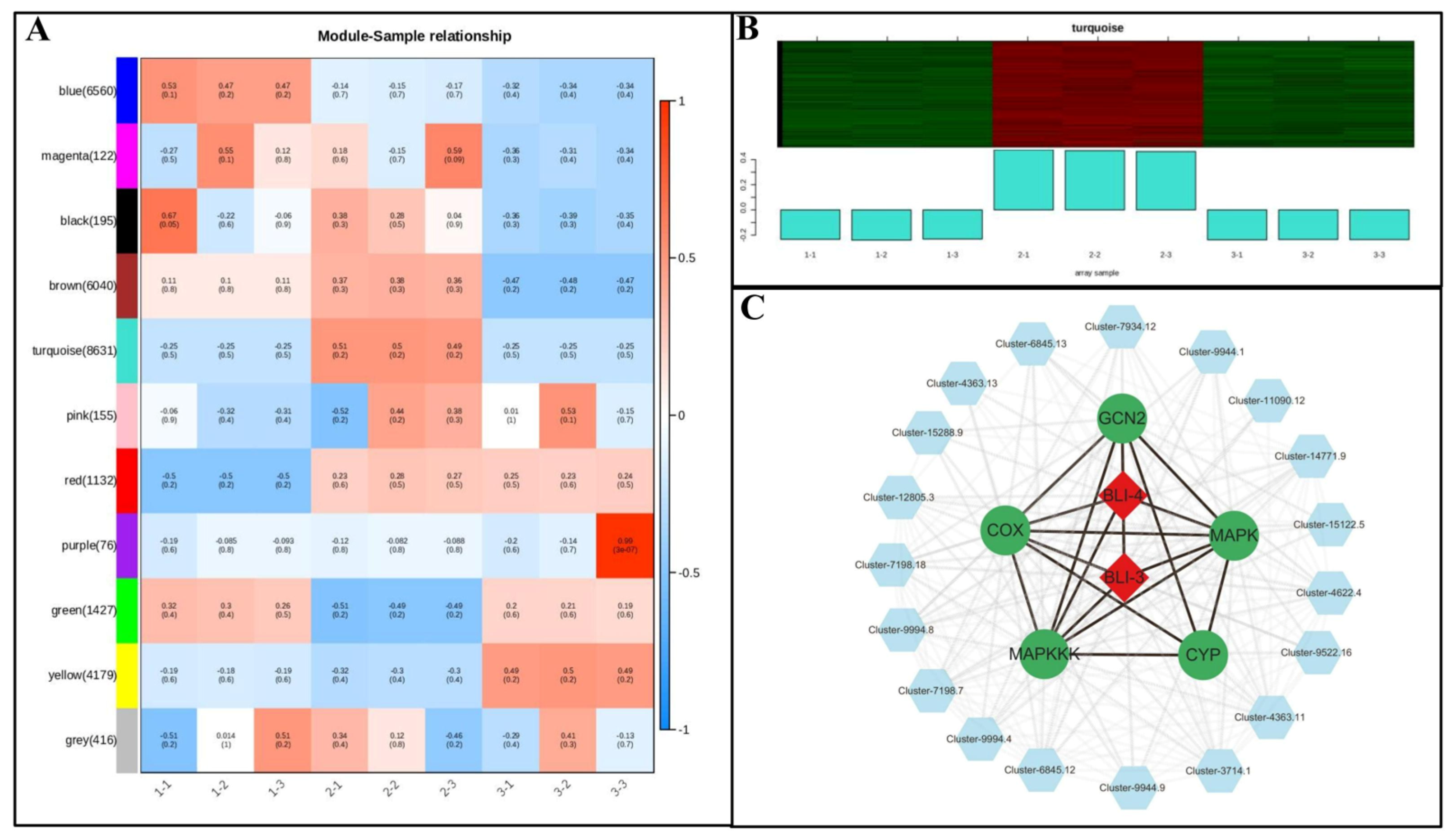
3.6. Analysis of Coexpression Network of DEG WGCNA Associated with Growth and Metabolism of Poria cocos
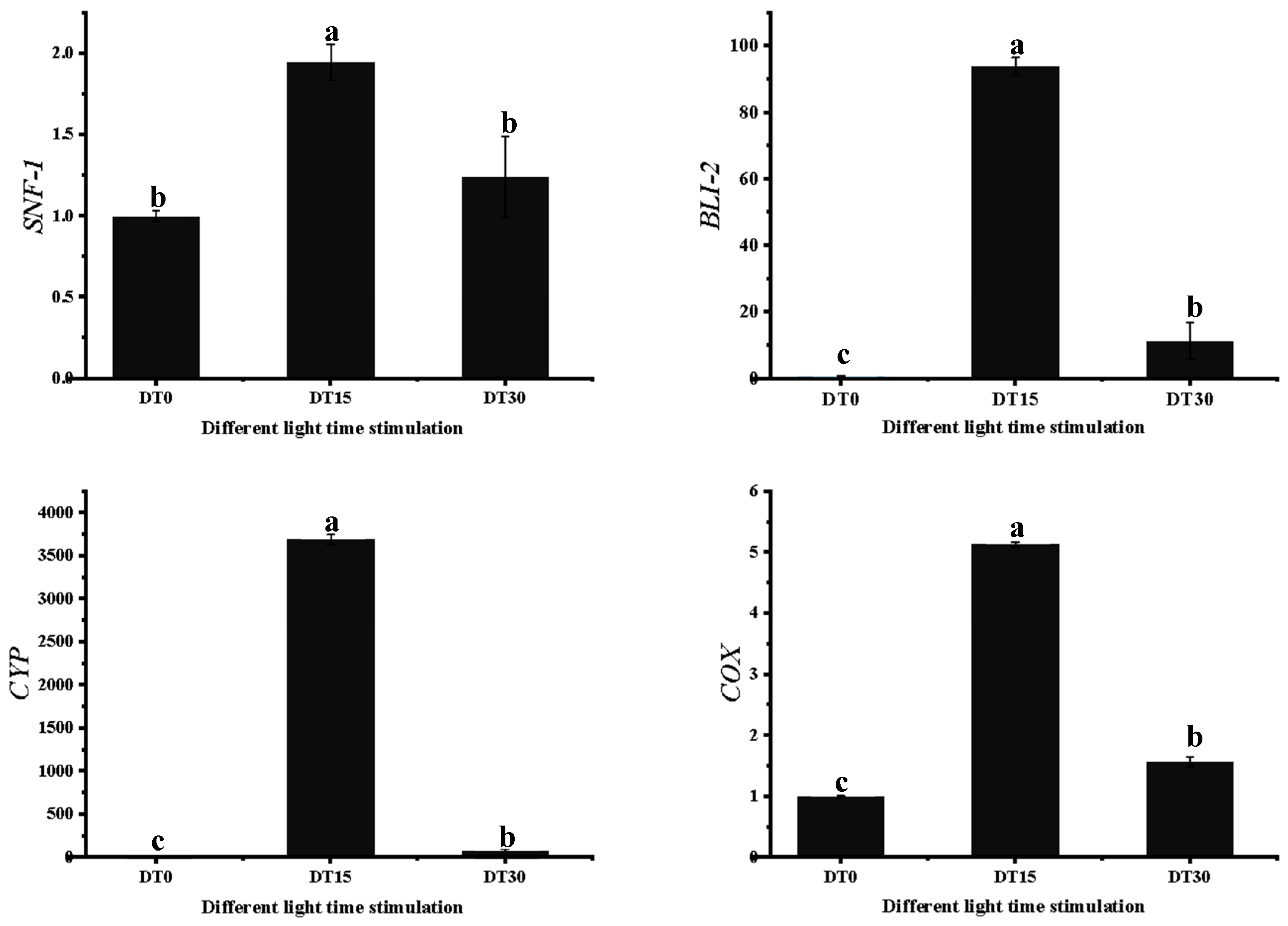
3.7. Some DEG Related to Growth and Metabolism of Poria cocos Was Verified by qRT-PCR
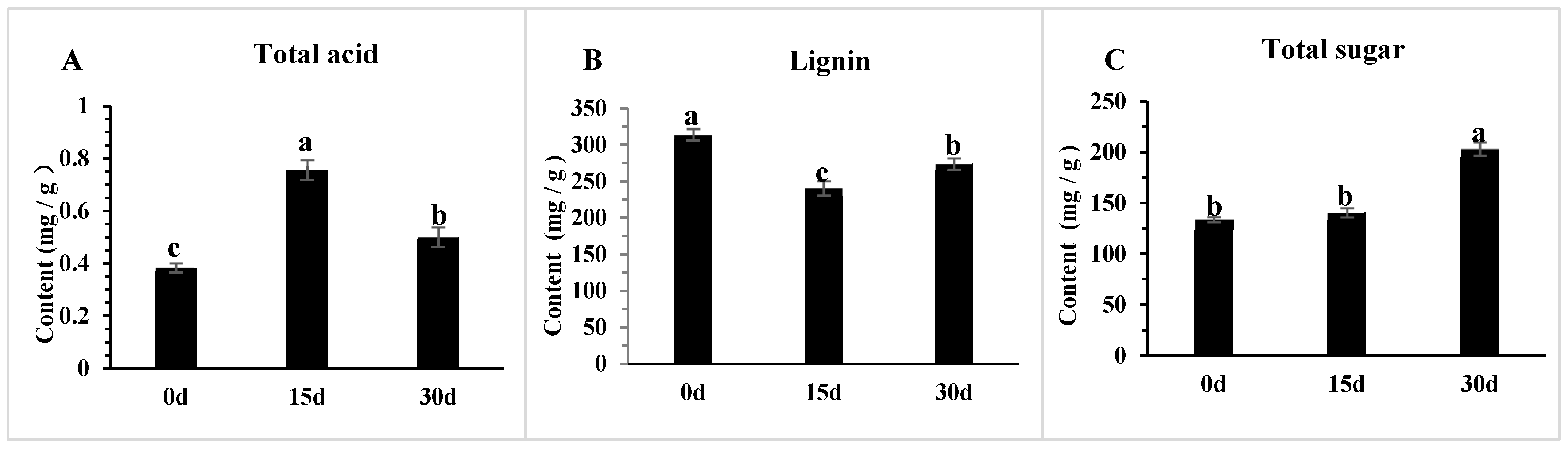
3.8. The Changes in Physiological Indicators of Poria cocos Under Different Light Exposure Times
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, X.; Ma, L.; Zhang, L. Molecular basis for Poria cocos mushroom polysaccharide used as an antitumor drug in China. Prog. Mol. Biol. Transl. Sci. 2019, 163, 263–296. [Google Scholar]
- Tian, S.-S.; Liu, X.Q.; Feng, W.; Zhang, Q.-W.; Yan, L.; Wang, A.M.; Gao, L. Quality evaluation of Poria based on specific chromatogram and quantitative analysis of multicomponents. China J. Chin. Mater. Medica 2019, 44, 1371–1380. [Google Scholar]
- Nie, A.; Chao, Y.; Zhang, X.; Jia, W.; Zhou, Z.; Zhu, C. Phytochemistry and Pharmacological Activities of Wolfiporia cocos (F.A. Wolf) Ryvarden & Gilb. Front. Pharmacol. 2020, 11, 11–49. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-Z.; Zhang, J.; Zhao, Y.-L.; Li, T.; Shen, T.; Li, J.-Q.; Li, W.-Y.; Liu, H.-G. Mycology, cultivation, traditional uses, phytochemistry and pharmacology of Wolfiporia cocos (Schwein.) Ryvarden et Gilb.: A review. J. Ethnopharmacol. 2013, 147, 265–276. [Google Scholar] [CrossRef]
- Cheng, Y.; Ding, Z.-X.; Zhang, Y.; Jiang, Y.H.; Wang, L.; Luo, J.P.; Peng, D.Y.; Yu, N.J.; Chen, W.D. Research progress on chemical structures and pharmacological activities of Poria cocos polysaccharide and its derivatives. China J. Chin. Mater. Medica 2020, 45, 4332–4340. [Google Scholar]
- Huang, Q.; Zhang, L. Preparation, chain conformation and anti-tumor activities of water-soluble phosphated (1 → 3)-α-d-glucan from Poria cocos mycelia. Carbohydr. Polym. 2011, 83, 1363–1369. [Google Scholar] [CrossRef]
- Zhao, J.; Niu, X.; Yu, J.; Xiao, X.; Li, W.; Zang, L.; Hu, Z.; Ip, P.S.-P.; Li, W. Poria cocos polysaccharides attenuated ox-LDL-induced inflammation and oxidative stress via ERK activated Nrf2/HO-1 signaling pathway and inhibited foam cell formation in VSMCs. Int. Immunopharmacol. 2020, 80, 106173. [Google Scholar] [CrossRef]
- Lu, M.-K.; Cheng, J.-J.; Lin, C.-Y.; Chang, C.-C. Purification, structural elucidation, and anti-inflammatory effect of a water-soluble 1,6-branched 1,3-α-d-galactan from cultured mycelia of Poria cocos. Food Chem. 2010, 118, 349–356. [Google Scholar] [CrossRef]
- Lin, T.-Y.; Lu, M.-K.; Chang, C.-C. Structural identification of a fucose-containing 1,3-β-mannoglucan from Poria cocos and its anti-lung cancer CL1-5 cells migration via inhibition of TGFβR-mediated signaling. Int. J. Biol. Macromol. 2020, 157, 311–318. [Google Scholar] [CrossRef]
- Lee, N.; Hwang, D.Y.; Gil Lee, H.; Hwang, H.; Kang, H.W.; Lee, W.; Choi, M.G.; Ahn, Y.J.; Lim, C.; Kim, J.-I.; et al. ASYMMETRIC LEAVES1 promotes leaf hyponasty in Arabidopsis by light-mediated auxin signaling. Plant Physiol. 2024, 197, kiae550. [Google Scholar] [CrossRef]
- DeOliveira, C.C.; Crane, B.R. A structural decryption of cryptochromes. Front. Chem. 2024, 12, 1436322. [Google Scholar] [CrossRef]
- Kalaitzoglou, P.; Taylor, C.; Calders, K.; Hogervorst, M.; van Ieperen, W.; Harbinson, J.; de Visser, P.; Nicole, C.C.; Marcelis, L.F. Unraveling the effects of blue light in an artificial solar background light on growth of tomato plants. Environ. Exp. Bot. 2021, 184, 104377. [Google Scholar] [CrossRef]
- Mancini, E.; Sanchez, S.E.; Romanowski, A.; Schlaen, R.G.; Sanchez-Lamas, M.; Cerdán, P.D.; Yanovsky, M.J. Acute Effects of Light on Alternative Splicing in Light-Grown Plants. Photochem. Photobiol. 2015, 92, 126–133. [Google Scholar] [CrossRef]
- Dou, K.; Pang, G.; Cai, F.; Chenthamara, K.; Zhang, J.; Liu, H.; Druzhinina, I.S.; Chen, J. Functional Genetics of Trichoderma Mycoparasitism. In Advances in Trichoderma Biology for Agricultural Applications; Fungal Biology; Springer: Berlin/Heidelberg, Germany, 2022; pp. 39–83. [Google Scholar]
- Fuller, K.K.; Loros, J.J.; Dunlap, J.C. Fungal photobiology: Visible light as a signal for stress, space and time. Curr. Genet. 2014, 61, 275–288. [Google Scholar] [CrossRef]
- Katherine, A.B.; Danielj, E. Cellular and Molecular Biology of Filamentous Fungi; ASM Press: Washington, DC, USA, 2010. [Google Scholar]
- Tisch, D.; Schmoll, M. Light regulation of metabolic pathways in fungi. Appl. Microbiol. Biotechnol. 2009, 85, 1259–1277. [Google Scholar] [CrossRef] [PubMed]
- Vinnikova, O.I. Long-lasting effects of red and blue light exposure on the growth of soil fungi. Acta Biol. Szeged. 2020, 64, 25–36. [Google Scholar] [CrossRef]
- Bellia, L.; Birolo, L.; Fragliasso, F.; Genovese, A.; Palella, B.; Petraretti, M.; Riccio, G.; Stanzione, I.; Corsaro, M. Does Light Affect Fungal Growth? Experimental Analysis Under Monochromatic Led Sources. In Proceedings of the CIE 2023 Conference, Ljubljana, Slovenia, 15–23 September 2023; pp. 697–705. [Google Scholar]
- Hu, Z.-H.; Sun, M.-Z.; Yang, K.-X.; Zhang, N.; Chen, C.; Xiong, J.-W.; Yang, N.; Chen, Y.; Liu, H.; Li, X.-H.; et al. High-Throughput Transcriptomic Analysis of Circadian Rhythm of Chlorophyll Metabolism Under Different Photoperiods in Tea Plants. Int. J. Mol. Sci. 2024, 25, 9270. [Google Scholar] [CrossRef] [PubMed]
- Velmurugan, P.; Lee, Y.H.; Venil, C.K.; Lakshmanaperumalsamy, P.; Chae, J.-C.; Oh, B.-T. Effect of light on growth, intracellular and extracellular pigment production by five pigment-producing filamentous fungi in synthetic medium. J. Biosci. Bioeng. 2010, 109, 346–350. [Google Scholar] [CrossRef]
- Neringa, R.; Brazaitytė, A.; Viktorija, V.-K.; Asta, K.; Pavelas, D.; Giedre, S.; Alma, V. The Effect of Monochromatic LED Light Wavelengths and Photoperiods on Botrytis cinerea. J. Fungi 2021, 7, 970. [Google Scholar] [CrossRef] [PubMed]
- Fuller, K.K.; Ringelberg, C.S.; Loros, J.J.; Dunlap, J.C. The Fungal Pathogen Aspergillus fumigatus Regulates Growth, Metabolism, and Stress Resistance in Response to Light. mBio 2013, 4, e00142-13. [Google Scholar] [CrossRef]
- Feng, Y.; Xu, H.; Sun, Y.; Xia, R.; Hou, Z.; Li, Y.; Wang, Y.; Pan, S.; Li, L.; Zhao, C.; et al. Effect of light on quality of preharvest and postharvest edible mushrooms and its action mechanism: A review. Trends Food Sci. Technol. 2023, 139, 104119. [Google Scholar] [CrossRef]
- Chen, L.; Liu, M.; Li, Y.; Guan, Y.; Ruan, J.; Mao, Z.; Wang, W.; Yang, H.-Q.; Guo, T. Arabidopsis cryptochromes interact with SOG1 to promote the repair of DNA double-strand breaks. Biochem. Biophys. Res. Commun. 2024, 32, 236–248. [Google Scholar] [CrossRef] [PubMed]
- Chaves, I.; Pokorny, R.; Byrdin, M.; Hoang, N.; Ritz, T.; Brettel, K.; Essen, L.-O.; van der Horst, G.T.J.; Batschauer, A.; Ahmad, M. The Cryptochromes: Blue Light Photoreceptors in Plants and Animals. Annu. Rev. Plant Biol. 2011, 62, 335–364. [Google Scholar] [CrossRef]
- Huang, R.; Liu, M.; Gong, G.; Wu, P.; Bai, M.; Qin, H.; Wang, G.; Liao, H.; Wang, X.; Li, Y.; et al. BLISTER promotes seed maturation and fatty acid biosynthesis by interacting with WRINKLED1 to regulate chromatin dynamics in Arabidopsis. Plant Cell 2022, 34, 2242–2265. [Google Scholar] [CrossRef]
- Sakamoto, Y. Influences of environmental factors on fruiting body induction, development and maturation in mushroom-forming fungi. Fungal Biol. Rev. 2018, 32, 236–248. [Google Scholar] [CrossRef]
- Li, Z.; Liu, N.; Yang, W.; Tu, J.; Huang, Y.; Wang, W.; Sheng, C.J. Controlling antifungal activity with light: Optical regulation of fungal ergosterol biosynthetic pathway with photo-responsive CYP51 inhibitors. Acta Pharm. Sin. B 2023, 13, 3080–3092. [Google Scholar] [CrossRef]
- Chakraborty, P.; Biswas, A.; Dey, S.; Bhattacharjee, T.; Chakrabarty, S. Cytochrome P450 Gene Families: Role in Plant Secondary Metabolites Production and Plant Defense. J. Xenobiot. 2023, 13, 402–423. [Google Scholar] [CrossRef]
- Lee, C.P.; Le, X.H.; Gawryluk, R.M.R.; Casaretto, J.A.; Rothstein, S.J.; Millar, A.H. EARLY NODULIN93 acts via cytochrome c oxidase to alter respiratory ATP production and root growth in plants. Plant Cell 2024, 36, 4716–4731. [Google Scholar] [CrossRef]
- Ge, X.; Li, R.; Zhang, X.; Zhao, J.; Zhang, Y.; Xin, Q. Transcriptome sequencing and global analysis of blue light-responsive genes provide clues for high carotenoid yields in Blakeslea trispora. Int. Microbiol. 2021, 25, 325–338. [Google Scholar] [CrossRef]
- Zhou, X.; Zhong, C.; Xie, J.; Jin, J.; Shen, B.; Chen, L.; Liu, H.; Zhang, S.-H. Optimization of the Solid-State Culture Conditions and Chemical Component Analysis of Poria cocos (Agaricomycetes). Int. J. Med. Mushrooms 2023, 25, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.-Y.; Dillies, M.-A. SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2014, 12, 59–60. [Google Scholar] [CrossRef]
- Tang, J.; Zhu, W.; Liu, H.; Su, Z.; Liu, J.; Shu, S. Cloning and sequence analysis of the cytochrome P450 gene PcCYP in Poria cocos. Chin. Herb. Med. 2023, 54, 3962–3970. (In Chinese) [Google Scholar]
- Ma, X.; Li, C.; Huang, R.; Zhang, K.; Wang, Q.; Fu, C.; Liu, W.; Sun, C.; Wang, P.; Wang, F.; et al. Rice Brittle Culm19 Encoding Cellulose Synthase Subunit CESA4 Causes Dominant Brittle Phenotype But has No Distinct Influence on Growth and Grain Yield. Rice 2021, 14, 95. [Google Scholar] [CrossRef]
- Yang, L.; Chen, Y.; Wang, M.; Hou, H.; Li, S.; Guan, L.; Yang, H.; Wang, W.; Hong, L. Metabolomic and transcriptomic analyses reveal the effects of grafting on blood orange quality. Front. Plant Sci. 2023, 14, 1169220. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Li, Z.; Liao, G.; Chen, L.; Xu, X. Variation in fruit quality within wild Actinidia eriantha germplasm. N. Z. J. Crop Hortic. Sci. 2020, 48, 153–163. [Google Scholar] [CrossRef]
- Segal, L.M.; Wilson, R.A. Reactive oxygen species metabolism and plant-fungal interactions. Fungal Genet. Biol. 2018, 110, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Gong, W.; Zhu, Z.; Yan, L.; Hu, Z.; Peng, Y. Comparative transcriptomics of Pleurotus eryngii reveals blue-light regulation of carbohydrate-active enzymes (CAZymes) expression at primordium differentiated into fruiting body stage. Genomics 2018, 110, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Bruchez, J.J.P.; Jürgen, E.; Kohler, W.; Volker, K.; Anthony, J.R.; Vincenzo, R. bli-4, a gene that is rapidly induced by blue light, encodes a novel mitochondrial, short-chain alcohol dehydrogenase-like protein in Neurospora crassa. Mol. Genet. Genom. 1996, 252, 223–229. [Google Scholar] [CrossRef]
- Pratiwi, R.A.; Yahya, N.S.W.; Chi, Y. Bio function of Cytochrome P450 on fungus: A review. IOP Conf. Ser. Earth Environ. Sci. 2022, 959, 012023. [Google Scholar] [CrossRef]
- Usaite, R.; Jewett, M.C.; Oliveira, A.P.; Yates, J.R.; Olsson, L.; Nielsen, J. Reconstruction of the yeast Snf1 kinase regulatory network reveals its role as a global energy regulator. Mol. Syst. Biol. 2009, 5, 319. [Google Scholar] [CrossRef]
- Lu, M.S.; Drubin, D.G. Unexplored Cdc42 functions at the budding yeast nucleus suggested by subcellular localization. Small GTPases 2021, 13, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.-M.; Liao, K. Saccharomyces cerevisiaeHog1 MAP kinase pathway is activated in response to honokiol exposure. J. Appl. Microbiol. 2018, 124, 754–763. [Google Scholar] [CrossRef]
- Kim, J.; Oh, J.; Sung, G.-H. MAP Kinase Hog1 Regulates Metabolic Changes Induced by Hyperosmotic Stress. Front. Microbiol. 2016, 7, 732. [Google Scholar] [CrossRef]
- Mårten, W.; Klaas, K.; Vivek, S. Oxygen Activation and Energy Conservation by Cytochrome c Oxidase. Chem. Rev. 2018, 118, 2469–2490. [Google Scholar] [CrossRef]
- Leng, Y.; Zhong, S. The Role of Mitogen-Activated Protein (MAP) Kinase Signaling Components in the Fungal Development, Stress Response and Virulence of the Fungal Cereal Pathogen Bipolaris sorokiniana. PLoS ONE 2015, 10, e0128291. [Google Scholar] [CrossRef]
- Fang, Y.-L.; Xia, L.-M.; Wang, P.; Zhu, L.-H.; Ye, J.-R.; Huang, L. The MAPKKK CgMck1 Is Required for Cell Wall Integrity, Appressorium Development, and Pathogenicity in Colletotrichum gloeosporioides. Genes 2018, 9, 543. [Google Scholar] [CrossRef]
- Wang, D.; Akhberdi, O.; Hao, X.; Yu, X.; Chen, L.; Liu, Y.; Zhu, X. Amino Acid Sensor Kinase Gcn2 Is Required for Conidiation, Secondary Metabolism, and Cell Wall Integrity in the Taxol-Producer Pestalotiopsis microspora. Front. Microbiol. 2017, 8, 1879. [Google Scholar] [CrossRef]
- Muharfiza, M.; Dimas Firmanda, A.-R.; Sen, N.; Yasushi, K.; Tetsuhito, S.; Makoto, K.; Yuichi, O.; Naoshi, K. Effect of relative humidity and light exposure on fluorescence compound dynamics, soluble solid and acidity of Japanese Citrus Iyokan during postharvest treatment. Adv. Food Sci. Sustain. Agric. Agroind. Eng. 2023, 6, 153–162. [Google Scholar] [CrossRef]
- Xie, M.; Zhang, J.; Tschaplinski, T.J.; Tuskan, G.A.; Chen, J.G.; Muchero, W. Regulation of Lignin Biosynthesis and Its Role in Growth-Defense Tradeoffs. Front. Plant Sci. 2018, 9, 1427. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.; Perez, V.C.; Dickinson, G.K.; Zhao, H.; Dai, R.; Tomiczek, B.; Cho, K.H.; Zhu, N.; Koh, J.; Grenning, A.; et al. Altered methionine metabolism impacts phenylpropanoid production and plant development in Arabidopsis thaliana. Plant J. 2023, 116, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Kula-Maximenko, M.; Hornyák, M.; Płażek, A. Measurement of the Light Intensity and Spectrum Influence on Plant Growth and Secondary Metabolites of Common Buckwheat. Methods Mol. Biol. 2024, 2791, 133–137. [Google Scholar]
- Proietti, S.; Moscatello, S.; Riccio, F.; Downey, P.; Battistelli, A. Continuous Lighting Promotes Plant Growth, Light Conversion Efficiency, and Nutritional Quality of Eruca vesicaria (L.) Cav. in Controlled Environment with Minor Effects Due to Light Quality. Front. Plant Sci. 2021, 12, 730119. [Google Scholar] [CrossRef]

| Sample | Raw Reads | Clean Reads | Clean Base (G) | Error Rate (%) | Q20 (%) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|---|---|
| DT0 1-1 | 53,215,642 | 47,486,284 | 7.12 | 0.01 | 97.86 | 94.03 | 57.83 |
| DT0 1-2 | 54,831,048 | 49,060,902 | 7.36 | 0.01 | 98.09 | 94.6 | 57.79 |
| DT0 1-3 | 56,642,570 | 50,319,860 | 7.55 | 0.01 | 98.04 | 94.47 | 57.83 |
| DT15 2-1 | 65,260,872 | 56,542,448 | 8.48 | 0.01 | 98.06 | 94.48 | 57.51 |
| DT15 2-2 | 55,131,772 | 48,6182,82 | 7.29 | 0.01 | 97.97 | 94.27 | 57.56 |
| DT15 2-3 | 54,250,752 | 47,533,692 | 7.13 | 0.01 | 98.21 | 94.86 | 57.52 |
| DT30 3-1 | 68,420,452 | 61,300,220 | 9.2 | 0.01 | 98.03 | 94.51 | 59.2 |
| DT30 3-2 | 44,341,606 | 39,574,124 | 5.94 | 0.01 | 97.9 | 94.18 | 59.01 |
| DT30 3-3 | 57,217,332 | 50,473,978 | 7.57 | 0.01 | 97.95 | 94.28 | 59.03 |
| Typle | Number | Mean Length | N 50 | N 90 |
|---|---|---|---|---|
| Transcript | 77,600 | 2610 | 4723 | 1348 |
| Unigene | 58,622 | 3198 | 5028 | 1612 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, C.; Ye, S.; Wei, X.; Zheng, R. Comparative Transcriptome Analysis Reveals the Regulation of Growth Activity in Poria cocos Under Different Light Duration Stimulations. Genes 2025, 16, 1404. https://doi.org/10.3390/genes16121404
Wu C, Ye S, Wei X, Zheng R. Comparative Transcriptome Analysis Reveals the Regulation of Growth Activity in Poria cocos Under Different Light Duration Stimulations. Genes. 2025; 16(12):1404. https://doi.org/10.3390/genes16121404
Chicago/Turabian StyleWu, Chengwen, Shanwen Ye, Xuhui Wei, and Rong Zheng. 2025. "Comparative Transcriptome Analysis Reveals the Regulation of Growth Activity in Poria cocos Under Different Light Duration Stimulations" Genes 16, no. 12: 1404. https://doi.org/10.3390/genes16121404
APA StyleWu, C., Ye, S., Wei, X., & Zheng, R. (2025). Comparative Transcriptome Analysis Reveals the Regulation of Growth Activity in Poria cocos Under Different Light Duration Stimulations. Genes, 16(12), 1404. https://doi.org/10.3390/genes16121404






