Genome-Wide Identification and Expression Profiling of the GRAS Gene Family in Taxus cuspidate
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of TcGRAS Gene Family Members
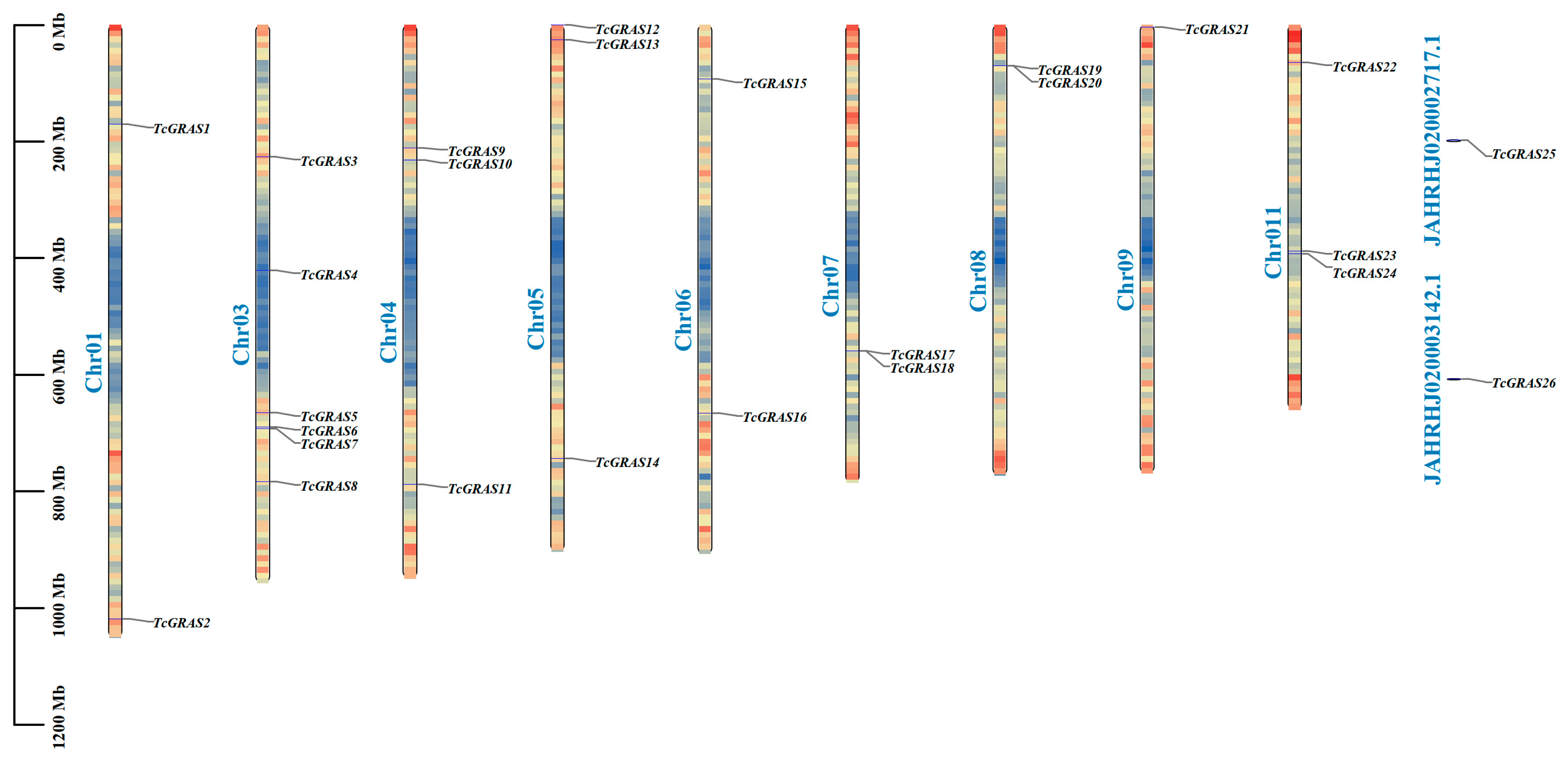
2.2. Chromosomal Localization of TcGRAS Genes
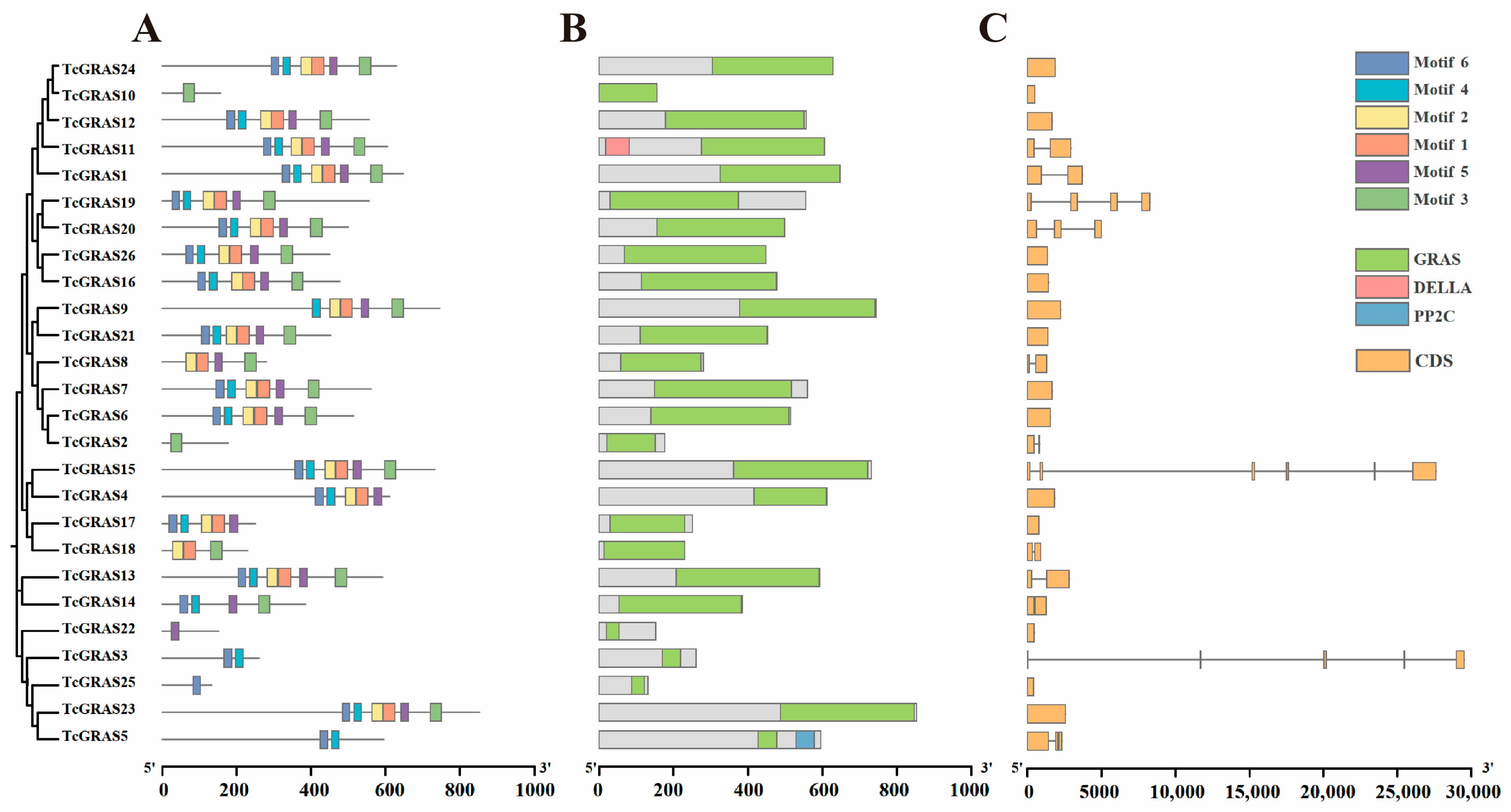
2.3. Gene Structure and Conserved Motif Analysis of TcGRAS Genes
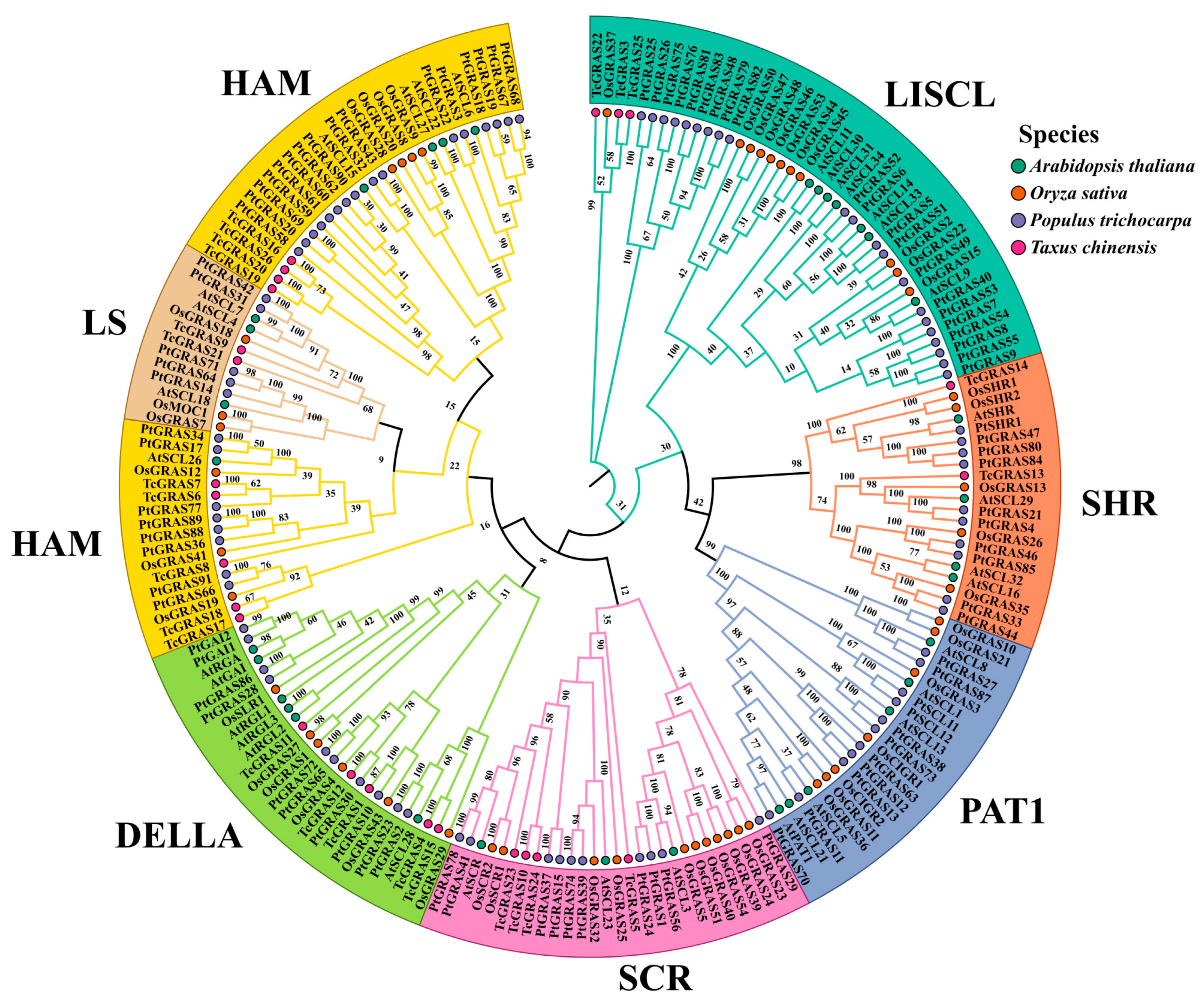
2.4. Phylogenetic Analysis of TcGRAS Genes
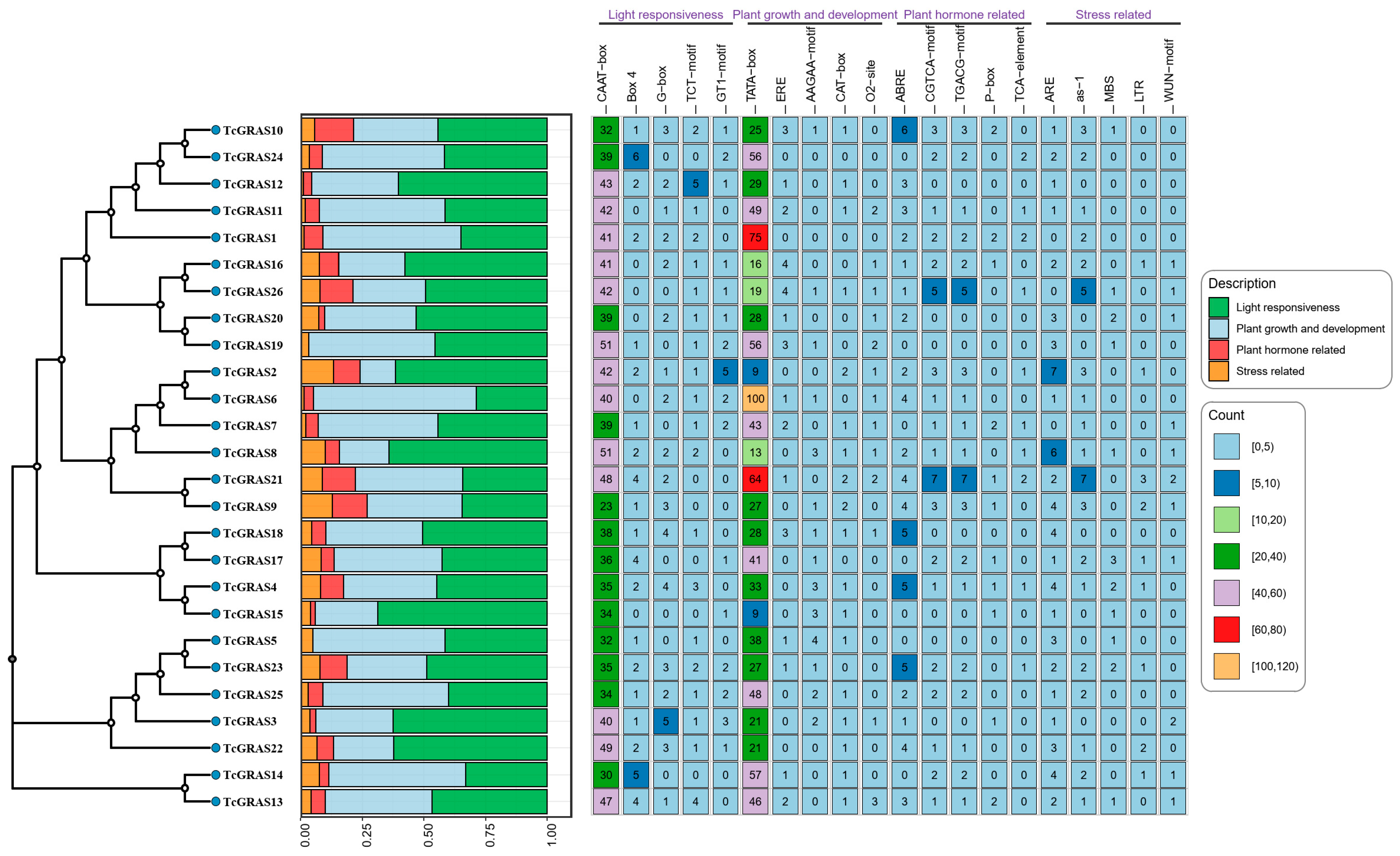
2.5. Cis-Acting Element Analysis of TcGRAS Genes
2.6. Expression Pattern and Functional Annotation Analysis of TcGRAS Genes
2.7. qRT-PCR Analysis
2.8. Data Analysis
3. Results
3.1. Genome-Wide Identification and Physicochemical Analysis of GRAS Genes in T. cuspidata
3.2. Chromosomal Localization of TcGRAS Gene Genes
3.3. Phylogenetic Analysis of TcGRAS Genes
3.4. Motif, Conserved Domain, and Gene Structure Analysis of TcGRAS Genes
3.5. Cis-Acting Elements Analysis of TcGRAS Genes
3.6. Expression Patterns, GO Enrichment, and Protein–Protein Interaction Network Analysis of TcGRAS Genes
3.7. qRT-PCR Analysis of Tissue-Specific Expression Patterns of TcGRAS Genes
4. Discussion
4.1. Structural and Evolutionary Features of the GRAS Gene Family in T. cuspidata Exhibit Distinct Characteristics
4.2. Cis-Regulatory Element and Go Analysis Reveals the Multifunctionality of TcGRAS Genes
4.3. Functional Divergence Potential of TcGRAS Genes in T. cuspidate
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, F.; Xi, M.; Liu, T.; Wu, X.; Ju, L.; Wang, D. The central role of transcription factors in bridging biotic and abiotic stress responses for plants’ resilience. New Crops 2024, 1, 100005. [Google Scholar] [CrossRef]
- Rabeh, K.; Hnini, M.; Oubohssaine, M. A comprehensive review of transcription factor-mediated regulation of secondary metabolites in plants under environmental stress. Stress Biol. 2025, 5, 15. [Google Scholar] [CrossRef]
- Lu, H.; Xu, J.; Li, G.; Zhong, T.; Chen, D.; Lv, J. Genome-wide identification and expression analysis of GRAS gene family in Eucalyptus grandis. BMC Plant Biol. 2024, 24, 573. [Google Scholar] [CrossRef]
- Fan, L.; Wang, R.; García Caparrós, P.; Meng, L.; Sun, X. Genome-wide identification and functional analysis of the GRAS gene family in Medicago lupulina L. BMC Genom. 2025, 26, 682. [Google Scholar] [CrossRef]
- Liu, X.; Widmer, A. Genome-wide Comparative Analysis of the GRAS Gene Family in Populus, Arabidopsis and Rice. Plant Mol. Biol. Rep. 2014, 32, 1129–1145. [Google Scholar] [CrossRef]
- Huang, Y.; Zheng, Q.; Zhang, M.; He, X.; Zhao, X.; Wang, L.; Lan, S.; Liu, Z. Genome-Wide Identification and Expression Analysis of the GRAS Gene Family and Their Responses to Heat Stress in Cymbidium goeringii. Int. J. Mol. Sci. 2024, 25, 6363. [Google Scholar] [CrossRef]
- Fu, X.; Richards, D.E.; Ait-ali, T.; Hynes, L.W.; Ougham, H.; Peng, J.; Harberd, N.P. Gibberellin-Mediated Proteasome-Dependent Degradation of the Barley DELLA Protein SLN1 Repressor. Plant Cell 2002, 14, 3191–3200. [Google Scholar] [CrossRef]
- Torres-Galea, P.; Hirtreiter, B.; Bolle, C. Two GRAS Proteins, SCARECROW-LIKE21 and PHYTOCHROME A SIGNAL TRANSDUCTION1, Function Cooperatively in Phytochrome A Signal Transduction. Plant Physiol. 2013, 161, 291–304. [Google Scholar] [CrossRef]
- Zhang, Z.; Ogawa, M.; Fleet, C.M.; Zentella, R.; Hu, J.; Heo, J.O.; Lim, J.; Kamiya, Y.; Yamaguchi, S.; Sun, T.P. SCARECROW-LIKE 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 2160–2165. [Google Scholar] [CrossRef] [PubMed]
- Helariutta, Y.; Fukaki, H.; Wysocka-Diller, J.; Nakajima, K.; Jung, J.; Sena, G.; Hauser, M.T.; Benfey, P.N. The SHORT-ROOT Gene Controls Radial Patterning of the Arabidopsis Root through Radial Signaling. Cell 2000, 101, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, K.; Hayashi, T.; Wu, S.; Gallagher, K.L. The SHORT-ROOT protein acts as a mobile, dose-dependent signal in patterning the ground tissue. Proc. Natl. Acad. Sci. USA 2012, 109, 13010–13015. [Google Scholar] [CrossRef]
- Bolle, C.; Koncz, C.; Chua, N.H. PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev. 2000, 14, 1269–1278. [Google Scholar] [CrossRef]
- Muntha, S.T.; Zhang, L.; Zhou, Y.; Zhao, X.; Hu, Z.; Yang, J.; Zhang, M. Phytochrome A signal transduction 1 and CONSTANS-LIKE 13 coordinately orchestrate shoot branching and flowering in leafy Brassica juncea. Plant Biotechnol. J. 2019, 17, 1333–1343. [Google Scholar] [CrossRef]
- Li, J.; Zhou, P.; Hu, Z.; Xiong, A.; Li, X.; Chen, X.; Zhuang, J. The transcription factor CsPAT1 from tea plant (Camellia sinensis) is involved in drought tolerance by modulating phenylpropanoid biosynthesis. J. Plant Physiol. 2025, 308, 154474. [Google Scholar] [CrossRef]
- Ming, R.; Fang, T.; Ling, W.; Geng, J.; Qu, J.; Zhang, Y.; Chen, J.; Yao, S.; Li, L.; Huang, D.; et al. The GRAS transcription factor PtrPAT1 of Poncirus trifoliata functions in cold tolerance and modulates glycine betaine content by regulating the BADH-like gene. Hortic. Res. 2025, 12, uhae296. [Google Scholar] [CrossRef]
- Feng, M.; Zhang, A.; Nguyen, V.; Bisht, A.; Almqvist, C.; De Veylder, L.; Carlsbecker, A.; Melnyk, C.W. A conserved graft formation process in Norway spruce and Arabidopsis identifies the PAT gene family as central regulators of wound healing. Nat. Plants 2024, 10, 53–65. [Google Scholar] [CrossRef]
- Jîjie, A.R.; Iliescu, D.; Sbârcea, L.; Boru, C.; Pătrașcu, D.; Iftode, O.A.; Minda, I.D.; Avram, Ș.; Trandafirescu, C.M.; Dehelean, C.A.; et al. A Deep Dive into the Botanical and Medicinal Heritage of Taxus. Plants 2025, 14, 1439. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Gao, L.; Wang, H.; Sun, Y.; Zhang, X.; Ke, H.; Liu, S.; Ma, P.; Liao, Q.; Wang, Y.; et al. Characterization and heterologous reconstitution of Taxus biosynthetic enzymes leading to baccatin III. Science 2024, 383, 622–629. [Google Scholar] [CrossRef]
- Xiong, X.; Gou, J.; Liao, Q.; Li, Y.; Zhou, Q.; Bi, G.; Li, C.; Du, R.; Wang, X.; Sun, T.; et al. The Taxus genome provides insights into paclitaxel biosynthesis. Nat. Plants 2021, 7, 1026–1036. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Sun, M.; Wang, X.; Zhou, X.; Di, Q.; Li, Y.; Wu, G.; Li, Y.; He, C.; Li, S. Genome-wide identification, expression analysis, and response to abiotic stress of the phosphate transporter gene family in cucumber (Cucumis sativus L.). BMC Plant Biol. 2025, 25, 677. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Wang, Y.; Dou, M.; Hu, Q.; Li, Q.; Gan, S.; Jing, M.; Wang, G.; Huang, X. Genome-wide identification and expression pattern analysis of NAC family in Taxus yunnanensis and the TyuNAC30 role in paclitaxel production. BMC Genom. 2025, 26, 705. [Google Scholar] [CrossRef]
- Waseem, M.; Nkurikiyimfura, O.; Niyitanga, S.; Jakada, B.H.; Shaheen, I.; Aslam, M.M. GRAS transcription factors emerging regulator in plants growth, development, and multiple stresses. Mol. Biol. Rep. 2022, 49, 9673–9685. [Google Scholar] [CrossRef]
- Ling, L.; Li, M.; Chen, N.; Ren, G.; Qu, L.; Yue, H.; Wu, X.; Zhao, J. Genome-Wide Analysis and Expression of the GRAS Transcription Factor Family in Avena sativa. Genes 2023, 14, 164. [Google Scholar] [CrossRef]
- Li, C.; Wang, K.; Chen, S.; Zhang, X.; Zhang, X.; Fan, L.; Dong, J.; Xu, L.; Wang, Y.; Li, Y.; et al. Genome-wide identification of RsGRAS gene family reveals positive role of RsSHRc gene in chilling stress response in radish (Raphanus sativus L.). Plant Physiol. Biochem. 2022, 192, 285–297. [Google Scholar] [CrossRef]
- Guo, Y.; Wu, H.; Li, X.; Li, Q.; Zhao, X.; Duan, X.; An, Y.; Lv, W.; An, H. Identification and expression of GRAS family genes in maize (Zea mays L.). PLoS ONE 2017, 12, e0185418. [Google Scholar] [CrossRef]
- Lee, M.; Kim, B.; Song, S.; Heo, J.; Yu, N.; Lee, S.A.; Kim, M.; Kim, D.G.; Sohn, S.O.; Lim, C.E.; et al. Large-scale analysis of the GRAS gene family in Arabidopsis thaliana. Plant Mol. Biol. 2008, 67, 659–670. [Google Scholar] [CrossRef]
- Niu, Y.; Zhao, T.; Xu, X.; Li, J. Genome-wide identification and characterization of GRAS transcription factors in tomato (Solanum lycopersicum). PeerJ 2017, 5, e3955. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Wan, P.; Sun, S.; Li, J.; Chen, M. Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol. Biol. 2004, 54, 519–532. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Chaudhary, R.; Pandey, B.; Singh, G.; Sharma, P. Genome-wide identification and expression analysis of the GRAS gene family under abiotic stresses in wheat (Triticum aestivum L.). Sci. Rep. 2023, 13, 18705. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, W. Characterization of the GRAS gene family reveals their contribution to the high adaptability of wheat. PeerJ 2021, 9, e10811. [Google Scholar] [CrossRef]
- Lu, X.; Liu, W.; Xiang, C.; Li, X.; Wang, Q.; Wang, T.; Liu, Z.; Zhang, J.; Gao, L.; Zhang, W. Genome-Wide Characterization of GRAS Family and Their Potential Roles in Cold Tolerance of Cucumber (Cucumis sativus L.). Int. J. Mol. Sci. 2020, 21, 3857. [Google Scholar] [CrossRef]
- Chang, J.; Fan, D.; Lan, S.; Cheng, S.; Chen, S.; Lin, Y.; Cao, S. Genome-Wide Identification, Expression and Stress Analysis of the GRAS Gene Family in Phoebe bournei. Plants 2023, 12, 2048. [Google Scholar] [CrossRef]
- Wang, P.; Yang, X.; Sun, S.; Wang, J.; Wang, J.; Li, X.; Li, D.; Wang, Y. The SCARECROW-LIKE transcription factor from Populus davidiana × P. bolleana simultaneously improved drought tolerance and plant growth through acetylation-dependent mechanisms. Plant Biotechnol. J. 2025, 23, 3650–3666. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhu, D.; Huang, X.; Li, S.; Gong, Y.; Yao, Q.; Fu, X.; Fan, L.; Deng, X.W. Biochemical Insights on Degradation of Arabidopsis DELLA Proteins Gained from a Cell-Free Assay System. Plant Cell 2009, 21, 2378–2390. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, M.; Zhang, W.; Wang, Y.; Zhang, H.; Yang, L.; Yu, L.; Fu, C. miR5298b regulated taxol biosynthesis by acting on TcNPR3, resulting in an alleviation of the strong inhibition of the TcNPR3-TcTGA6 complex in Taxus chinensis. Int. J. Biol. Macromol. 2023, 248, 125909. [Google Scholar] [CrossRef]
- Yu, C.; Luo, X.; Zhang, C.; Xu, X.; Huang, J.; Chen, Y.; Feng, S.; Zhan, X.; Zhang, L.; Yuan, H.; et al. Tissue-specific study across the stem of Taxus media identifies a phloem-specific TmMYB3 involved in the transcriptional regulation of paclitaxel biosynthesis. Plant J. 2020, 103, 95–110. [Google Scholar] [CrossRef]
- Khan, Y.; Xiong, Z.; Zhang, H.; Liu, S.; Yaseen, T.; Hui, T. Expression and roles of GRAS gene family in plant growth, signal transduction, biotic and abiotic stress resistance and symbiosis formation—A review. Plant Biol. 2021, 24, 404–416. [Google Scholar] [CrossRef] [PubMed]
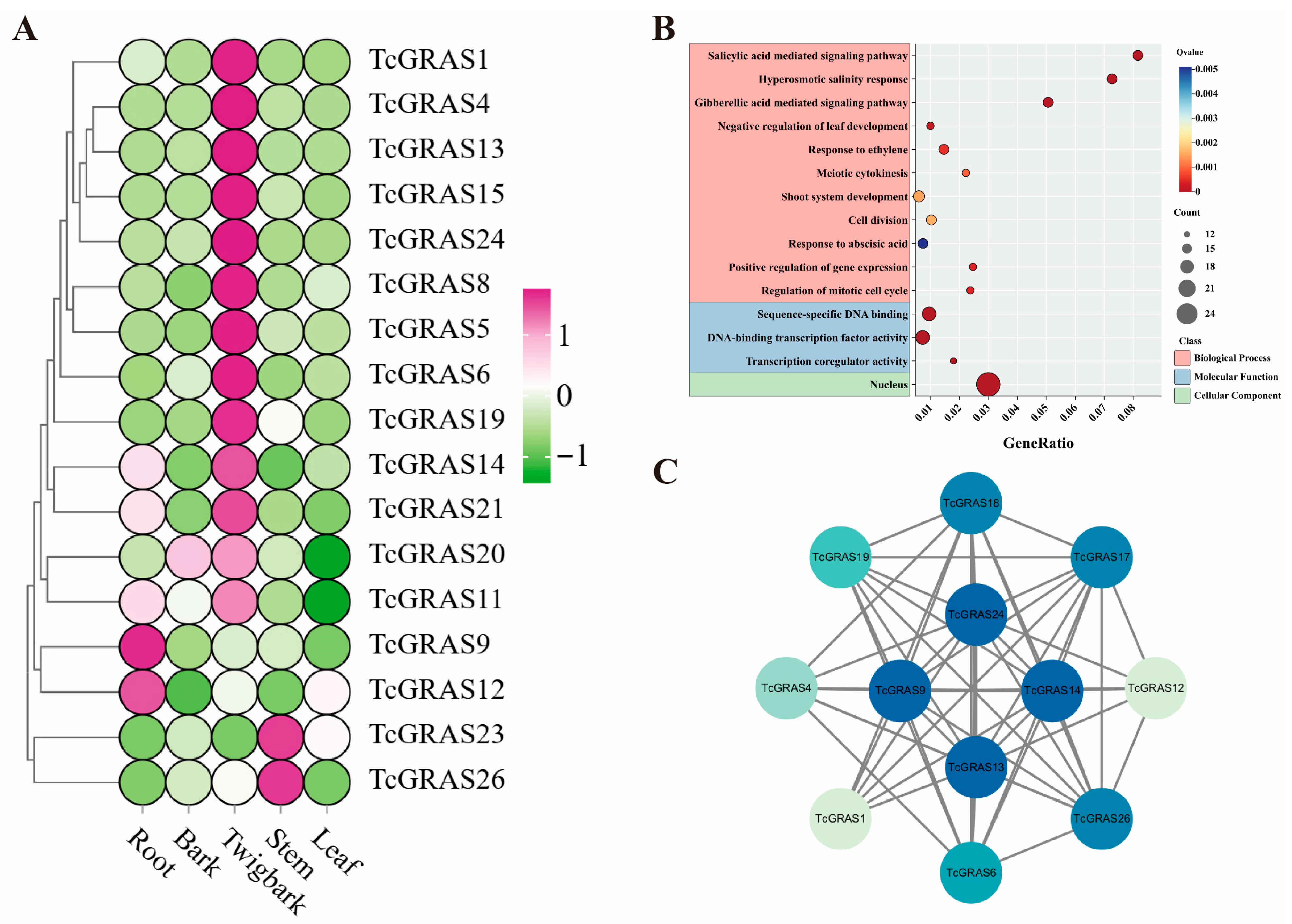
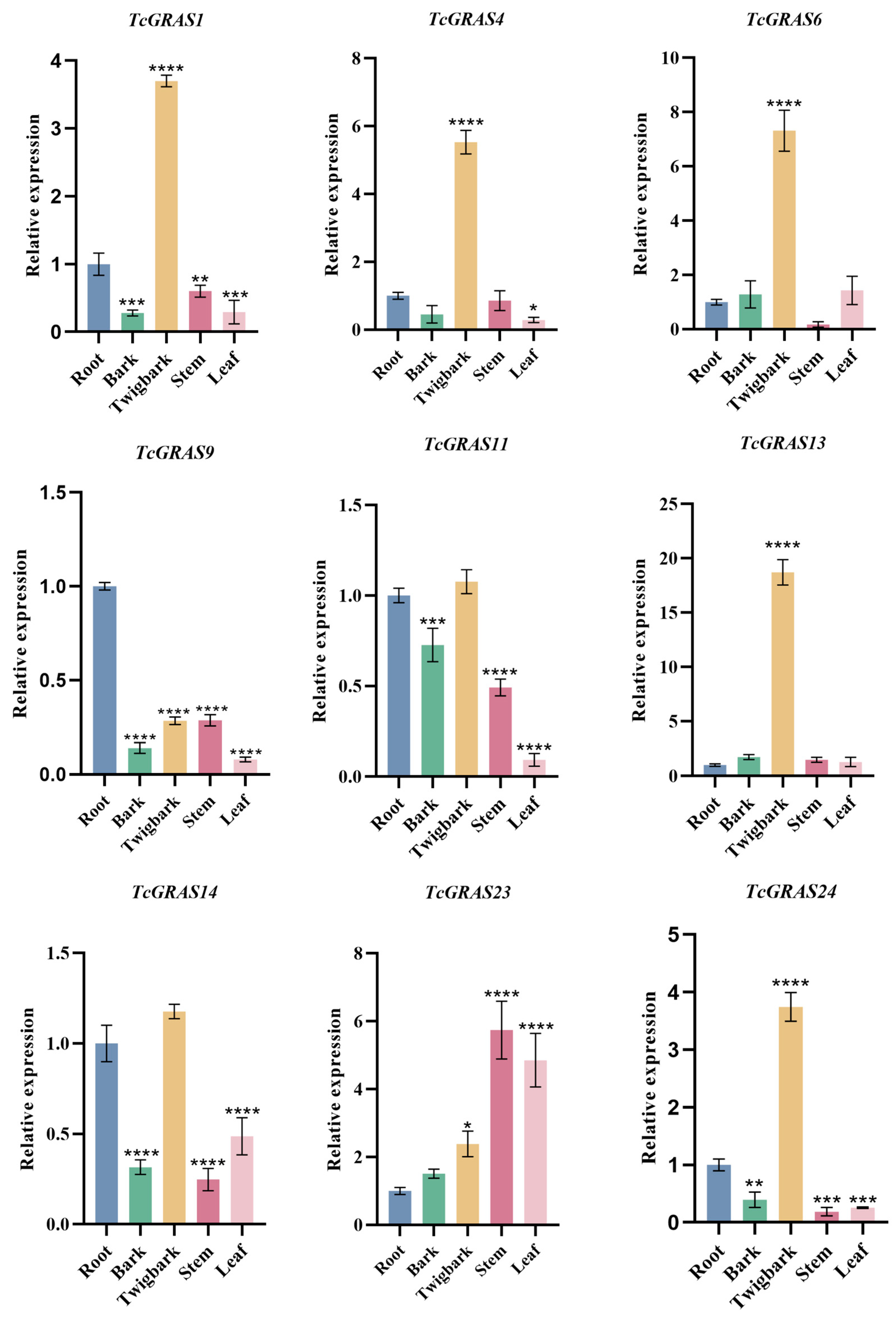
| Sequence ID | Number of Amino Acids | Molecular Weight | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity |
|---|---|---|---|---|---|---|
| TcGRAS1 | 647 | 71,235.42 | 5.81 | 58.8 | 77.68 | −0.378 |
| TcGRAS2 | 177 | 19,725.61 | 5.15 | 34.31 | 87.01 | 0.081 |
| TcGRAS3 | 261 | 28,649.41 | 4.73 | 40.51 | 95.98 | −0.067 |
| TcGRAS4 | 612 | 67,175.21 | 5.95 | 58.12 | 77.52 | −0.468 |
| TcGRAS5 | 596 | 63,669.06 | 5.62 | 49.64 | 68.15 | −0.552 |
| TcGRAS6 | 515 | 57,609.72 | 4.61 | 39.35 | 80.1 | −0.33 |
| TcGRAS7 | 561 | 63,439.37 | 4.56 | 46.86 | 77.02 | −0.361 |
| TcGRAS8 | 280 | 31,481.6 | 7.17 | 51.79 | 111.43 | 0.026 |
| TcGRAS9 | 745 | 81,673.17 | 5.33 | 47.87 | 74.07 | −0.306 |
| TcGRAS10 | 156 | 17,703 | 5.96 | 51.48 | 86.22 | −0.212 |
| TcGRAS11 | 605 | 67,384.64 | 4.73 | 49.58 | 84.99 | −0.375 |
| TcGRAS12 | 557 | 62,434.98 | 5.47 | 52.91 | 81.83 | −0.273 |
| TcGRAS13 | 592 | 65,868.94 | 5.34 | 37.26 | 76.49 | −0.507 |
| TcGRAS14 | 385 | 43,992.3 | 5.53 | 42.25 | 81.06 | −0.386 |
| TcGRAS15 | 732 | 82,508.3 | 5.81 | 52.95 | 87.68 | −0.243 |
| TcGRAS16 | 477 | 54,182.12 | 5.78 | 37.72 | 89.37 | −0.129 |
| TcGRAS17 | 251 | 28,369.17 | 5.42 | 41.07 | 86.73 | −0.223 |
| TcGRAS18 | 230 | 26,586.59 | 6.67 | 53.56 | 90.26 | −0.239 |
| TcGRAS19 | 556 | 62,685.07 | 6.25 | 41.16 | 90.29 | −0.036 |
| TcGRAS20 | 499 | 56,327.17 | 4.97 | 45.77 | 87.33 | −0.24 |
| TcGRAS21 | 453 | 50,918.65 | 4.74 | 51.84 | 80.55 | −0.189 |
| TcGRAS22 | 152 | 17,072.55 | 5.93 | 48.32 | 95.53 | −0.122 |
| TcGRAS23 | 853 | 92,140.23 | 6 | 45.62 | 75.77 | −0.446 |
| TcGRAS24 | 629 | 69,349.67 | 5.54 | 48.57 | 73.66 | −0.372 |
| TcGRAS25 | 132 | 14,348.34 | 4.23 | 63.13 | 110.76 | 0.168 |
| TcGRAS26 | 449 | 50,692.78 | 5.23 | 41.03 | 89.53 | −0.125 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, L.; Shi, A.; Wang, M.; Tian, H.; Zhang, Y. Genome-Wide Identification and Expression Profiling of the GRAS Gene Family in Taxus cuspidate. Genes 2025, 16, 1345. https://doi.org/10.3390/genes16111345
Gao L, Shi A, Wang M, Tian H, Zhang Y. Genome-Wide Identification and Expression Profiling of the GRAS Gene Family in Taxus cuspidate. Genes. 2025; 16(11):1345. https://doi.org/10.3390/genes16111345
Chicago/Turabian StyleGao, Li, Aokun Shi, Mian Wang, Hui Tian, and Yanwen Zhang. 2025. "Genome-Wide Identification and Expression Profiling of the GRAS Gene Family in Taxus cuspidate" Genes 16, no. 11: 1345. https://doi.org/10.3390/genes16111345
APA StyleGao, L., Shi, A., Wang, M., Tian, H., & Zhang, Y. (2025). Genome-Wide Identification and Expression Profiling of the GRAS Gene Family in Taxus cuspidate. Genes, 16(11), 1345. https://doi.org/10.3390/genes16111345




