Genome-Wide Identification of Juglans regia GABA Transcription Factors and Expression Pattern Analysis in Response to Abiotic Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Identification and Analysis of J. regia GABA Branch Gene Family Members
2.3. Phylogenetic Study of the J. regia Genes Encoding GABA
2.4. Structure Analysis of J. regia GABA Branch Gene Family
2.5. Genomic Mapping and Synteny Assessment
2.6. Cis-Regulatory Element Survey in Promoters
2.7. Gene Expression Analysis
3. Results
3.1. Physicochemical Characterization of the GABA Pathway Gene Family in J. regia
3.2. Protein Secondary Structure Prediction for J. regia GABA Receptor Gene Family
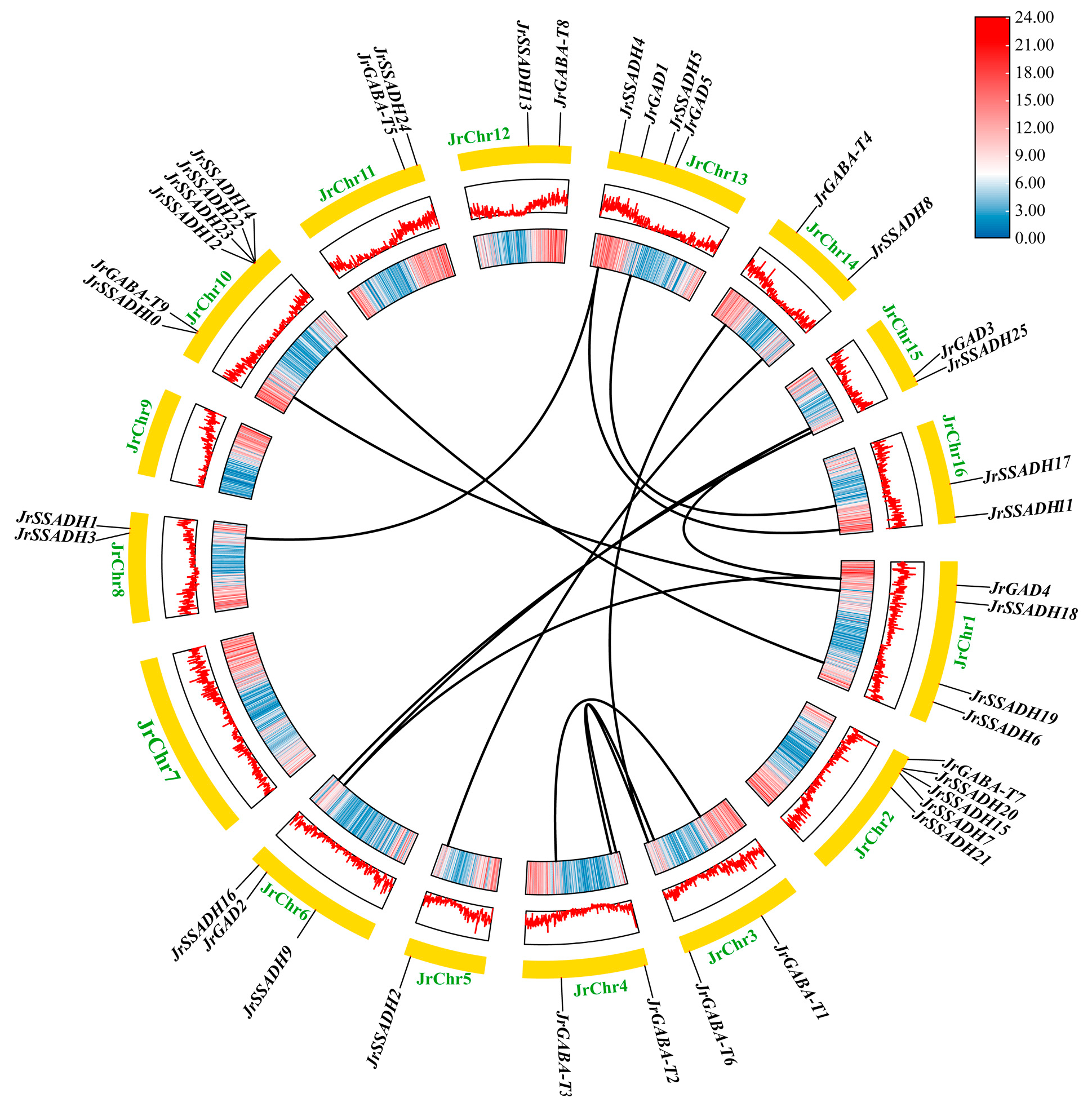
3.3. Genetic Mapping of J. regia GABA-Related Genes
3.4. Interspecific and Intraspecific Collinearity Analysis of J. regia GABA Branch Gene Family
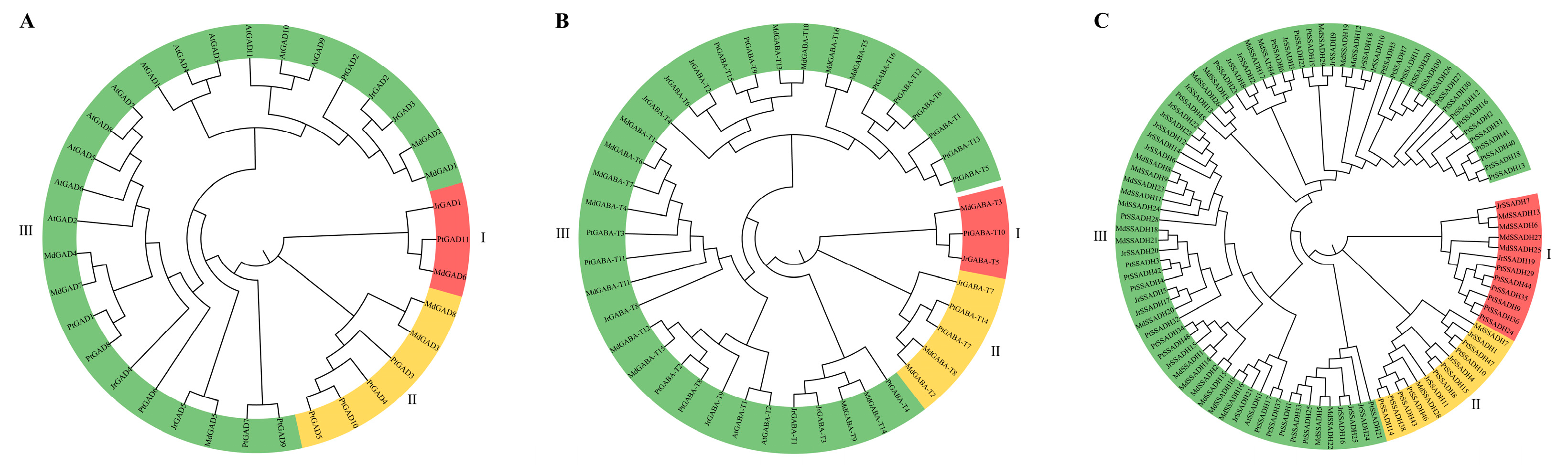
3.5. Evolutionary Analysis of J. regia GABA Branch Gene Family
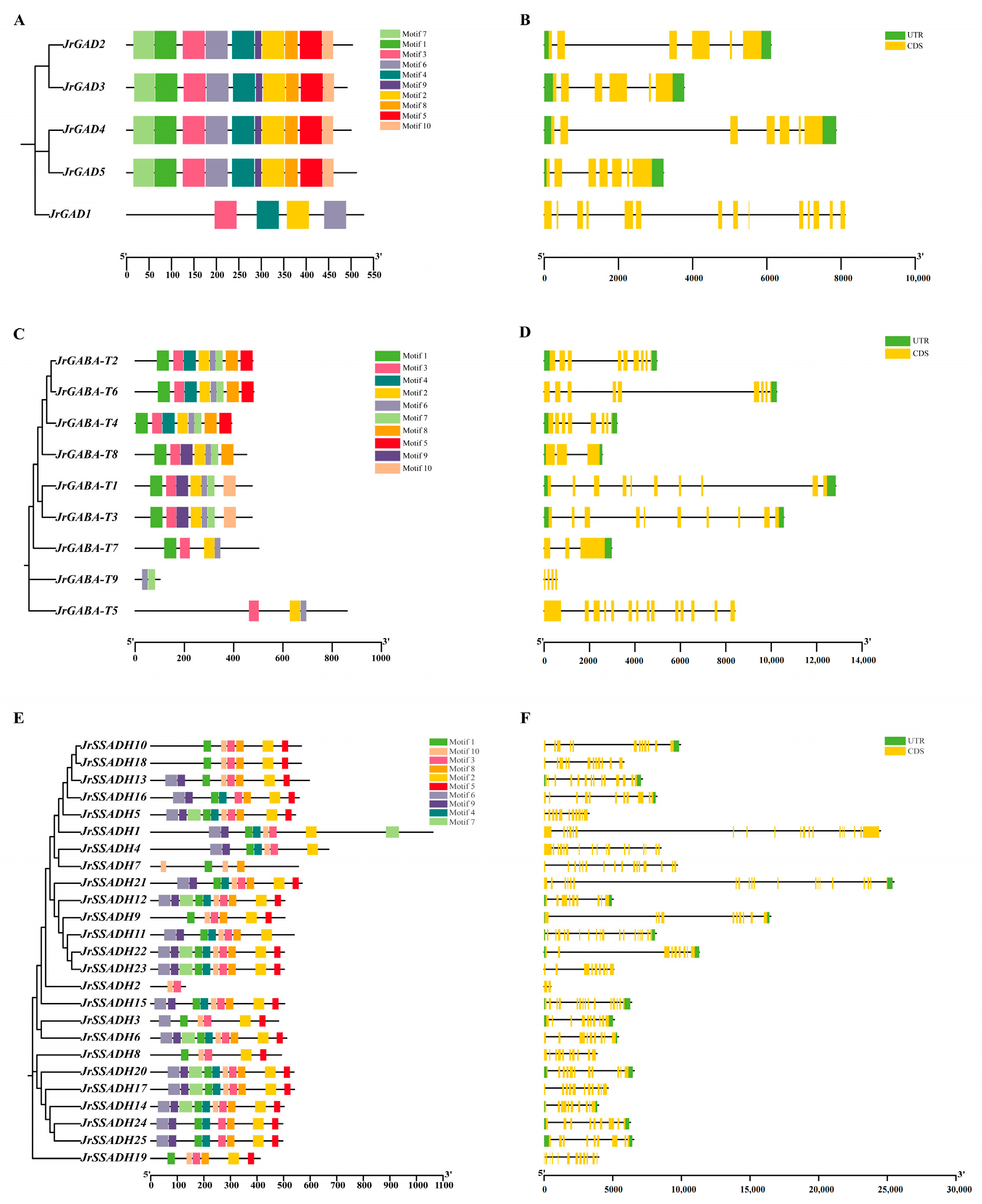
3.6. Analysis of Conserved Motifs and Gene Architecture in J. regia GABA Genes
3.7. Promoter Cis-Acting Element Analysis of GABA Branch Gene Family in J. regia
3.8. Expression Profiling of J. regia GABA Pathway Genes During Abiotic Stress
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Hussain, M.; Farooq, S.; Hasan, W.; Ul-Allah, S.; Tanveer, M.; Farooq, M.; Nawaz, A. Drought stress in sunflower: Physiological effects and its management through breeding and agronomic alternatives. Agric. Water Manag. 2018, 201, 152–166. [Google Scholar] [CrossRef]
- Fu, Z.W.; Feng, Y.R.; Gao, X.; Ding, F.; Li, J.H.; Yuan, T.T.; Lu, Y.T. Salt stress-induced chloroplastic hydrogen peroxide stimulates pdTPI sulfenylation and methylglyoxal accumulation. Plant Cell 2023, 35, 1593–1616. [Google Scholar] [CrossRef] [PubMed]
- Jan, R.; Asaf, S.; Numan, M.; Lubna; Kim, K.-M. Plant secondary metabolite biosynthesis and transcriptional regulation in response to biotic and abiotic stress conditions. Agronomy 2021, 11, 968. [Google Scholar] [CrossRef]
- Gabira, M.M.; Bergeron, Y.; Duarte, M.M.; Aguiar, N.S.d.; Kratz, D.; Silva, M.R.d.; Wendling, I.; Girona, M.M. Morphological, physiological, and biochemical responses of yerba mate (Ilex paraguariensis) genotypes to water deficit. New For. 2024, 55, 1771–1785. [Google Scholar] [CrossRef]
- Wang, K.; Yang, M.; Wu, S.; Liu, Q.; Cao, S.; Chen, W.; Shi, L. Identification of laccase gene family members in peach and its relationship with chilling induced browning. Chin. J. Biotechnol. 2022, 38, 264–274. (In Chinese) [Google Scholar] [CrossRef]
- Hu, Y.; Huang, X.; Xiao, Q.; Wu, X.; Tian, Q.; Ma, W.; Shoaib, N.; Liu, Y.; Zhao, H.; Feng, Z.; et al. Advances in plant GABA research: Biological functions, synthesis mechanisms and regulatory pathways. Plants 2024, 13, 2891. [Google Scholar] [CrossRef]
- Zhang, Y.Y. γ-Aminobutyric acid (GABA) in Fresh-Cut fruits and Vegetables. Master’s Thesis, Cornell University, Ithaca, NY, USA, 2016. [Google Scholar] [CrossRef]
- Ramesh, S.A.; Tyerman, S.D.; Gilliham, M.; Xu, B. γ-aminobutyric acid (GABA) signalling in plants. Cell. Mol. Life Sci. 2017, 74, 1577–1603. [Google Scholar] [CrossRef]
- Ramos-Ruiz, R.; Martinez, F.; Knauf-Beiter, G. The effects of GABA in plants. Cogent Food Agric. 2019, 5, 1670553. [Google Scholar] [CrossRef]
- Hayat, F.; Khan, U.; Li, J.; Ahmed, N.; Khanum, F.; Iqbal, S.; Altaf, M.A.; Ahmad, J.; Javed, H.U.; Peng, Y.; et al. γ aminobutyric acid (GABA): A key player in alleviating abiotic stress resistance in horticultural crops: Current insights and future directions. Horticulturae 2023, 9, 647. [Google Scholar] [CrossRef]
- Ahmad, S.; Fariduddin, Q. Deciphering the enigmatic role of gamma-aminobutyric acid (GABA) in plants: Synthesis, transport, regulation, signaling, and biological roles in interaction with growth regulators and abiotic stresses. Plant Physiol. Biochem. 2024, 208, 108502. [Google Scholar] [CrossRef]
- Jiang, X.; Xu, Q.; Zhang, A.; Liu, Y.; Zhao, L.; Gu, L.; Yuan, J.; Jia, H.; Shen, X.; Li, Z.; et al. Optimization of γ-aminobutyric acid (GABA) accumulation in germinating adzuki beans (Vigna angularis) by vacuum treatment and monosodium glutamate, and the molecular mechanisms. Front. Nutr. 2021, 8, 693862. [Google Scholar] [CrossRef]
- Bown, A.W.; Shelp, B.J. Does the GABA shunt regulate cytosolic GABA? Trends Plant Sci. 2020, 25, 422–424. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Cheng, C.; Fang, W. Effects of the inhibitor of glutamate decarboxylase on the development and GABA accumulation in germinating fava beans under hypoxia-NaCl stress. RSC Adv. 2018, 8, 20456–20461. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Hassan, M.J.; Peng, Y.; Liu, L.; Liu, W.; Zhang, Y.; Li, Z. γ-aminobutyric acid (GABA) priming improves seed germination and seedling stress tolerance associated with enhanced antioxidant metabolism, DREB expression, and dehydrin accumulation in white clover under water stress. Front. Plant Sci. 2021, 12, 776939. [Google Scholar] [CrossRef]
- AL-Quraan, N.A.; Sartawe, F.A.; Qaryouti, M.M. Characterization of γ-aminobutyric acid metabolism and oxidative damage in wheat (Triticum aestivum L.) seedlings under salt and osmotic stress. J. Plant Physiol. 2013, 170, 1003–1009. [Google Scholar] [CrossRef] [PubMed]
- Akçay, N.; Bor, M.; Karabudak, T.; Özdemir, F.; Türkan, İ. Contribution of gamma amino butyric acid (GABA) to salt stress responses of Nicotiana sylvestris CMSII mutant and wild type plants. J. Plant Physiol. 2012, 169, 452–458. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, Y.J.; Jeong, D.Y.; Sathiyaraj, G.; Pulla, R.K.; Shim, J.S.; In, J.G.; Yang, D.C. Isolation and characterization of a glutamate decarboxylase (GAD) gene and their differential expression in response to abiotic stresses from Panax ginseng C. A. Meyer. Mol. Biol. Rep. 2010, 37, 3455–3463. [Google Scholar] [CrossRef]
- Carillo, P. GABA shunt in durum Wheat. Front. Plant Sci. 2018, 9, 100. [Google Scholar] [CrossRef]
- Jalil, S.U.; Ahmad, I.; Ansari, M.I. Functional loss of GABA transaminase (GABA-T) expressed early leaf senescence under various stress conditions in Arabidopsis thaliana. Curr. Plant Biol. 2017, 9, 11–22. [Google Scholar] [CrossRef]
- Renault, H.; Roussel, V.; El Amrani, A.; Arzel, M.; Renault, D.; Bouchereau, A.; Deleu, C. The Arabidopsis pop2-1 mutant reveals the involvement of GABA transaminase in salt stress tolerance. BMC Plant Biol. 2010, 10, 20. [Google Scholar] [CrossRef]
- Menduti, G.; Vitaliti, A.; Capo, C.R.; Lettieri-Barbato, D.; Aquilano, K.; Malaspina, P.; Rossi, L. SSADH variants increase susceptibility of U87 cells to mitochondrial pro-oxidant insult. Int. J. Mol. Sci. 2020, 21, 4374. [Google Scholar] [CrossRef] [PubMed]
- Bouche, N. GABA signaling: A conserved and ubiquitous mechanism. Trends Cell Biol. 2003, 13, 607–610. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wang, P.; Chen, Z.; Gu, Z.; Yang, R. GABA enhances physio-biochemical metabolism and antioxidant capacity of germinated hulless barley under NaCl stress. J. Plant Physiol. 2018, 231, 192–201. [Google Scholar] [CrossRef]
- AL-Quraan, N.A.; Samarah, N.H.; AL-Fawaz, A.M. The physiological effect of GABA priming on pepper (Capsicum annuum L.) during seed germination under salt stress. Plant Growth Regul. 2024, 104, 953–974. [Google Scholar] [CrossRef]
- Rezaei-Chiyaneh, E.; Seyyedi, S.M.; Ebrahimian, E.; Moghaddam, S.S.; Damalas, C.A. Exogenous application of gamma-aminobutyric acid (GABA) alleviates the effect of water deficit stress in black cumin (Nigella sativa L.). Ind. Crops Prod. 2018, 112, 741–748. [Google Scholar] [CrossRef]
- Guo, W.; Chen, J.; Li, J.; Huang, J.; Wang, Z.; Lim, K.J. Portal of Juglandaceae: A comprehensive platform for Juglandaceae study. Hortic. Res. 2020, 7, 35. [Google Scholar] [CrossRef]
- Keller, I.; Rodrigues, C.M.; Neuhaus, H.E.; Pommerrenig, B. Improved resource allocation and stabilization of yield under abiotic stress. J. Plant Physiol. 2021, 257, 153336. [Google Scholar] [CrossRef]
- Jiang, Z.; Van Zanten, M.; Sasidharan, R. Mechanisms of plant acclimation to multiple abiotic stresses. Commun. Biol. 2025, 8, 655. [Google Scholar] [CrossRef]
- Abdullah, W.; Wani, K.I.; Naeem, M.; Aftab, T. From neurotransmitter to plant protector: The intricate world of GABA signaling and its diverse functions in stress mitigation. J. Plant Growth Regul. 2025, 44, 403–418. [Google Scholar] [CrossRef]
- Zheng, J.; Zhang, Z.; Zhang, N.; Liang, Y.; Gong, Z.; Wang, J.; Ditta, A.; Sang, Z.; Wang, J.; Li, X. Identification and function analysis of GABA branch three gene families in the cotton related to abiotic stresses. BMC Plant Biol. 2024, 24, 57. [Google Scholar] [CrossRef]
- Zheng, Q.; Su, S.; Wang, Z.; Wang, Y.; Xu, X. Comprehensive genome-wide identification and transcript profiling of GABA pathway gene family in apple (Malus domestica). Genes 2021, 12, 1973. [Google Scholar] [CrossRef]
- Chen, W.; Cheng, T.L.; Ji, J.; Wu, Y.Y.; Xie, T.T.; Jiang, Z.P. Identification of three gene families in the GABA shunt and their expression analysis in poplar. J. Nanjing For. Univ./Nat. Sci. Ed. 2020, 44, 67–77. (In Chinese) [Google Scholar] [CrossRef]
- Dove, A. Arabidopsis database. Nat. Biotechnol. 2000, 18, 701. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular evolutionary genetic analysis version 12 for adaptive and green computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Wang, X.; Sun, Y.; Joseph, P.V.; Paterson, A.H. Detection of colinear blocks and synteny and evolutionary analyses based on utilization of MCScanX. Nat. Protoc. 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhou, H.; Xiong, C.; Peng, Z.; Du, W.; Li, H.; Wang, L.; Ruan, C. Genome-wide analysis R2R3-MYB transcription factors in Xanthoceras sorbifolium Bunge and functional analysis of XsMYB30 in drought and salt stresses tolerance. Ind. Crop. Prod. 2022, 178, 114597. [Google Scholar] [CrossRef]
- Jiao, W.H.; Wei, Q.L.; Chen, J.H.; Cao, S.F.; Luo, M.; Qian, Y.; Chen, Y.; Wei, Y.Y.; Shao, X.F.; Xu, F. Identification analysis of GAD gene family, and the role of BoGAD5 in GABA enrichment in broccoli sprouts. Plant Growth Regul. 2024, 104, 1643–1655. [Google Scholar] [CrossRef]
- Benidickson, K.H.; Raytek, L.M.; Hoover, G.J.; Flaherty, E.J.; Shelp, B.J.; Snedden, W.A.; Plaxton, W.C. Glutamate decarboxylase-1 is essential for efficient acclimation of Arabidopsis thaliana to nutritional phosphorus deprivation. New Phytol. 2023, 240, 2372–2385. [Google Scholar] [CrossRef]
- Shimajiri, Y.; Ozaki, K.; Kainou, K.; Akama, K. Differential subcellular localization, enzymatic properties and expression patterns of γ-aminobutyric acid transaminases (GABA-ts) in rice (Oryza sativa). J. Plant Physiol. 2013, 170, 196–201. [Google Scholar] [CrossRef]
- Faës, P.; Niogret, M.F.; Montes, E.; Cahérec, F.L.; Bouchereau, A.; Deleu, C. Transcriptional profiling of genes encoding GABA-transaminases in Brassica napus reveals their regulation by water deficit. Environ. Exp. Bot. 2015, 116, 20–31. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, Z.; Fan, Y.; Liu, C.; Wang, H.; Li, Y.; Xin, Y.; Gai, Y.; Ji, X. Characterization of GABA-transaminase gene from mulberry (Morus multicaulis) and its role in salt stress tolerance. Genes 2022, 13, 501. [Google Scholar] [CrossRef]
- Guo, X.; Yang, F.; Zhang, X.; Tang, M.; Wan, K.; Lai, C.; Lai, Z.; Lin, Y. Genome-wide identification and expression analysis unveil the involvement of the succinic semialdehyde dehydrogenase (SSADH) gene family in banana low temperature stress. Int. J. Mol. Sci. 2025, 26, 3006. [Google Scholar] [CrossRef]
- Guo, Y.; Bao, Z.; Deng, Y.; Li, Y.; Wang, P. Protein subcellular localization and functional studies in horticultural research: Problems, solutions, and new approaches. Hortic. Res. 2023, 10, uhac271. [Google Scholar] [CrossRef]
- Zhang, Z.; Kumar, V.; Dybkov, O.; Will, C.L.; Zhong, J.Y.; Ludwig, S.E.J.; Urlaub, H.; Kastner, B.; Stark, H.; Lührmann, R. Structural insights into the cross-exon to cross-intron spliceosome switch. Nature 2024, 630, 1012–1019. [Google Scholar] [CrossRef] [PubMed]
- Tajo, S.M.; Pan, Z.; Jia, Y.; He, S.; Chen, B.; Sadau, S.B.; Km, Y.; Ajadi, A.A.; Nazir, M.F.; Auta, U.; et al. Silencing of GhORP_A02 enhances drought tolerance in Gossypium hirsutum. BMC Genomics 2023, 24, 7. [Google Scholar] [CrossRef] [PubMed]
- Gut, H.; Dominici, P.; Pilati, S.; Astegno, A.; Petoukhov, M.V.; Svergun, D.I.; Grütter, M.G.; Capitani, G. A common structural basis for pH- and calmodulin-mediated regulation in plant glutamate decarboxylase. J. Mol. Biol. 2009, 392, 334–351. [Google Scholar] [CrossRef]
- Shahjahan, M.; Rahman, S. Insilico analysis of γ-aminobutyric acid transaminase (GABA-T) of Brassica napus (Rape). J. Adv. Biotechnol. Exp. Ther. 2019, 2, 4–9. [Google Scholar] [CrossRef]
- Song, X.; Wang, L.Y.; Fu, B.X.; Li, S.D.; Wei, Y.Y.; Hong, Y.; Dai, S.L. Advances in identification and synthesis of promoter elements in higher plants. Chin. J. Bot. 2024, 59, 691–708. (In Chinese) [Google Scholar] [CrossRef]
- Hong, L.; Liu, X.; Li, L. Plant AREB/ABF transcription factors and their involvement in ABA signal transduction. Plant Physiol. J. 2011, 47, 211–217. (In Chinese) [Google Scholar] [CrossRef]
- Huang, H.; He, Y.; Cui, A.; Sun, L.; Han, M.; Wang, J.; Rui, C.; Lei, Y.; Liu, X.; Xu, N.; et al. Genome-wide identification of GAD family genes suggests GhGAD6 functionally respond to Cd2+ stress in cotton. Front. Genet. 2022, 13, 965058. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.; Wang, J.; Ma, F.; Wang, L.; Zhan, X.; Li, G.; Hu, S.; Khan, A.; Dang, H.; et al. Promoting γ-aminobutyric acid accumulation to enhances saline-alkali tolerance in tomato. Plant Physiol. 2024, 196, 2089–2104. [Google Scholar] [CrossRef]




| Gene ID | Gene Name | Number of Amino Acid | Molecular Weight | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Predicted Location(s) |
|---|---|---|---|---|---|---|---|---|
| LOC109003131 | JrGAD1 | 527 | 57,972.18 | 8.29 | 41.26 | 95.43 | −0.02 | plas |
| LOC109012088 | JrGAD2 | 502 | 56,537.07 | 5.91 | 37.29 | 89.08 | −0.172 | cysk |
| LOC108993168 | JrGAD3 | 490 | 55,133.44 | 5.78 | 40.77 | 89.33 | −0.167 | cysk |
| LOC108988130 | JrGAD4 | 499 | 56,626.09 | 5.62 | 31.54 | 90.02 | −0.23 | cyto |
| LOC108997089 | JrGAD5 | 511 | 57,674.08 | 5.59 | 35.41 | 89.45 | −0.255 | cyto |
| LOC109011059 | JrGABA-T1 | 474 | 52,156.99 | 6.65 | 36.02 | 89.32 | −0.172 | mito |
| LOC108990021 | JrGABA-T2 | 477 | 51,739.45 | 6.43 | 32.11 | 89.12 | 0.004 | pero |
| LOC108990262 | JrGABA-T3 | 474 | 52,020.95 | 6.8 | 35.87 | 91.81 | −0.155 | chlo |
| LOC108996966 | JrGABA-T4 | 391 | 42,294.75 | 6.31 | 21.71 | 92.79 | 0.099 | pero |
| LOC108998892 | JrGABA-T5 | 860 | 94,738.09 | 6.08 | 41.52 | 85.99 | −0.061 | chlo |
| LOC108985282 | JrGABA-T6 | 481 | 52,078.73 | 7.59 | 34.87 | 86.15 | −0.062 | pero |
| LOC108998090 | JrGABA-T7 | 501 | 53,932.96 | 7.23 | 39.28 | 81.06 | −0.086 | chlo |
| LOC109009895 | JrGABA-T8 | 452 | 48,609.57 | 6.43 | 38.76 | 94.76 | 0.032 | chlo |
| LOC108996782 | JrGABA-T9 | 101 | 11,061.99 | 6.8 | 41.12 | 108.02 | 0.489 | cyto |
| LOC108986827 | JrSSADH1 | 1061 | 116,054.54 | 7.29 | 51.31 | 75.57 | −0.391 | nucl |
| LOC108985392 | JrSSADH2 | 130 | 13,817.1 | 8.84 | 33.99 | 104.31 | 0.272 | chlo |
| LOC108983652 | JrSSADH3 | 480 | 53,271.88 | 8.38 | 40.77 | 101.6 | −0.009 | cysk |
| LOC108983908 | JrSSADH4 | 669 | 73,056.89 | 8.83 | 38.28 | 87.13 | −0.182 | cyto |
| LOC118344148 | JrSSADH5 | 544 | 59,635.3 | 6.3 | 29.66 | 87.72 | −0.135 | chlo |
| LOC108992687 | JrSSADH6 | 511 | 55,778.21 | 5.81 | 29.94 | 90.84 | −0.048 | cyto |
| LOC108999249 | JrSSADH7 | 555 | 61,844.14 | 6.48 | 37.56 | 89.24 | −0.151 | mito |
| LOC108992688 | JrSSADH8 | 503 | 54,693.07 | 5.57 | 30.5 | 92.15 | 0.013 | cysk |
| LOC108998675 | JrSSADH9 | 491 | 54,689.42 | 8.93 | 43.44 | 95.78 | −0.055 | cyto |
| LOC108994687 | JrSSADH10 | 504 | 54,974.99 | 7.66 | 35.06 | 104.64 | 0.122 | plas |
| LOC108983383 | JrSSADH11 | 566 | 62,522.22 | 8.11 | 37.37 | 98.89 | 0.002 | chlo |
| LOC108995743 | JrSSADH12 | 539 | 57,994.52 | 8.27 | 39.16 | 85.96 | −0.044 | mito |
| LOC109004656 | JrSSADH13 | 504 | 54,805.97 | 6.11 | 28.73 | 93.89 | −0.021 | cyto |
| LOC108994823 | JrSSADH14 | 596 | 66,033.3 | 7.18 | 38.99 | 91.12 | 0.015 | vacu |
| LOC109004652 | JrSSADH15 | 501 | 54,194.62 | 5.99 | 25.18 | 94.05 | 0.064 | cyto |
| LOC108999250 | JrSSADH16 | 503 | 54,539.68 | 5.44 | 27.77 | 93.34 | −0.01 | pero |
| LOC109012892 | JrSSADH17 | 558 | 60,823.29 | 8.05 | 45.82 | 88.75 | −0.109 | cyto |
| LOC108996365 | JrSSADH18 | 566 | 62,483.16 | 8.53 | 38.45 | 94.75 | −0.091 | chlo |
| LOC109009030 | JrSSADH19 | 411 | 44,362.14 | 6.04 | 33.54 | 92.26 | 0.106 | chlo |
| LOC108999220 | JrSSADH20 | 538 | 58,649.98 | 7.59 | 33.54 | 89.11 | −0.074 | mito |
| LOC108983566 | JrSSADH21 | 569 | 61,110.39 | 8.63 | 33.67 | 93.25 | −0.032 | mito |
| LOC109004654 | JrSSADH22 | 502 | 54,439.69 | 5.83 | 26.94 | 92.53 | 0.029 | cyto |
| LOC108997881 | JrSSADH23 | 502 | 54,422.71 | 6 | 27.59 | 93.86 | 0.027 | cyto |
| LOC109005232 | JrSSADH24 | 496 | 53,190.69 | 7.08 | 37.99 | 94.33 | 0.053 | cyto |
| LOC109000833 | JrSSADH25 | 496 | 53,276.73 | 7.47 | 39.32 | 91.41 | 0.015 | cyto |
| Gene Name | Alpha Helix (Hh) | Extended Strand (Ee) | Beta Turn (Tt) | Random Coil (Cc) |
|---|---|---|---|---|
| JrGAD1 | 47.63% | 12.90% | 4.93% | 34.54% |
| JrGAD2 | 41.43% | 13.55% | 5.58% | 39.44% |
| JrGAD3 | 40.41% | 15.92% | 6.73% | 36.94% |
| JrGAD4 | 40.48% | 14.23% | 6.01% | 39.28% |
| JrGAD5 | 38.36% | 15.07% | 6.07% | 40.51% |
| JrGABA-T1 | 38.19% | 17.09% | 10.76% | 33.97% |
| JrGABA-T2 | 40.25% | 17.61% | 7.76% | 34.38% |
| JrGABA-T3 | 38.19% | 15.82% | 9.92% | 36.08% |
| JrGABA-T4 | 41.18% | 17.90% | 8.95% | 31.97% |
| JrGABA-T5 | 39.88% | 13.14% | 5.23% | 41.74% |
| JrGABA-T6 | 42.62% | 16.22% | 8.32% | 32.85% |
| JrGABA-T7 | 40.52% | 18.36% | 9.18% | 31.94% |
| JrGABA-T8 | 40.04% | 19.47% | 9.07% | 31.42% |
| JrGABA-T9 | 41.58% | 15.84% | 13.86% | 28.71% |
| JrSSADH1 | 24.98% | 15.55% | 7.26% | 52.21% |
| JrSSADH2 | 46.92% | 18.46% | 10.00% | 24.62% |
| JrSSADH3 | 44.79% | 15.62% | 6.67% | 32.92% |
| JrSSADH4 | 36.17% | 17.34% | 7.47% | 39.01% |
| JrSSADH5 | 40.81% | 15.81% | 7.90% | 35.48% |
| JrSSADH6 | 40.12% | 18.20% | 8.02% | 33.66% |
| JrSSADH7 | 38.74% | 15.86% | 5.23% | 40.18% |
| JrSSADH8 | 44.40% | 16.90% | 7.74% | 30.96% |
| JrSSADH9 | 47.02% | 15.87% | 6.35% | 30.75% |
| JrSSADH10 | 40.46% | 18.90% | 6.36% | 34.28% |
| JrSSADH11 | 38.59% | 18.00% | 7.98% | 35.44% |
| JrSSADH12 | 40.08% | 18.06% | 9.13% | 32.74% |
| JrSSADH13 | 42.79% | 12.75% | 6.54% | 37.92% |
| JrSSADH14 | 39.52% | 18.76% | 7.78% | 33.93% |
| JrSSADH15 | 43.74% | 15.90% | 6.76% | 33.60% |
| JrSSADH16 | 42.65% | 16.67% | 6.63% | 34.05% |
| JrSSADH17 | 41.48% | 16.11% | 7.78% | 34.63% |
| JrSSADH18 | 42.93% | 15.72% | 7.07% | 34.28% |
| JrSSADH19 | 38.20% | 20.19% | 6.08% | 35.52% |
| JrSSADH20 | 41.08% | 16.91% | 8.18% | 33.83% |
| JrSSADH21 | 48.15% | 14.06% | 7.73% | 30.05% |
| JrSSADH22 | 41.04% | 17.53% | 6.97% | 34.46% |
| JrSSADH23 | 40.95% | 17.69% | 6.76% | 34.50% |
| JrSSADH24 | 40.73% | 18.55% | 6.05% | 34.68% |
| JrSSADH25 | 41.33% | 18.55% | 6.25% | 33.87% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Wang, B.; Chen, W.; Wang, B.; Li, T.; Luo, X.; Xue, J.; Wang, X.; He, J.; Wang, X. Genome-Wide Identification of Juglans regia GABA Transcription Factors and Expression Pattern Analysis in Response to Abiotic Stress. Genes 2025, 16, 1290. https://doi.org/10.3390/genes16111290
Wang Y, Wang B, Chen W, Wang B, Li T, Luo X, Xue J, Wang X, He J, Wang X. Genome-Wide Identification of Juglans regia GABA Transcription Factors and Expression Pattern Analysis in Response to Abiotic Stress. Genes. 2025; 16(11):1290. https://doi.org/10.3390/genes16111290
Chicago/Turabian StyleWang, Yulian, Bin Wang, Wei Chen, Bin Wang, Tianlei Li, Xiang Luo, Jia Xue, Xinyi Wang, Jing He, and Xiujuan Wang. 2025. "Genome-Wide Identification of Juglans regia GABA Transcription Factors and Expression Pattern Analysis in Response to Abiotic Stress" Genes 16, no. 11: 1290. https://doi.org/10.3390/genes16111290
APA StyleWang, Y., Wang, B., Chen, W., Wang, B., Li, T., Luo, X., Xue, J., Wang, X., He, J., & Wang, X. (2025). Genome-Wide Identification of Juglans regia GABA Transcription Factors and Expression Pattern Analysis in Response to Abiotic Stress. Genes, 16(11), 1290. https://doi.org/10.3390/genes16111290





