Integrated Transcriptomic and Metabolomic Analyses Reveal Key Genes Involved in Phenylpropanoid Metabolism in Lonicera macranthoides Flowers
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. RNA Extraction and Quality Control
2.3. cDNA Library Construction and Sequencing
2.4. Read Processing and De Novo Transcriptome Assembly
2.5. Functional Annotation of Genes
2.6. Expression Quantification and Differential Expression Analysis
2.7. Sample Preparation and UPLC-MS/MS Analysis
2.8. Data Preprocessing and Quality Control
2.9. Metabolite Identification and Dataset Construction
2.10. Statistical Analysis and Differential Metabolite Screening
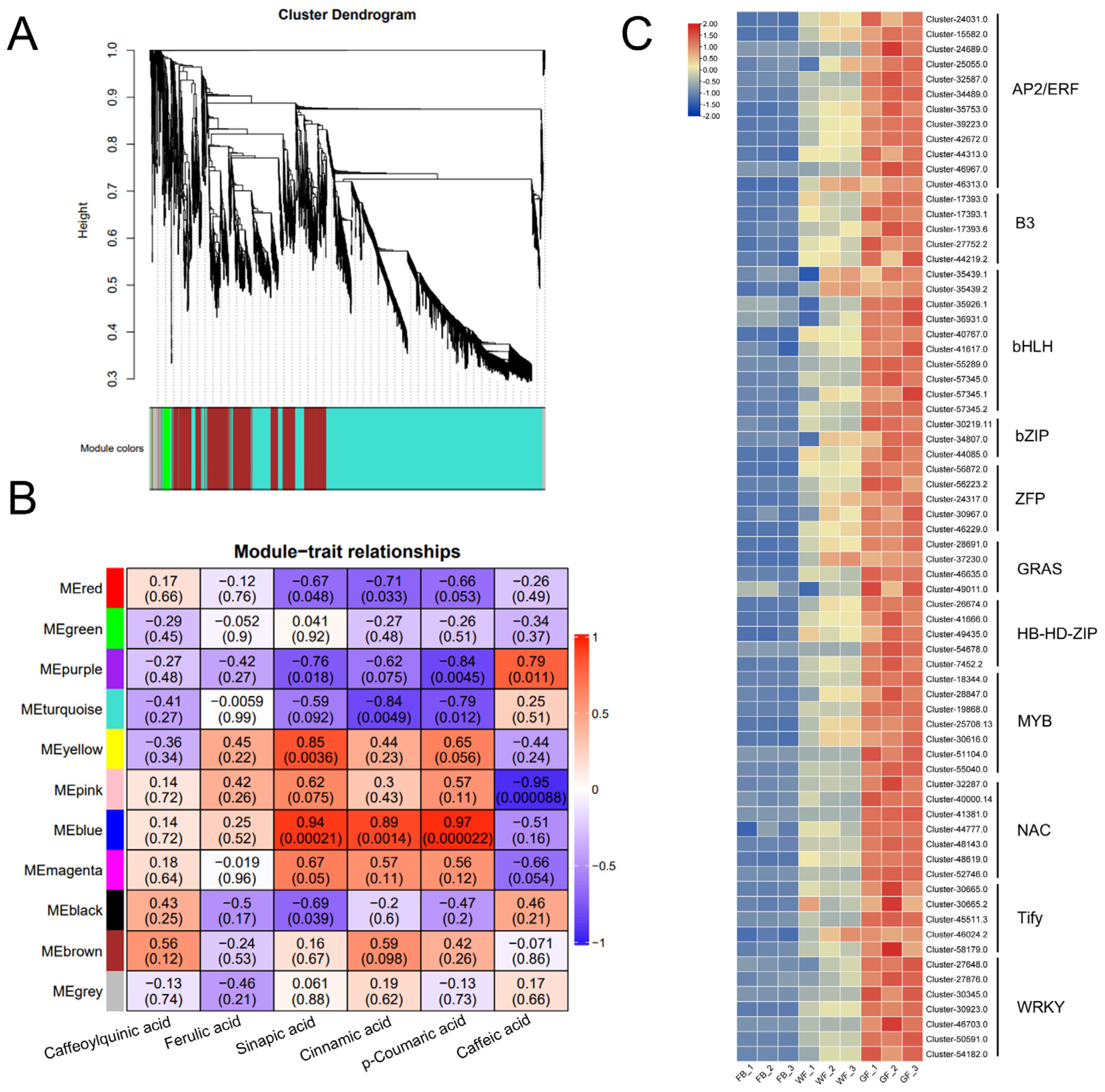
2.11. Weighted Gene Co-Expression Network Analysis
3. Results
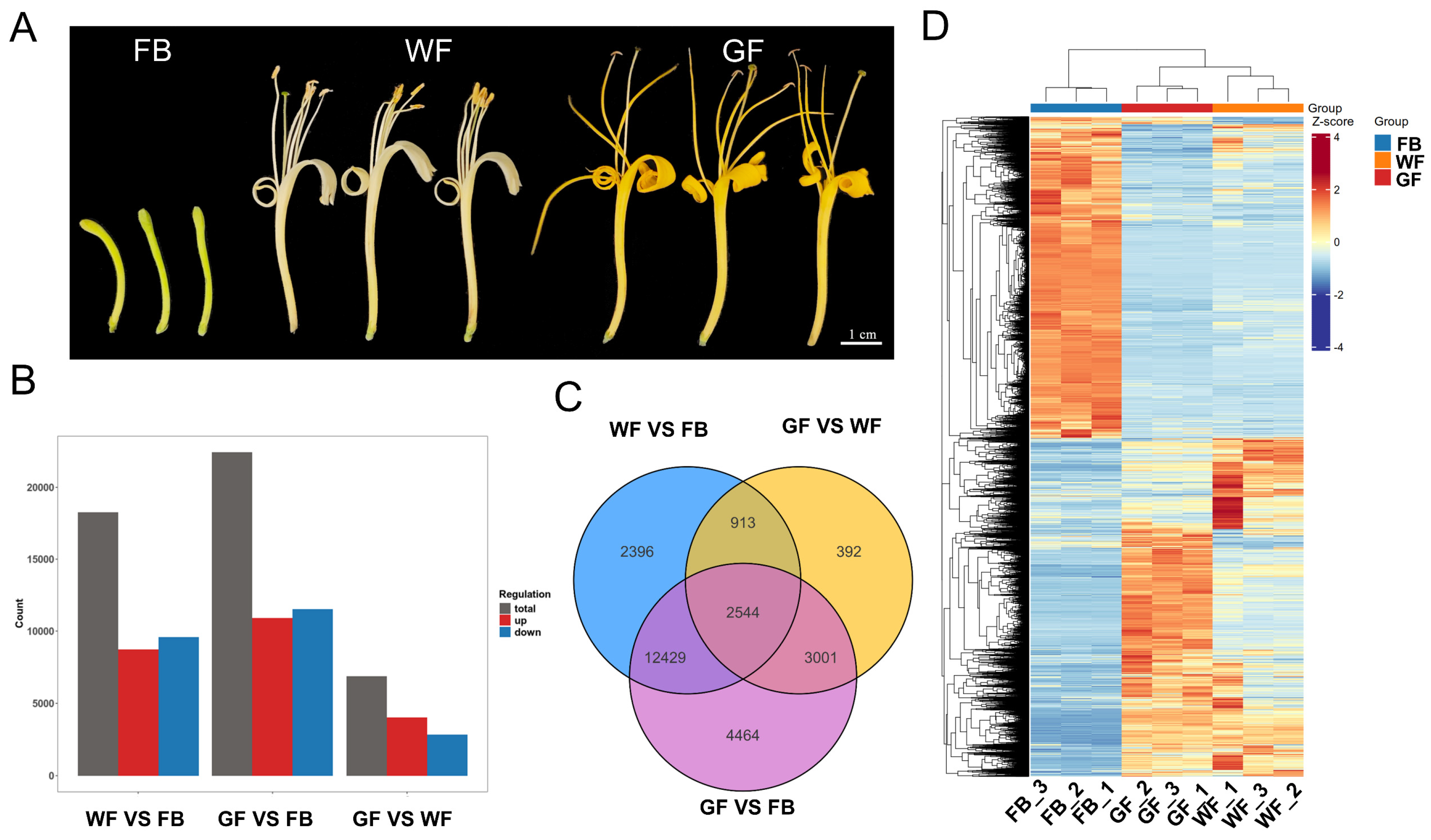
3.1. Transcriptome Analysis of Floral Developmental Stages
3.2. Functional Enrichment Analysis of DEGs at Three Floral Developmental Stages
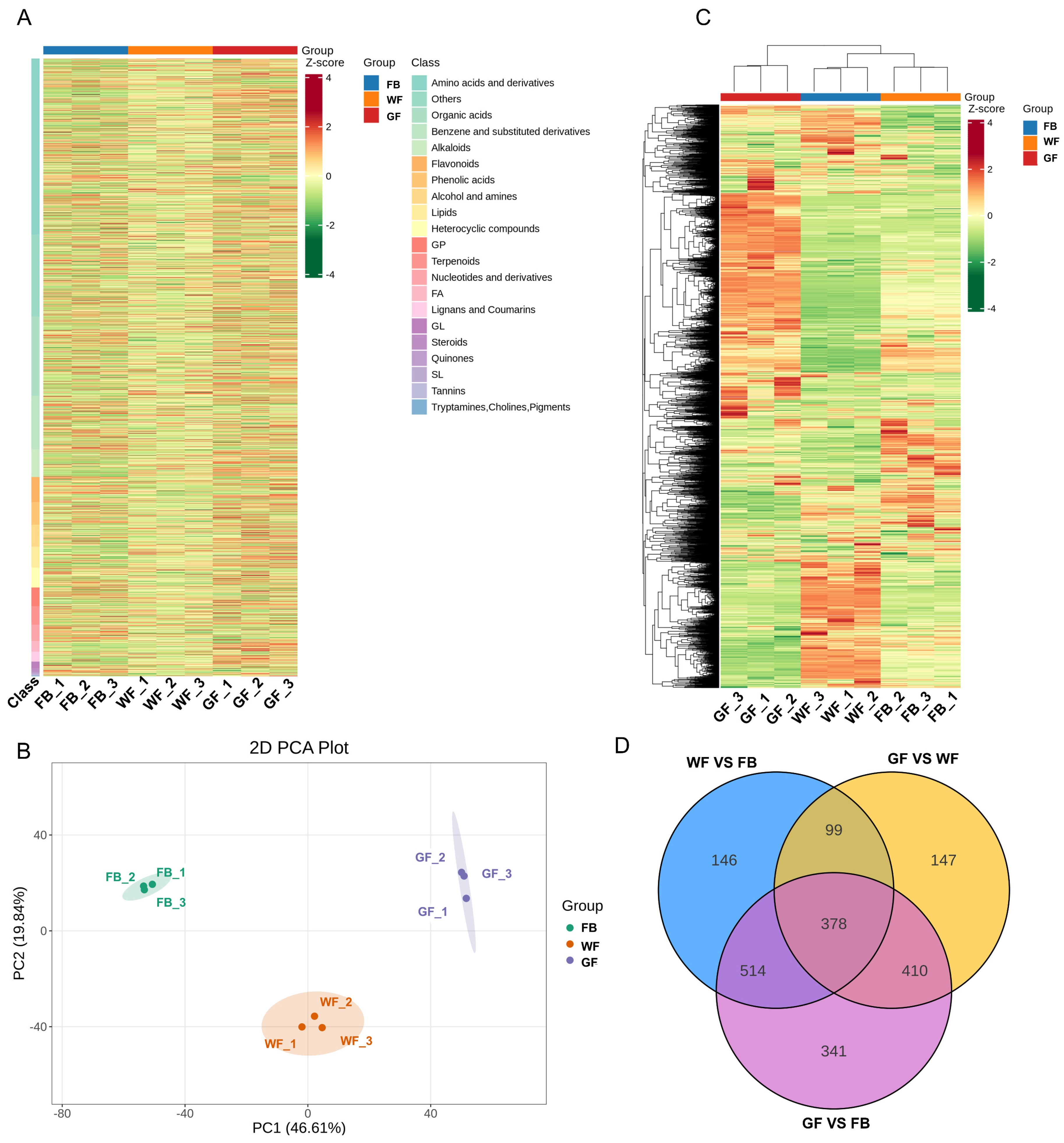
3.3. Metabolome Analysis of Three Floral Developmental Stages
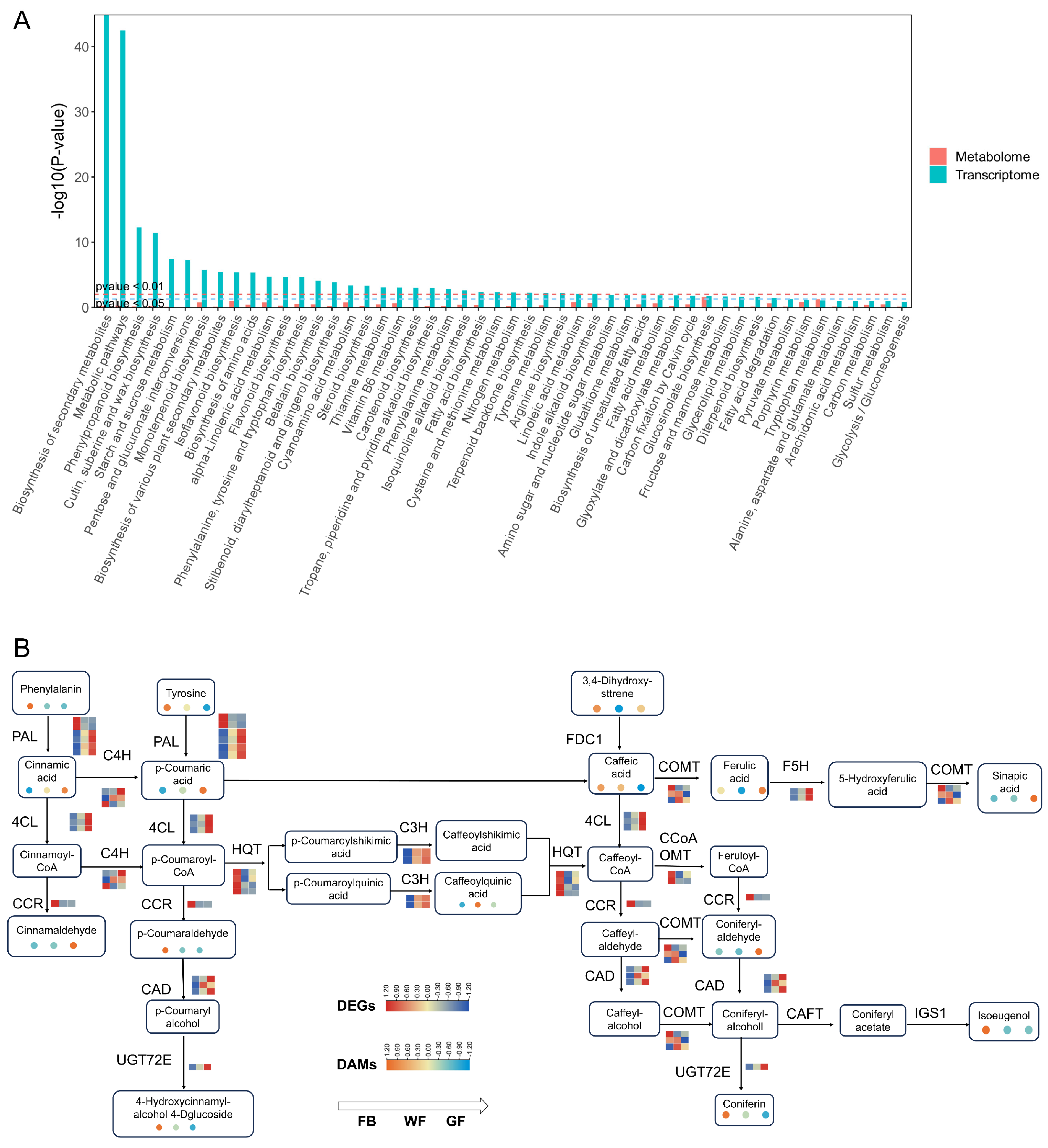
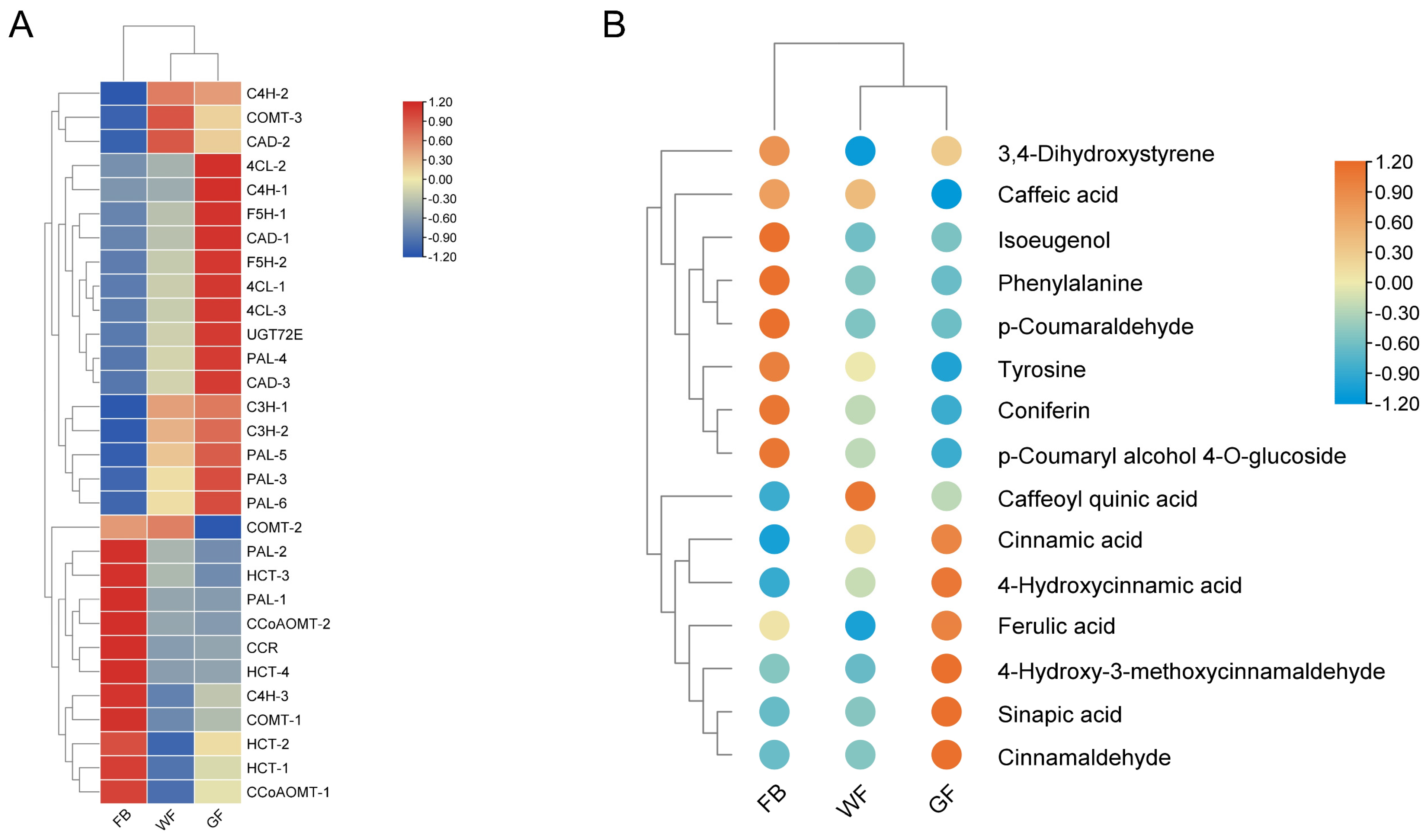
3.4. Integrated Analysis of Genes and Metabolites in the Phenylpropanoid Biosynthesis Pathway
3.5. Identification of Candidate Transcription Factors Involved in Chlorogenic Acid Biosynthesis
4. Discussion
4.1. Transcriptome and Metabolome Data Reveal Stage-Specific Molecular and Metabolic Variations in L. macranthoides Flowers
4.2. Gene–Metabolite Coordination in the Phenylpropanoid Biosynthesis Pathway
4.3. Transcription Factors Regulate CGA Biosynthesis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, Q.; Yu, H.; Dong, Y.; Quan, W.; Su, Z.; Li, L. Quality Evaluation of Lonicerae Flos Produced in Southwest China Based on HPLC Analysis and Antioxidant Activity. Molecules 2024, 29, 2560. [Google Scholar] [CrossRef]
- Lyu, H.; Liu, W.; Bai, B.; Shan, Y.; Paetz, C.; Feng, X.; Chen, Y. Prenyleudesmanes and A Hexanorlanostane from the Roots of Lonicera macranthoides. Molecules 2019, 24, 4276. [Google Scholar] [CrossRef]
- Yu, M.; Yang, L.; Xue, Q.; Yin, P.; Sun, L.; Liu, Y. Comparison of Free, Esterified, and Insoluble-Bound Phenolics and Their Bioactivities in Three Organs of Lonicera japonica and L. macranthoides. Molecules 2019, 24, 970. [Google Scholar] [CrossRef]
- Ding, J.-X.; Liu, C.; Liu, X.-W.; Yan, W.-N.; Li, W.-P.; Shi, H.; Li, J.-X.; Tang, C.-L.; Zhou, Y. Identification of Compounds with Antipyretic Effects and Anti-endotoxin Activity in Different Species of Lonicera japonica Using Spectrum-effect Correlation. Exp. Ther. Med. 2021, 22, 665. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, M.; Ahmed, K.; Yang, J.; Song, L.; Cui, Z.-G.; Hiraku, Y. Mechanistic Study of Macranthoside B Effects on Apoptotic Cell Death in Human Cervical Adenocarcinoma Cells. Folia Biol. 2022, 68, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.; Yun, S.; Yinxue, M.; Xin, Y.; Kunming, Q. Chemical Composition and Antibacterial Activity of the Essential Oil Isolated From Flos Lonicerae (Flower Buds of Lonicera macranthoides Hand.-Mazz.). Nat. Prod. Commun. 2021, 16, 4. [Google Scholar] [CrossRef]
- Zhou, R.; Tong, Q. Comparative study on content of chlorogenic acid in Lonicera japonica and L. macranthoides. Zhong Yao Cai 2003, 26, 399–400. (In Chinese) [Google Scholar] [PubMed]
- Liu, Y.; Su, W.; Wang, L.; Lei, J.; Chai, S.; Zhang, W.; Yang, X. Integrated Transcriptome, Small RNA and Degradome Sequencing Approaches Proffer Insights into Chlorogenic Acid Biosynthesis in Leafy Sweet Potato. PLoS ONE 2021, 16, e0245266. [Google Scholar] [CrossRef]
- Yang, Y.; Xu, J.; Li, Y.; He, Y.; Yang, Y.; Liu, D.; Wu, C. Effects of Coumarin on Rhizosphere Microbiome and Metabolome of Lolium multiflorum. Plants 2023, 12, 1096. [Google Scholar] [CrossRef]
- Xu, X.; Li, C.; Wu, F.; Zhao, S.; Chen, T.; You, H.; Lin, Y.; Zou, X. Integrated Transcriptomic and Targeted Metabolomic Analysis Reveals the Key Genes Involved in Triterpenoid Biosynthesis of Ganoderma lucidum. J. Fungi 2025, 11, 57. [Google Scholar] [CrossRef]
- Yang, Y.; Zhu, W.; Jin, N.; Liu, W.; Lie, Y.; Wang, L.; Jin, L.; Wang, S.; Yu, J.; Lyu, J. Exogenous Silicon Applied at Appropriate Concentrations Is Effective at Improving Tomato Nutritional and Flavor Qualities. Food Chem. X 2024, 22, 101306. [Google Scholar] [CrossRef]
- Kumar, S.; Pandey, A.K. Chemistry and Biological Activities of Flavonoids: An Overview. Sci. World J. 2013, 2013, 162750. [Google Scholar] [CrossRef] [PubMed]
- Spanò, R.; Gena, P.; Linsalata, V.; Sini, V.; D’Antuono, I.; Cardinali, A.; Cotugno, P.; Calamita, G.; Mascia, T. Spotlight on Secondary Metabolites Produced by an Early-Flowering Apulian Artichoke Ecotype Sanitized from Virus Infection by Meristem-Tip-Culture and Thermotherapy. Antioxidants 2024, 13, 852. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Kader, M.S.; Radwan, M.M.; Metwaly, A.M.; Eissa, I.H.; Hazekamp, A.; ElSohly, M.A. Chemistry and Biological Activities of Cannflavins of the Cannabis Plant. Cannabis Cannabinoid Res. 2023, 8, 974–985. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Xie, J.; Sun, M.; Xiong, S.; Xu, C.; Zhang, Z.; Li, M.; Li, C.; Lin, L. Plant-Derived Caffeic Acid and Its Derivatives: An Overview of Their NMR Data and Biosynthetic Pathways. Molecules 2024, 29, 1625. [Google Scholar] [CrossRef]
- Lu, C.; Yan, X.; Zhang, H.; Zhong, T.; Gui, A.; Liu, Y.; Pan, L.; Shao, Q. Integrated Metabolomic and Transcriptomic Analysis Reveals Biosynthesis Mechanism of Flavone and Caffeoylquinic Acid in Chrysanthemum. BMC Genom. 2024, 25, 759. [Google Scholar] [CrossRef]
- Wang, L.; Stegemann, J.P. Extraction of High Quality RNA from Polysaccharide Matrices Using Cetlytrimethylammonium Bromide. Biomaterials 2010, 31, 1612–1618. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Davidson, N.M.; Oshlack, A. Corset: Enabling Differential Gene Expression Analysis for de Novoassembled Transcriptomes. Genome Biol. 2014, 15, 410. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De Novo Transcript Sequence Reconstruction from RNA-Seq Using the Trinity Platform for Reference Generation and Analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Boccia, A. DG-CST (Disease Gene Conserved Sequence Tags), a Database of Human-Mouse Conserved Elements Associated to Disease Genes. Nucleic Acids Res. 2004, 33, D505–D510. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Kiryutin, B.; Koonin, E.V.; Krylov, D.M.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; et al. The COG Database: An Updated Version Includes Eukaryotes. BMC Bioinform. 2003, 4, 41. [Google Scholar] [CrossRef]
- Boeckmann, B. The SWISS-PROT Protein Knowledgebase and Its Supplement TrEMBL in 2003. Nucleic Acids Res. 2003, 31, 365–370. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and Sensitive Protein Alignment Using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Bateman, A. The Pfam Protein Families Database. Nucleic Acids Res. 2004, 32, D138–D141. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and Quantifying Mammalian Transcriptomes by RNA-Seq. Nat Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.L.; Li, L.Y.; Pi, J.H. Carotenoid Metabolomic and Transcriptomic Analyses Provide Insights into the Flower Color Transition in Lonicera macranthoides. BMC Biotechnol 2025, 25, 69. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.L.; Li, L.Y.; Xiao, L.Q.; Pi, J.H. Transcriptomic and Targeted Metabolomic Analyses Provide Insights into the Flavonoids Biosynthesis in the Flowers of Lonicera macranthoides. BMC Biotechnol 2024, 24, 19. [Google Scholar] [CrossRef]
- Liu, Z.; Mohsin, A.; Wang, Z.; Zhu, X.; Zhuang, Y.; Cao, L.; Guo, M.; Yin, Z. Enhanced Biosynthesis of Chlorogenic Acid and Its Derivatives in Methyl-Jasmonate-Treated Gardenia Jasminoides Cells: A Study on Metabolic and Transcriptional Responses of Cells. Front. Bioeng. Biotechnol. 2021, 8, 604957. [Google Scholar] [CrossRef]
- Yang, K.; Zhou, G.; Chen, C.; Liu, X.; Wei, L.; Zhu, F.; Liang, Z.; Chen, H. Joint Metabolomic and Transcriptomic Analysis Identify Unique Phenolic Acid and Flavonoid Compounds Associated with Resistance to Fusarium Wilt in Cucumber (Cucumis sativus L.). Front. Plant Sci. 2024, 15, 1447860. [Google Scholar] [CrossRef]
- He, Y.; Mao, S.; Zhao, Y.; Yang, J. Research Advances in the Synthesis, Metabolism, and Function of Chlorogenic Acid. Foods 2025, 14, 1914. [Google Scholar] [CrossRef]
- Gu, J.; Sohail, H.; Qiu, L.; Chen, C.; Yue, H.; Li, Z.; Yang, X.; Zhang, L. Genome-Wide Characterization and Expression Analysis of CsPALs in Cucumber (Cucumis sativus L.) Reveal Their Potential Roles in Abiotic Stress and Aphid Stress Tolerance. Plants 2024, 13, 2537. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, Y.; Yu, Y.; Ma, P.; Jia, Z.; Guo, X.; Xie, Y.; Bian, X. Overexpression of IbPAL1 Promotes Chlorogenic Acid Biosynthesis in Sweetpotato. Crop J. 2021, 9, 204–215. [Google Scholar] [CrossRef]
- Wang, P.; Li, M.; Ma, X.; Zhao, B.; Jin, X.; Chen, S.; Zhang, X.; Wu, X.; Zhang, H. Integrated Physiological, Transcriptomic and Metabolomic Analysis Revealed Heterosis for Cadmium Tolerance in Maize. Plant Physiol. Biochem. 2025, 228, 110265. [Google Scholar] [CrossRef]
- Meyer, K.; Cusumano, J.C.; Somerville, C.; Chapple, C.C. Ferulate-5-Hydroxylase from Arabidopsis thaliana Defines a New Family of Cytochrome P450-Dependent Monooxygenases. Proc. Natl. Acad. Sci. USA 1996, 93, 6869–6874. [Google Scholar] [CrossRef]
- Nair, R.B.; Joy, R.W.; Kurylo, E.; Shi, X.; Schnaider, J.; Datla, R.S.S.; Keller, W.A.; Selvaraj, G. Identification of a CYP84 Family of Cytochrome P450-Dependent Mono-Oxygenase Genes in Brassica Napus and Perturbation of Their Expression for Engineering Sinapine Reduction in the Seeds. Plant Physiol. 2000, 123, 1623–1634. [Google Scholar] [CrossRef]
- Kocábek, T.; Mishra, A.K.; Matoušek, J.; Patzak, J.; Lomnická, A.; Khare, M.; Krofta, K. The R2R3 Transcription Factor HlMYB8 and Its Role in Flavonoid Biosynthesis in Hop (Humulus lupulus L.). Plant Sci. 2018, 269, 32–46. [Google Scholar] [CrossRef]
- Orduña, L.; Li, M.; Navarro-Payá, D.; Zhang, C.; Santiago, A.; Romero, P.; Ramšak, Ž.; Magon, G.; Höll, J.; Merz, P.; et al. Direct Regulation of Shikimate, Early Phenylpropanoid, and Stilbenoid Pathways by Subgroup 2 R2R3-MYBS in Grapevine. Plant J. 2022, 110, 529–547. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lou, J.; Liu, G.; Li, Q.; Cao, Z.; Wu, P.; Mashu, H.; Liu, Z.; Deng, J.; Yang, Z.; et al. A R2R3-MYB Transcription Factor LmMYB111 Positively Regulates Chlorogenic Acid and Luteoloside Biosynthesis in Lonicera macranthoides. Plant Sci. 2025, 358, 112556. [Google Scholar] [CrossRef] [PubMed]
- Tang, N.; Cao, Z.; Yang, C.; Ran, D.; Wu, P.; Gao, H.; He, N.; Liu, G.; Chen, Z. A R2R3-MYB Transcriptional Activator LmMYB15 Regulates Chlorogenic Acid Biosynthesis and Phenylpropanoid Metabolism in Lonicera macranthoides. Plant Sci. 2021, 308, 110924. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Li, L.; Cheng, H.; Yao, L.; Wu, J.; Huang, H.; Ning, W.; Kai, G. The Basic Helix-Loop-Helix Transcription Factor TabHLH1 Increases Chlorogenic Acid and Luteolin Biosynthesis in Taraxacum antungense Kitag. Hortic. Res. 2021, 8, 195. [Google Scholar] [CrossRef]
- Wang, Z.; Ma, L.; Liu, P.; Luo, Z.; Li, Z.; Wu, M.; Xu, X.; Pu, W.; Huang, P.; Yang, J. Transcription Factor NtWRKY33a Modulates the Biosynthesis of Polyphenols by Targeting NtMYB4 and NtHCT Genes in Tobacco. Plant Sci. 2023, 326, 111522. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, J.; Gao, Q.; He, S.; Xu, Y.; Luo, Z.; Liu, P.; Wu, M.; Xu, X.; Ma, L.; et al. The Transcription Factor NtERF13a Enhances Abiotic Stress Tolerance and Phenylpropanoid Compounds Biosynthesis in Tobacco. Plant Sci. 2023, 334, 111772. [Google Scholar] [CrossRef]
- Su, Z.; Xu, J.; Cai, Z.; Ma, R.; Shen, Z.; Yan, J.; Yu, M. PpMYB and PpbHLH Transcription Factors Activate PpHCT5 to Regulate Chlorogenic Acid Biosynthesis in Peach Fruit. Hortic. Plant J. 2025, S2468014125000329. [Google Scholar] [CrossRef]
- Ahmad, N.; Zhang, K.; Ma, J.; Yuan, M.; Zhao, S.; Wang, M.; Deng, L.; Ren, L.; Gangurde, S.S.; Pan, J.; et al. Transcriptional Networks Orchestrating Red and Pink Testa Color in Peanut. BMC Plant Biol 2023, 23, 44. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Zhou, Z.; Feng, H.; Wang, Y.; Zeng, Z.; Hou, Q.; Shen, L. Integrated Transcriptomic and Metabolomic Analyses Reveal Key Genes Involved in Phenylpropanoid Metabolism in Lonicera macranthoides Flowers. Genes 2025, 16, 1339. https://doi.org/10.3390/genes16111339
Li Z, Zhou Z, Feng H, Wang Y, Zeng Z, Hou Q, Shen L. Integrated Transcriptomic and Metabolomic Analyses Reveal Key Genes Involved in Phenylpropanoid Metabolism in Lonicera macranthoides Flowers. Genes. 2025; 16(11):1339. https://doi.org/10.3390/genes16111339
Chicago/Turabian StyleLi, Zhengchun, Zijing Zhou, Hua Feng, Yong Wang, Zhaohua Zeng, Qiandong Hou, and Luonan Shen. 2025. "Integrated Transcriptomic and Metabolomic Analyses Reveal Key Genes Involved in Phenylpropanoid Metabolism in Lonicera macranthoides Flowers" Genes 16, no. 11: 1339. https://doi.org/10.3390/genes16111339
APA StyleLi, Z., Zhou, Z., Feng, H., Wang, Y., Zeng, Z., Hou, Q., & Shen, L. (2025). Integrated Transcriptomic and Metabolomic Analyses Reveal Key Genes Involved in Phenylpropanoid Metabolism in Lonicera macranthoides Flowers. Genes, 16(11), 1339. https://doi.org/10.3390/genes16111339





