Chloroplast Genome Features and Phylogeny of Two Nationally Protected Medicinal Plants, Euchresta tubulosa and Euchresta japonica: Molecular Resources for Identification and Conservation
Abstract
1. Introduction
2. Materials and Methods
2.1. Material Collection and DNA Extraction
2.2. Chloroplast Genome Assembly and Annotation
2.3. Repetitive Sequences and SSRs
2.4. Codon Preference
2.5. Structural Alignment and Variation Hotspots
2.6. Phylogenetic Analysis
3. Results
3.1. Composition and Features of the Chloroplast Genome
3.2. IR Boundaries and Structural Variations
3.3. Collinearity Analysis of the Complete Chloroplast Genome
3.4. Relative Synonymous Codon Usage (RSCU)
3.5. Repetitive Sequences and Simple Sequence Repeats (SSRs)
3.6. Nucleotide Diversity
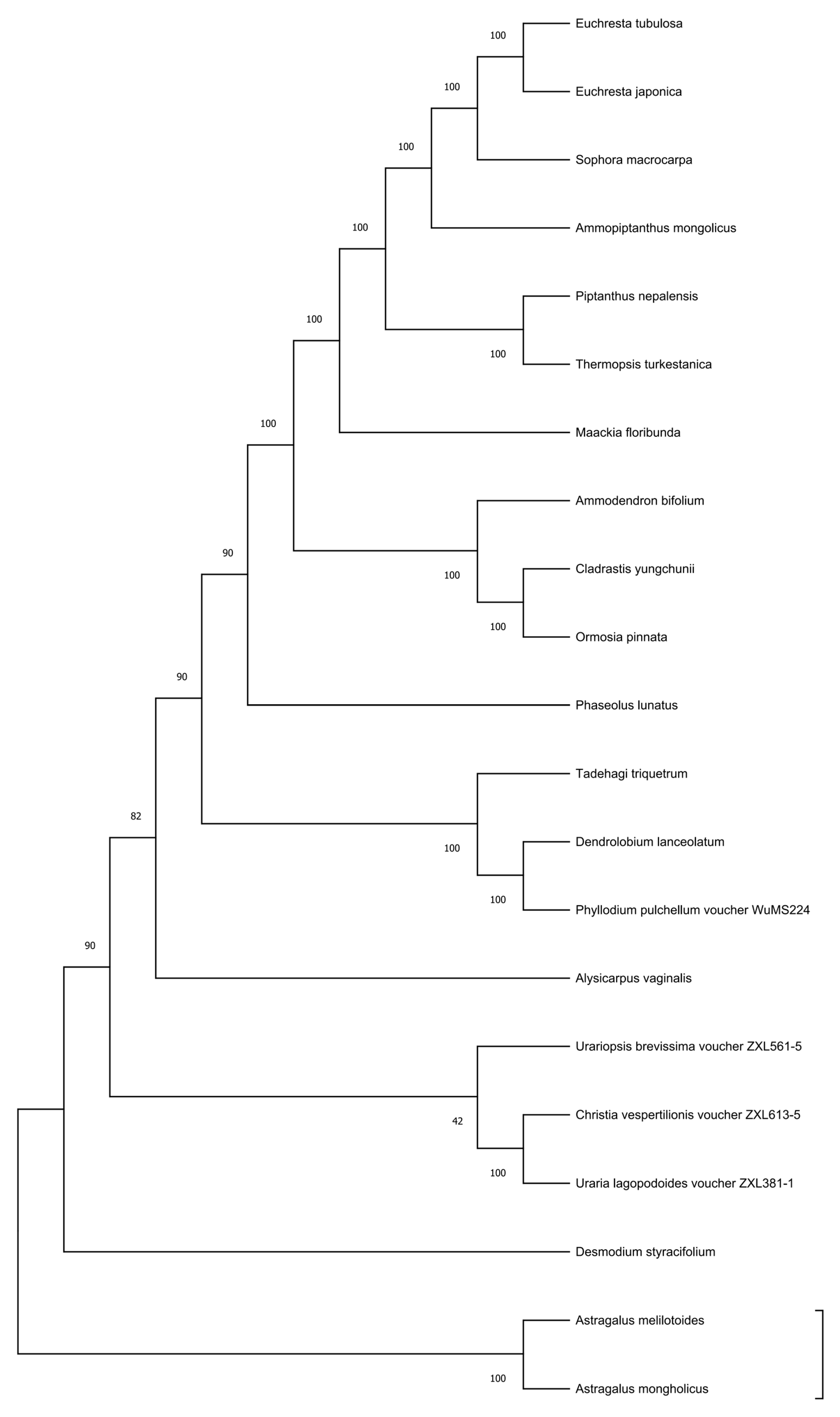
3.7. Phylogenetic Analysis Based on Complete Chloroplast Genomes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, Z.; Gong, L.M.; Guo, J.M.; Liu, X.R.; Zheng, H.; Li, S.X. Study on Artificial Propagation of Rare and Endangered Tujia Medicine Euchresta japonica. J. Hunan Univ. Chin. Med. 2019, 39, 1018–1020. [Google Scholar]
- Yang, H.; Qin, X. HPLC-DAD Detection of Multiple Active Components in Euchresta japonica. Chem. Reag. 2025, 47, 85–91. [Google Scholar]
- Wu, Y.Q.; Li, J.P.; Wang, L.S. Synthesis and Antitumor Activity of Novel Matrine C-14 Hydrazone Derivatives. Chem. Reag. 2024, 46, 121–129. [Google Scholar]
- Zhao, J.; Cai, X.H. Structural Modification of D-Ring for Antitumor Activity Based on Matrine-Type Alkaloids. Chem. Reag. 2022, 44, 136–141. [Google Scholar]
- Shizuo, T.; Yoshiaki, S. Inhibitory Effect of Prenylated Flavonoid in Euchresta japonica and Artocarpus heterophyllus on Lipid Peroxidation by Interaction of Hemoglobin and Hydrogen Peroxide. Pharm. Biol. 2006, 44, 261–263. [Google Scholar] [CrossRef]
- Lei, J.X. Extraction, Isolation and Identification of Flavonoids from Euchresta tubulosa Dunn. Master’s Thesis, Jishou University, Jishou, China, 2024. [Google Scholar]
- Lin, M.X.; Han, F.; Han, R.G.; Ren, M.B.; Deng, C.F.; Zhang, W.W. Preliminary Observation on Biological Characteristics of Euchresta tubulosa Dunn. Hunan Agric. Sci. 2013, 3, 204–214. [Google Scholar] [CrossRef]
- Deng, C.F.; Zhang, W.W.; Lin, M.X. Investigation on Resources and Ecological Environment of Euchresta japonica Sieb. et Zucc., a Folk Analgesic Medicine. Lishizhen Med. Mater. Med. Res. 2008, 19, 627–628. [Google Scholar]
- State Forestry Administration; Ministry of Agriculture. List of National Key Protected Wild Plants (First Batch); State Forestry Administration: Beijing, China; Ministry of Agriculture: New Delhi, India, 1999.
- Wang, S.; Xie, Y. China Species Red List (Volume 1); Higher Education Press: Beijing, China, 2004. [Google Scholar]
- Hua, Y.S. Identification of Sophora tonkinensis and Its Seven Common Adulterants. West. J. Tradit. Chin. Med. 2015, 28, 74–76. [Google Scholar]
- Liu, J.; Zhang, H.; Li, M. Pharmacological Effects and Quality Control of Euchresta Species: A Review. Chin. J. Exp. Tradit. Med. Formulae 2020, 26, 215–222. [Google Scholar]
- Lin, M.X.; Han, F.; Liu, Z.Y.; Zhang, J.; Shen, J.; Ren, M. Textual Research on Folk Anti-Cancer Drug Euchresta japonica. Res. Pract. Mod. Chin. Med. 2009, 23, 26–28. [Google Scholar]
- Chen, S.L.; Yao, H.; Han, J.P.; Liu, C.; Song, J.Y.; Shi, L.C.; Zhu, Y.J.; Ma, X.Y.; Gao, T.; Pang, X.H.; et al. Validation of the ITS2 Region as a Novel DNA Barcode for Identifying Medicinal Plant Species. Genes 2010, 1, 212–228. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.X.; Liu, Z.Y.; Ren, M.B.; Liu, X.; Zhang, J. Euchresta japonica: An Endangered Anti-Cancer Chinese Herbal Medicine in Jinfo Mountain. For. By-Product Spec. China 2007, 6, 72–73. [Google Scholar] [CrossRef]
- Zhang, J.; Shu, G.M.; Liu, Z.Y.; Lin, M.X.; Ren, M.B.; Shen, J. Pharmacognostic Study on Endangered Chinese Herbal Medicine Euchresta japonica in Jinfo Mountain. Lishizhen Med. Mater. Med. Res. 2009, 20, 532–533. [Google Scholar]
- Lin, P.; Yin, H.; Wang, K.; Gao, H.; Liu, L.; Yao, X. Comparative Genomic Analysis Uncovers the Chloroplast Genome Variation and Phylogenetic Relationships of Camellia Species. Biomolecules 2022, 12, 1474. [Google Scholar] [CrossRef]
- Wang, A.; Wu, H.; Zhu, X.; Lin, J. Species Identification of Conyza bonariensis Assisted by Chloroplast Genome Sequencing. Front. Genet. 2018, 9, 374. [Google Scholar] [CrossRef]
- Kelchner, S. The Evolution of Non-Coding Chloroplast DNA and Its Application in Plant Systematics. Ann. Mo. Bot. Gard. 2000, 87, 482–498. [Google Scholar] [CrossRef]
- Heinke, L. Chilling Paternal Chloroplasts. Nat. Rev. Mol. Cell Biol. 2023, 24, 166. [Google Scholar] [CrossRef]
- Dong, W.; Xu, C.; Wen, J.; Zhou, S. Evolutionary Directions of Single Nucleotide Substitutions and Structural Mutations in the Chloroplast Genomes of the Family Calycanthaceae. BMC Evol. Biol. 2020, 20, 96. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, Y.; Chen, T.; Chen, Q.; Wang, L.; Liu, Z.S.; Wang, H.; Xie, R.; He, W.; Li, M.; et al. Evolution of Rosaceae Plastomes Highlights Unique Cerasus Diversification and Independent Origins of Fruiting Cherry. Front. Plant Sci. 2021, 12, 736053. [Google Scholar] [CrossRef]
- Zheng, H.Z.; Peng, G.X.; Zhao, L.C.; Dai, W.; Xu, M.H.; Xu, X.G.; Tang, M. Comparative and Evolutionary Analysis of Chloroplast Genomes from Five Rare Styrax Species. BMC Genom. 2025, 26, 450. [Google Scholar] [CrossRef] [PubMed]
- Linh, N.N.; Hang, P.L.B.; Hue, H.T.T.; Ha, N.H.; Hanh, H.H.; Ton, N.D.; Hien, L.T.T. Species discrimination of novel chloroplast DNA barcodes and their application for identification of Panax (Aralioideae, Araliaceae). PhytoKeys 2022, 188, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Sahin, C.E.; Aydin, Y.; Uncuoglu, A.A. Assessing the accuracy of cp-DNA barcodes in Colchicum species identification. Mol. Biol. Rep. 2025, 52, 584. [Google Scholar] [CrossRef]
- Dufourmantel, N.; Pelissier, B.; Garco, N.F.; Peltier, G.; Ferullo, J.-M.; Tissot, G. Generation of Fertile Transplastomic Soybean. Plant Mol. Biol. 2004, 55, 479–489. [Google Scholar] [CrossRef]
- Hama, I.; Saito, Y.; Umehara, C.; Lian, L.; Ide, Y. Development of mi- crosatellite markers for Euchresta japonica and E. formosana (Leguminosae). Mol. Ecol. Resour. 2009, 9, 1188–1190. [Google Scholar] [CrossRef]
- Priyadi, A.; Feng, C.; Kang, M.; Huang, H. Development of 10 single- copy nuclear DNA markers for Euchresta horsfieldii (Fabaceae), a rare medicinal plant. Appl. Plant Sci. 2018, 6, e1178. [Google Scholar] [CrossRef]
- Zhuo, W.; Ren, F.; Wang, L.; Chen, S.; Chen, Y.; Huang, H. Characterization of the first chloroplast genome of Euchresta tubulosa Dunn and its phylogenetic analysis. Mitochondrial DNA. Part B Resour. 2021, 6, 2884–2885. [Google Scholar] [CrossRef]
- Arseneau, J.R.; Steeves, R.; Laflamme, M. Modified Low-Salt CTAB Extraction of High-Quality DNA from Contaminant-Rich Tissues. Mol. Ecol. Resour. 2017, 17, 686–693. [Google Scholar] [CrossRef] [PubMed]
- Twyford, A.D.; Ness, R.W. Strategies for Complete Plastid Genome Sequencing. Mol. Ecol. Resour. 2017, 17, 858–868. [Google Scholar] [CrossRef]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive Visualization of De Novo Genome Assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Chen, H.; Jiang, M.; Wang, L.; Wu, X.; Huang, L.; Liu, C. CPGAVAS2, an Integrated Plastome Sequence Annotator and Analyzer. Nucleic Acids Res. 2019, 47, W65–W73. [Google Scholar] [CrossRef]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved Detection and Functional Classification of Transfer RNA Genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.H.; Zhang, M.; Zhao, K.; Zhang, X.H.; Ge, C.L. Polypleurum chinense (Podostemaceae), a New Species from Fujian, China, Based on Morphological and Genomic Evidence. PhytoKeys 2022, 199, 167–186. [Google Scholar] [CrossRef]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The Manifold Applications of Repeat Analysis on a Genomic Scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef] [PubMed]
- Du, L.M.; Sun, D.L.; Chen, J.H.; Zhou, X.; Zhao, K.; Zeng, Q.; Yang, N. Pytrf: A Python Package for Finding Tandem Repeats from Genomic Sequences. BMC Bioinform. 2025, 26, 151–159. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Munch, T.; Scholz, U.; Mascher, M. MISA-Web: A Web Server for Microsatellite Prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Jansen, R.K.; Cai, Z.; Raubeson, L.A.; Daniell, H.; Depamphilis, C.W.; Leebens-Mack, J.; Müller, K.F.; Guisinger-Bellian, M.; Haberle, R.C.; Hansen, A.K.; et al. Analysis of 81 Genes from 64 Plastid Genomes Resolves Relationships in Angiosperms and Identifies Genome-Scale Evolutionary Patterns. Proc. Natl. Acad. Sci. USA 2007, 104, 19369–19374. [Google Scholar] [CrossRef]
- Sharp, P.M.; Li, W.H. The Codon Adaptation Index—A Measure of Directional Synonymous Codon Usage Bias, and Its Potential Applications. Nucleic Acids Res. 1987, 15, 1281–1295. [Google Scholar] [CrossRef]
- Amiryousefi, A.; Hyvönen, P.; Poczai, P. IRscope: An Online Program to Visualize the Junction Sites of Chloroplast Genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Chen, Z.Y.; Liu, Q.; Ying, X.; Zhou, G.; Yu, P.; Bai, J.; Huang, H.; Gong, Y. Correction to: Complete Chloroplast Genome Sequence of Camellia sinensis: Genome Structure, Adaptive Evolution, and Phylogenetic Relationships. J. Appl. Genet. 2023, 64, 601. [Google Scholar] [CrossRef] [PubMed]
- Song, W.C.; Chen, Z.M.; Shi, W.B.; Han, W.; Feng, Q.; Shi, C.; Engel, M.S.; Wang, S. Comparative Analysis of Complete Chloroplast Genomes of Nine Species of Litsea (Lauraceae): Hypervariable Regions, Positive Selection, and Phylogenetic Relationships. Genes 2022, 13, 1550. [Google Scholar] [CrossRef]
- Kim, G.B.; Lim, C.E.; Kim, J.S.; Kim, K.; Lee, J.H.; Yu, H.-J.; Mun, J.-H. Comparative Chloroplast Genome Analysis of Artemisia (Asteraceae) in East Asia: Insights into Evolutionary Divergence and Phylogenomic Implications. BMC Genom. 2020, 21, 415. [Google Scholar] [CrossRef]
- Liu, H. Assembly and Evolutionary Analysis of Soybean Mitochondrial and Chloroplast Genomes. Master’s Thesis, Northwest University, Xi’an, China, 2022. [Google Scholar]
- Li, Y.C.; Wang, R.; Wang, H.H.; Pu, F.; Feng, X.; Jin, L.; Ma, Z.; Ma, X.-X. Codon Usage Bias in Autophagy-Related Gene 13 in Eukaryotes: Uncovering the Genetic Divergence by the Interplay Between Nucleotides and Codon Usages. Front. Cell. Infect. Microbiol. 2021, 11, 771010. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Luckow, M.A. Chloroplast Genome Evolution in Legumes: Inversions, Expansion/Contraction of Inverted Repeats, and Gene Loss. Mol. Phylogenet. Evol. 1999, 12, 305–319. [Google Scholar]
- Wang, Y.; Li, L.; Zhang, X.; Liu, H.; Li, M. Comparative Chloroplast Genomics of Sophora (Fabaceae): Insights into IR Boundary Stability and Species Diversification. Int. J. Mol. Sci. 2022, 23, 8472. [Google Scholar]
- Raubeson, L.A.; Jansen, R.K. Plastid Genome Evolution in Angiosperms. Annu. Rev. Plant Biol. 2005, 56, 237–264. [Google Scholar]
- Palmer, J.D.; Thompson, W.F. Chloroplast DNA Rearrangements Are More Frequent When a Large Inverted Repeat Sequence Is Lost. Cell 1982, 29, 537–544. [Google Scholar] [CrossRef]
- Smith, S.A.; Donoghue, M.J. Rates of Molecular Evolution Are Linked to Life-History Traits in Flowering Plants. Science 2008, 322, 862–864. [Google Scholar] [CrossRef]
- Wicke, S.; Naumann, J.; Martens, K.M. The Evolution of the Plastid Chromosome in Land Plants: Gene Content, Gene Order, Gene Function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef]
- Nickrent, D.L.; Blarer, A.; Wicke, S. Plastid Genome Evolution in Parasitic Plants. Annu. Rev. Plant Biol. 2010, 61, 143–165. [Google Scholar]
- Rumeau, D.; Suorsa, M.; Aro, E. The Chloroplast NAD(P)H Dehydrogenase Complex: Structure, Function and Photoprotection. Trends Plant Sci. 2007, 12, 511–518. [Google Scholar]
- Yamori, W.; Hikosaka, K.; Way, D.A. Temperature Response of Photosynthesis in C3, C4, and CAM Plants: Temperature Acclimation and Temperature Adaptation. Photosynth. Res. 2014, 119, 101–117. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.; Lickey, E.B.; Schilling, E.E.; Small, R.L. Comparison of Whole Chloroplast Genome Sequences to Choose Noncoding Regions for Phylogenetic Studies in Angiosperms: The Tortoise and the Hare III. Am. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef] [PubMed]
- CBOL Plant Working Group. A DNA Barcode for Land Plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, S.; Yao, H.; Han, J. Development of DNA Barcodes for the Identification of Medicinal Plants in Euchresta (Fabaceae) and Their Adulterants. J. Ethnopharmacol. 2020, 259, 112968. [Google Scholar]

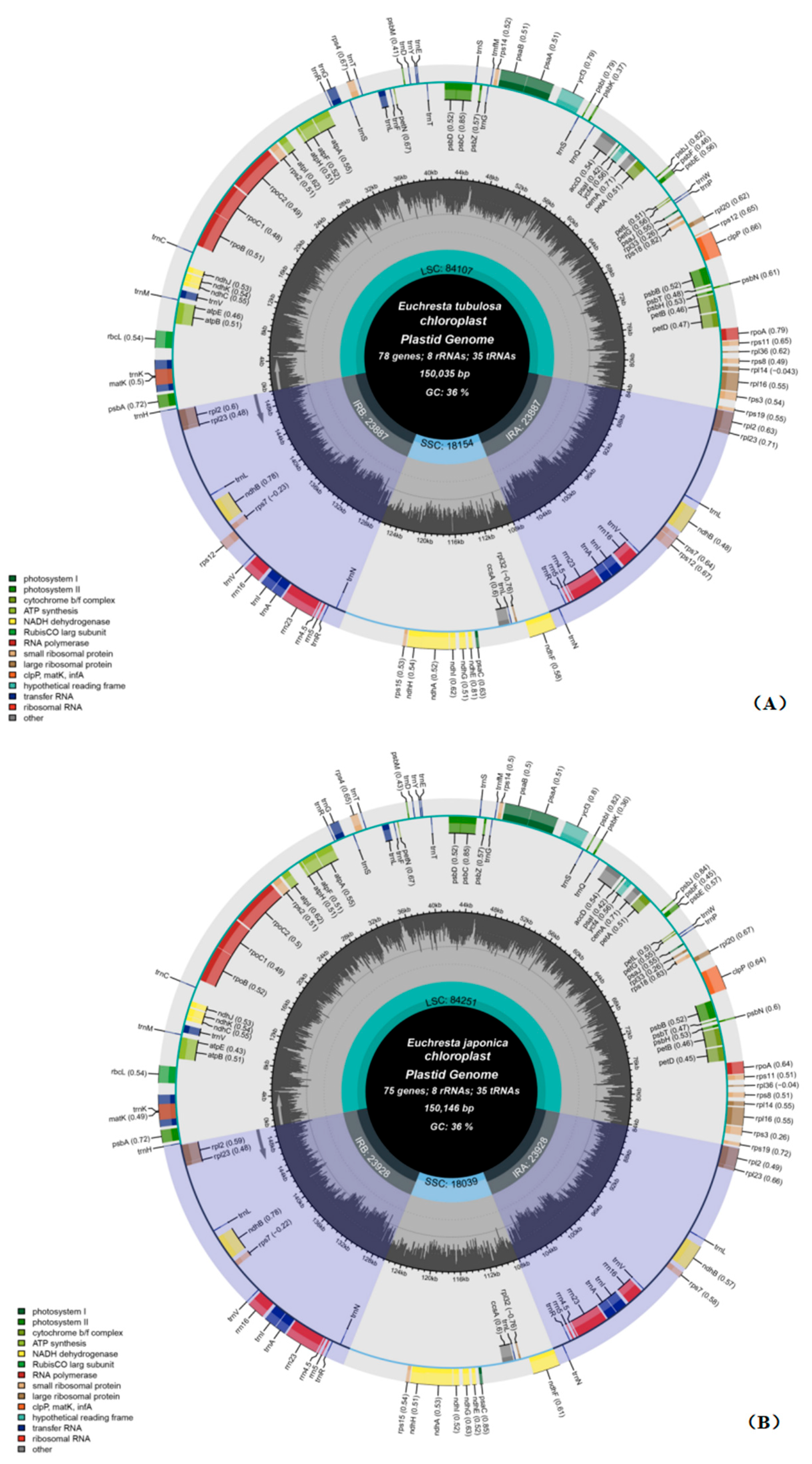
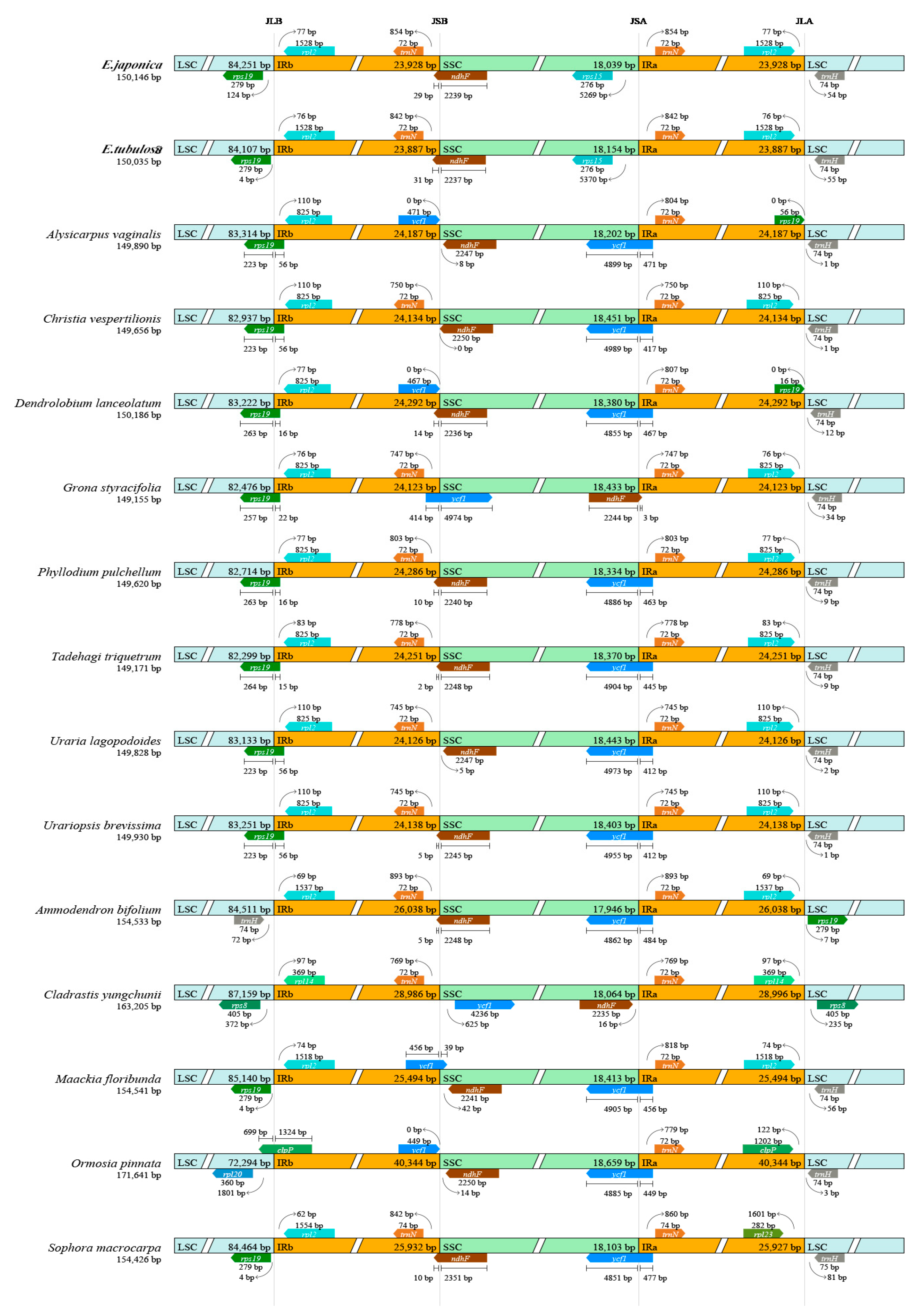
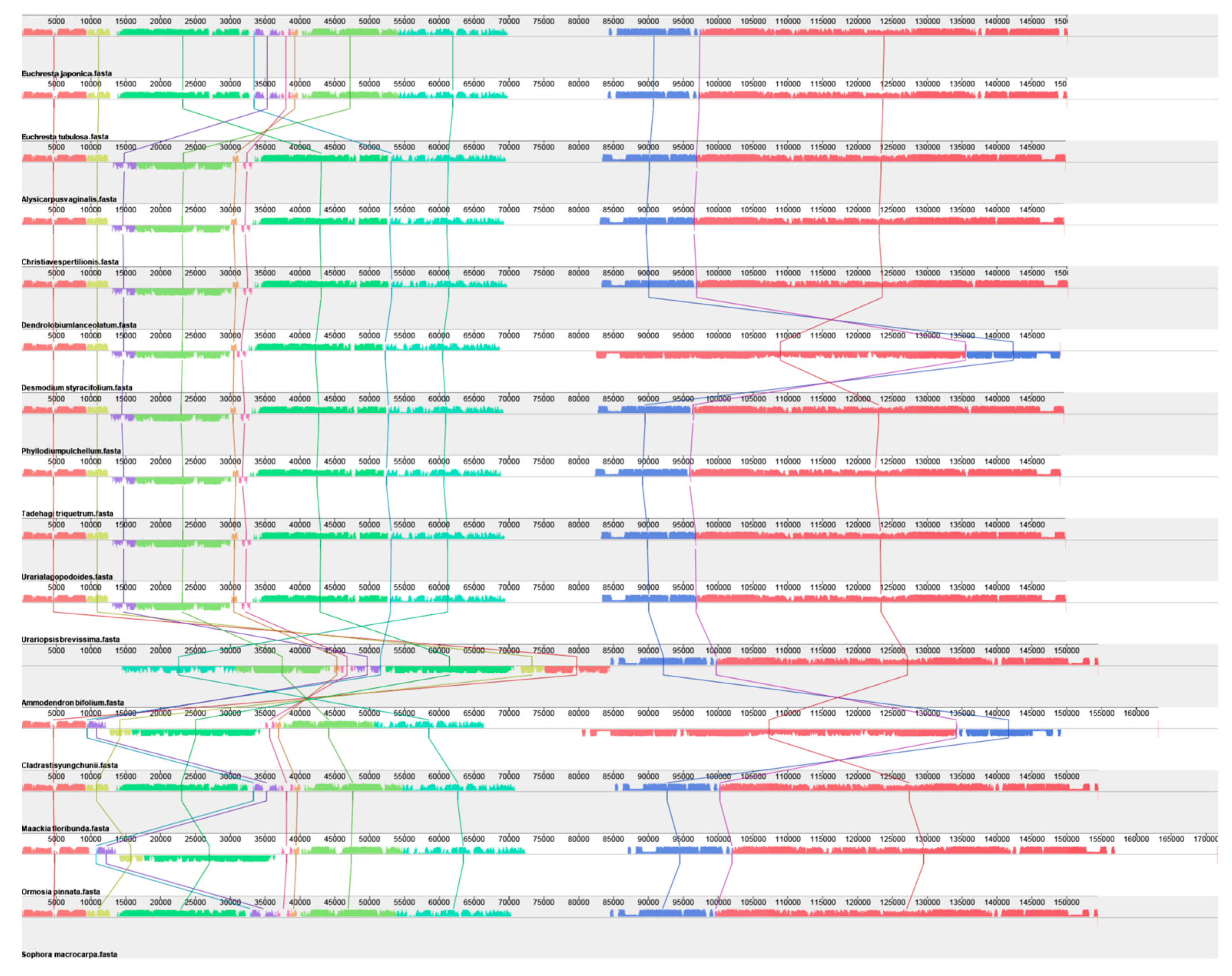


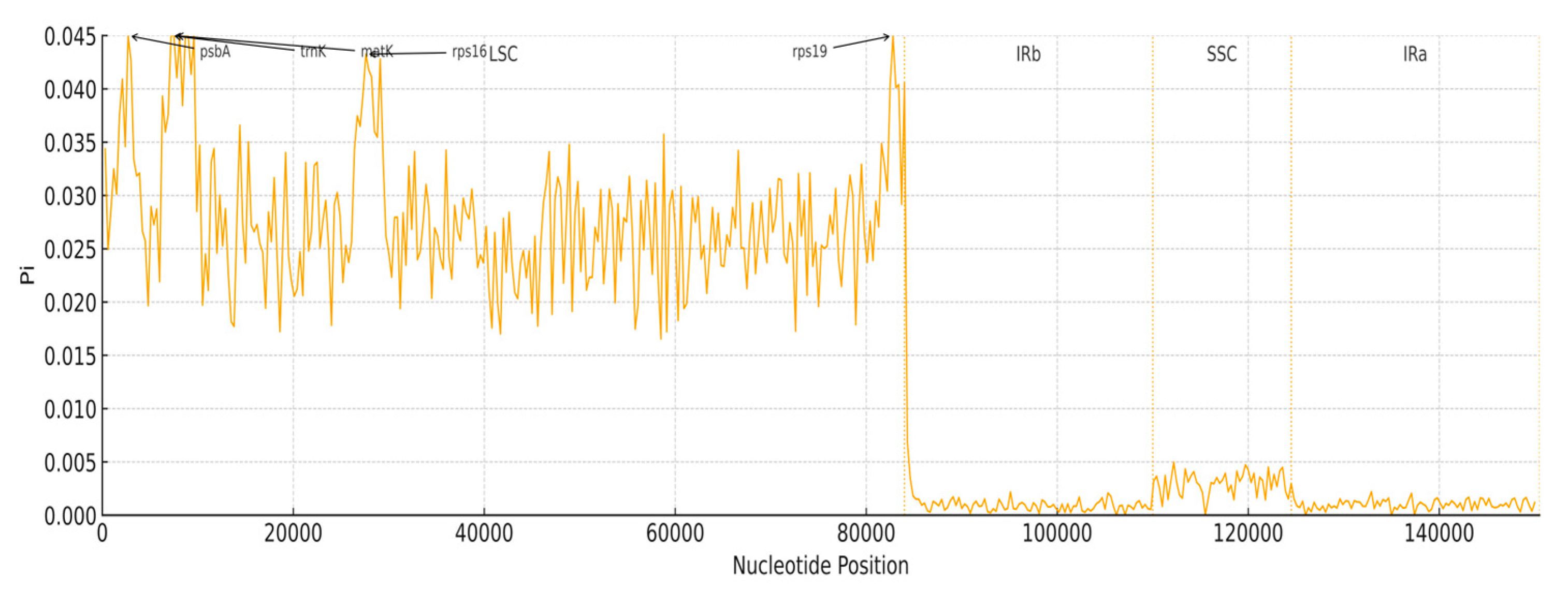
| Gene Category | Gene Group | Gene Name |
|---|---|---|
| Genes for photosynthesis | Subunits of photosystemI | psaa, psaB, psaC, psaI, psaJ |
| Subunits of photosystemII | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbM, psbN, psbT, psbZ | |
| Subunits of NADH dehydrogenase | ndhA *, ndhB *(2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Subunit of cytochromeb/f complex | petA, petB *, petD *, petG, petL, petN | |
| Subunits of ATP synthase | atpA, atpB, atpE, atpF *, atpH, atpI | |
| Large subunit of Rubisco | rbcL | |
| Self-replication | Proteins of large ribosomal subunit | rpl14, rpl16 *, rpl2 *(2), rpl20, rpl23(2), rpl32, rpl33, rpl36 |
| Proteins of small ribosomal subunit | rps11, rps12 **(2), rps14, rps15, rps16, rps18, rps19, rps2, rps3, rps4, rps7(2), rps8 | |
| Subunits of RNA polymerase | rpoA, rpoB, rpoC1 *, rpoC2 | |
| Ribosomal RNAs | rrn16S(2), rrn23S(2), rrn4.5S(2), rrn5S(2) | |
| Transfer RNAs | trnA-UGC *(2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCC*, trnH-GUG, trnI-CAU(2), trnI-GAU *(2), trnL-CAA(2), trnL-UAA *, trnL-UAG, trnM-CAU, trnN-GUU(2), trnP-GGG, trnQ-UUG, trnR-ACG(2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(2), trnV-UAC*, trnW-CCA, trnY-GUA, trnfM-CAU | |
| Other genes | Maturase | matK |
| Protease | clpP ** | |
| Envelop membrane proteinn | cemA | |
| acetyl-CoA-carboxylase | accD | |
| c-type cytochrome synthesis gene | ccsA | |
| Genes of unknown function | Conserved hypothetical chloroplast ORF | ycf1(2), ycf2 *(2), ycf3 **, ycf4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, D.; Li, X.; Xiao, Z.; Zhou, L. Chloroplast Genome Features and Phylogeny of Two Nationally Protected Medicinal Plants, Euchresta tubulosa and Euchresta japonica: Molecular Resources for Identification and Conservation. Genes 2025, 16, 1286. https://doi.org/10.3390/genes16111286
Yin D, Li X, Xiao Z, Zhou L. Chloroplast Genome Features and Phylogeny of Two Nationally Protected Medicinal Plants, Euchresta tubulosa and Euchresta japonica: Molecular Resources for Identification and Conservation. Genes. 2025; 16(11):1286. https://doi.org/10.3390/genes16111286
Chicago/Turabian StyleYin, Dabao, Xue Li, Zhongchun Xiao, and Li Zhou. 2025. "Chloroplast Genome Features and Phylogeny of Two Nationally Protected Medicinal Plants, Euchresta tubulosa and Euchresta japonica: Molecular Resources for Identification and Conservation" Genes 16, no. 11: 1286. https://doi.org/10.3390/genes16111286
APA StyleYin, D., Li, X., Xiao, Z., & Zhou, L. (2025). Chloroplast Genome Features and Phylogeny of Two Nationally Protected Medicinal Plants, Euchresta tubulosa and Euchresta japonica: Molecular Resources for Identification and Conservation. Genes, 16(11), 1286. https://doi.org/10.3390/genes16111286




