Analysis of the Dirigent Pan-Gene Family in 26 Diverse Inbred Lines Reveals Genomic Diversity in Maize
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of Dirigent Genes in the Maize Pan-Genome
2.2. Orthogroup Classification of the Dirigent Pan-Gene Family
2.3. Analysis of Gene Duplication Events
2.4. Analysis of Cis-Acting Regulatory Elements in Dirigent Genes
2.5. Ka/Ks Calculation
2.6. Expression Profile Analysis
2.7. GWAS for Dirigent Genes
3. Results
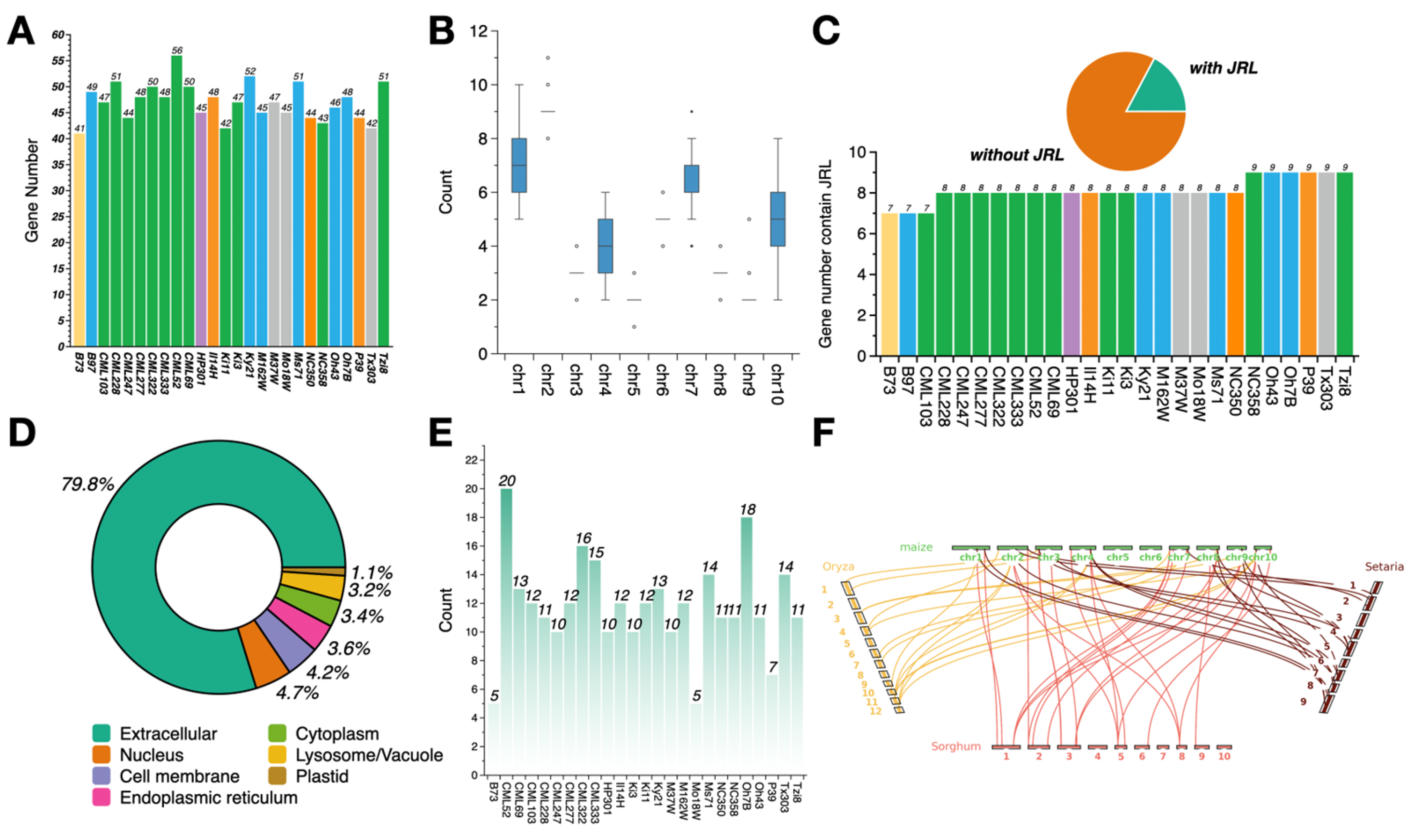
3.1. Identification of Dirigent Genes in 26 Maize NAM Founder Lines
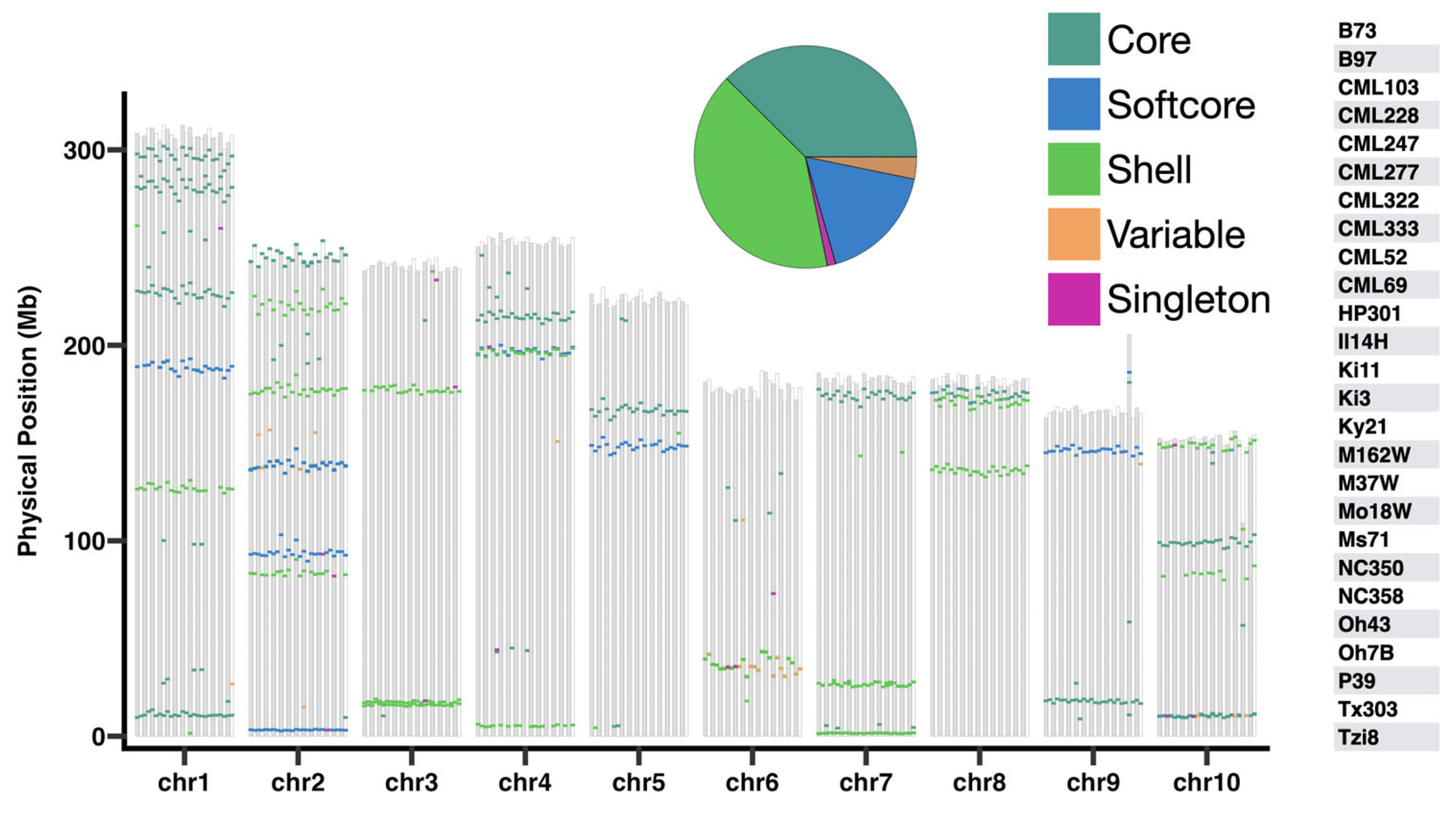
3.2. Construction of the Dirigent Pan-Gene Family
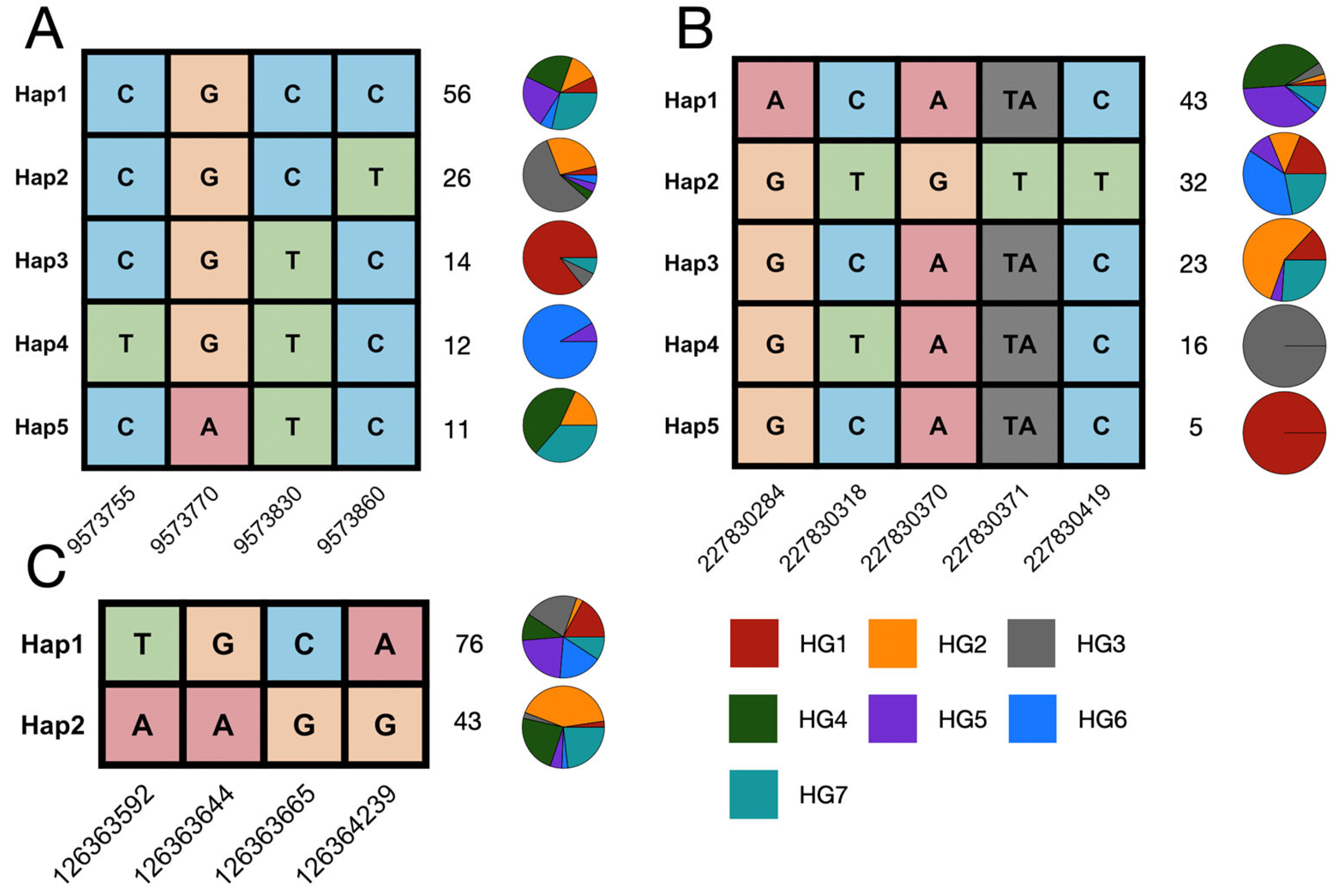
3.3. Maize Germplasm Haplotypes of the Dirigent Pan-Gene Family
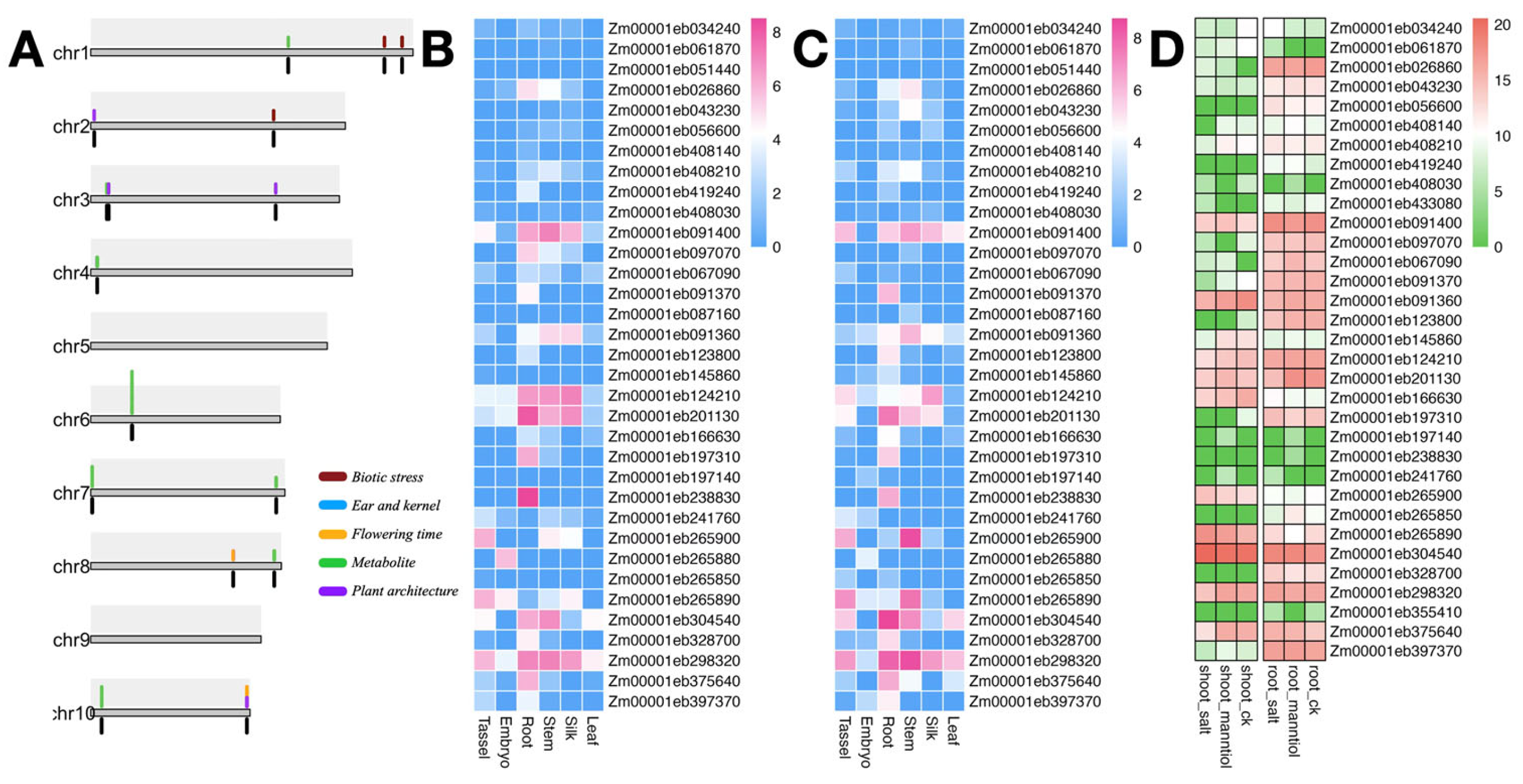
3.4. Association of Dirigent Genes with Agronomic Traits and Their Expression Profiles
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Davin, L.B.; Wang, H.-B.; Crowell, A.L.; Bedgar, D.L.; Martin, D.M.; Sarkanen, S.; Lewis, N.G. Stereoselective Bimolecular Phenoxy Radical Coupling by an Auxiliary (Dirigent) Protein Without an Active Center. Science 1997, 275, 362–367. [Google Scholar] [CrossRef]
- Davin, L.B.; Jourdes, M.; Patten, A.M.; Kim, K.-W.; Vassão, D.G.; Lewis, N.G. Dissection of Lignin Macromolecular Configuration and Assembly: Comparison to Related Biochemical Processes in Allyl/Propenyl Phenol and Lignan Biosynthesis. Nat. Prod. Rep. 2008, 25, 1015. [Google Scholar] [CrossRef] [PubMed]
- Halls, S.C.; Lewis, N.G. Secondary and Quaternary Structures of the (+)-Pinoresinol-Forming Dirigent Protein. Biochemistry 2002, 41, 9455–9461. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.E.; Collier, C.C.; Nemacheck, J.A.; Liang, C.; Cambron, S.E. A Lectin-Like Wheat Gene Responds Systemically to Attempted Feeding by Avirulent First-Instar Hessian Fly Larvae. J. Chem. Ecol. 2002, 28, 1411–1428. [Google Scholar] [CrossRef] [PubMed]
- Paniagua, C.; Bilkova, A.; Jackson, P.; Dabravolski, S.; Riber, W.; Didi, V.; Houser, J.; Gigli-Bisceglia, N.; Wimmerova, M.; Budínská, E.; et al. Dirigent Proteins in Plants: Modulating Cell Wall Metabolism during Abiotic and Biotic Stress Exposure. J. Exp. Bot. 2017, 68, 3287–3301. [Google Scholar] [CrossRef]
- Zhou, J.; Lee, C.; Zhong, R.; Ye, Z.-H. MYB58 and MYB63 Are Transcriptional Activators of the Lignin Biosynthetic Pathway during Secondary Cell Wall Formation in Arabidopsis. Plant Cell 2009, 21, 248–266. [Google Scholar] [CrossRef]
- Corbin, C.; Drouet, S.; Markulin, L.; Auguin, D.; Lainé, É.; Davin, L.B.; Cort, J.R.; Lewis, N.G.; Hano, C. A Genome-Wide Analysis of the Flax (Linum usitatissimum L.) Dirigent Protein Family: From Gene Identification and Evolution to Differential Regulation. Plant Mol. Biol. 2018, 97, 73–101. [Google Scholar] [CrossRef]
- Khan, A.; Li, R.-J.; Sun, J.-T.; Ma, F.; Zhang, H.-X.; Jin, J.-H.; Ali, M.; Haq, S.U.; Wang, J.-E.; Gong, Z.-H. Genome-Wide Analysis of Dirigent Gene Family in Pepper (Capsicum annuum L.) and Characterization of CaDIR7 in Biotic and Abiotic Stresses. Sci. Rep. 2018, 8, 5500. [Google Scholar] [CrossRef]
- Liao, Y.; Liu, S.; Jiang, Y.; Hu, C.; Zhang, X.; Cao, X.; Xu, Z.; Gao, X.; Li, L.; Zhu, J.; et al. Genome-Wide Analysis and Environmental Response Profiling of Dirigent Family Genes in Rice (Oryza sativa). Genes Genomics 2017, 39, 47–62. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, X.; Sun, Z.; Zhang, Y.; Meng, C.; Chen, B.; Wang, G.; Ke, H.; Wu, J.; Yan, Y.; et al. Evolution, Expression and Functional Analysis of Cultivated Allotetraploid Cotton DIR Genes. BMC Plant Biol. 2021, 21, 89. [Google Scholar] [CrossRef]
- Ma, Q.-H.; Liu, Y.-C. TaDIR13, a Dirigent Protein from Wheat, Promotes Lignan Biosynthesis and Enhances Pathogen Resistance. Plant Mol. Biol. Report. 2015, 33, 143–152. [Google Scholar] [CrossRef]
- Wang, W.; Mauleon, R.; Hu, Z.; Chebotarov, D.; Tai, S.; Wu, Z.; Li, M.; Zheng, T.; Fuentes, R.R.; Zhang, F.; et al. Genomic Variation in 3,010 Diverse Accessions of Asian Cultivated Rice. Nature 2018, 557, 43–49. [Google Scholar] [CrossRef]
- Wang, B.; Hou, M.; Shi, J.; Ku, L.; Song, W.; Li, C.; Ning, Q.; Li, X.; Li, C.; Zhao, B.; et al. De Novo Genome Assembly and Analyses of 12 Founder Inbred Lines Provide Insights into Maize Heterosis. Nat. Genet. 2023, 55, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Alonge, M.; Wang, X.; Benoit, M.; Soyk, S.; Pereira, L.; Zhang, L.; Suresh, H.; Ramakrishnan, S.; Maumus, F.; Ciren, D.; et al. Major Impacts of Widespread Structural Variation on Gene Expression and Crop Improvement in Tomato. Cell 2020, 182, 145–161.e23. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Du, H.; Li, P.; Shen, Y.; Peng, H.; Liu, S.; Zhou, G.-A.; Zhang, H.; Liu, Z.; Shi, M.; et al. Pan-Genome of Wild and Cultivated Soybeans. Cell 2020, 182, 162–176.e13. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Jiang, W.; Hui, Y.; Kong, M.; Feng, L.-Y.; Gao, L.-Z.; Li, P.; Lu, S. Gapless Indica Rice Genome Reveals Synergistic Contributions of Active Transposable Elements and Segmental Duplications to Rice Genome Evolution. Mol. Plant 2021, 14, 1745–1756. [Google Scholar] [CrossRef]
- Zhang, A.; Kong, T.; Sun, B.; Qiu, S.; Guo, J.; Ruan, S.; Guo, Y.; Guo, J.; Zhang, Z.; Liu, Y.; et al. A Telomere-to-Telomere Genome Assembly of Zhonghuang 13, a Widely-Grown Soybean Variety from the Original Center of Glycine Max. Crop J. 2024, 12, 142–153. [Google Scholar] [CrossRef]
- Shang, L.; Li, X.; He, H.; Yuan, Q.; Song, Y.; Wei, Z.; Lin, H.; Hu, M.; Zhao, F.; Zhang, C.; et al. A Super Pan-Genomic Landscape of Rice. Cell Res. 2022, 32, 878–896. [Google Scholar] [CrossRef]
- Guo, D.; Li, Y.; Lu, H.; Zhao, Y.; Kurata, N.; Wei, X.; Wang, A.; Wang, Y.; Zhan, Q.; Fan, D.; et al. A Pangenome Reference of Wild and Cultivated Rice. Nature 2025, 642, 662–671. [Google Scholar] [CrossRef]
- Cao, S.; Sawettalake, N.; Shen, L. Lactuca Super-Pangenome Provides Insights into Lettuce Genome Evolution and Domestication. Nat. Commun. 2025, 16, 7257. [Google Scholar] [CrossRef]
- Li, N.; He, Q.; Wang, J.; Wang, B.; Zhao, J.; Huang, S.; Yang, T.; Tang, Y.; Yang, S.; Aisimutuola, P.; et al. Super-Pangenome Analyses Highlight Genomic Diversity and Structural Variation across Wild and Cultivated Tomato Species. Nat. Genet. 2023, 55, 852–860. [Google Scholar] [CrossRef]
- Chen, W.; Chen, L.; Zhang, X.; Yang, N.; Guo, J.; Wang, M.; Ji, S.; Zhao, X.; Yin, P.; Cai, L.; et al. Convergent Selection of a WD40 Protein That Enhances Grain Yield in Maize and Rice. Science 2022, 375, eabg7985. [Google Scholar] [CrossRef]
- Hu, Y.; Colantonio, V.; Müller, B.S.F.; Leach, K.A.; Nanni, A.; Finegan, C.; Wang, B.; Baseggio, M.; Newton, C.J.; Juhl, E.M.; et al. Genome Assembly and Population Genomic Analysis Provide Insights into the Evolution of Modern Sweet Corn. Nat. Commun. 2021, 12, 1227. [Google Scholar] [CrossRef]
- Sun, B.; Peng, Y.; Yang, H.; Li, Z.; Gao, Y.; Wang, C.; Yan, Y.; Liu, Y. Alfalfa (Medicago sativa L.)/Maize (Zea mays L.) Intercropping Provides a Feasible Way to Improve Yield and Economic Incomes in Farming and Pastoral Areas of Northeast China. PLoS ONE 2014, 9, e110556. [Google Scholar] [CrossRef]
- Chen, P.; Xie, F.; Zhao, L.; Qiao, Q.; Liu, X. Effect of Acid Hydrolysis on the Multi-Scale Structure Change of Starch with Different Amylose Content. Food Hydrocoll. 2017, 69, 359–368. [Google Scholar] [CrossRef]
- Luo, N.; Meng, Q.; Feng, P.; Qu, Z.; Yu, Y.; Liu, D.L.; Müller, C.; Wang, P. China Can Be Self-Sufficient in Maize Production by 2030 with Optimal Crop Management. Nat. Commun. 2023, 14, 2637. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, Z.; Tan, K.; Huang, W.; Shi, J.; Li, T.; Hu, J.; Wang, K.; Wang, C.; Xin, B.; et al. A Complete Telomere-to-Telomere Assembly of the Maize Genome. Nat. Genet. 2023, 55, 1221–1231. [Google Scholar] [CrossRef] [PubMed]
- Hufford, M.B.; Seetharam, A.S.; Woodhouse, M.R.; Chougule, K.M.; Ou, S.; Liu, J.; Ricci, W.A.; Guo, T.; Olson, A.; Qiu, Y.; et al. De Novo Assembly, Annotation, and Comparative Analysis of 26 Diverse Maize Genomes. Science 2021, 373, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic Orthology Inference for Comparative Genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, İ.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An Information Aesthetic for Comparative Genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xiao, J.; Wu, J.; Zhang, H.; Liu, G.; Wang, X.; Dai, L. ParaAT: A Parallel Tool for Constructing Multiple Protein-Coding DNA Alignments. Biochem. Biophys. Res. Commun. 2012, 419, 779–781. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A Toolkit Incorporating Gamma-Series Methods and Sliding Window Strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Brown, P.J.; Upadyayula, N.; Mahone, G.S.; Tian, F.; Bradbury, P.J.; Myles, S.; Holland, J.B.; Flint-Garcia, S.; McMullen, M.D.; Buckler, E.S.; et al. Distinct Genetic Architectures for Male and Female Inflorescence Traits of Maize. PLoS Genet. 2011, 7, e1002383. [Google Scholar] [CrossRef]
- Zhao, H.; Sun, Z.; Wang, J.; Huang, H.; Kocher, J.-P.; Wang, L. CrossMap: A Versatile Tool for Coordinate Conversion between Genome Assemblies. Bioinformatics 2014, 30, 1006–1007. [Google Scholar] [CrossRef]
- Gong, L.; Lu, Y.; Wang, Y.; He, F.; Zhu, T.; Xue, B. Comparative Analysis of the JRL Gene Family in the Whole-Genome of Five Gramineous Plants. Front. Plant Sci. 2024, 15, 1501975. [Google Scholar] [CrossRef]
- Song, X.; Zhang, Y.; Li, C.; Li, N.; Shen, S.; Yu, T.; Liu, Z.; Zhou, R.; Cao, R.; Ma, X.; et al. Two Major Duplication Events Shaped the Transcription Factor Repertoires in Solanaceae Species. Sci. Hortic. 2024, 323, 112484. [Google Scholar] [CrossRef]
- Yang, Z.; Nielsen, R. Estimating Synonymous and Nonsynonymous Substitution Rates Under Realistic Evolutionary Models. Mol. Biol. Evol. 2000, 17, 32–43. [Google Scholar] [CrossRef]
- Xue, Y.; Zhao, Y.; Zhang, Y.; Wang, R.; Li, X.; Liu, Z.; Wang, W.; Zhu, S.; Fan, Y.; Xu, L.; et al. Insights into the Genomic Divergence of Maize Heterotic Groups in China. J. Integr. Plant Biol. 2025, 67, 1467–1486. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, Y.; Liang, X.; Zhuang, J.; Wang, X.; Qin, F.; Jiang, C. A Dirigent Family Protein Confers Variation of Casparian Strip Thickness and Salt Tolerance in Maize. Nat. Commun. 2022, 13, 2222. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Xue, Y.; Xie, Y.; Zhao, Y.; Yang, W.; Yang, W.; Wang, F.; Ren, X. Analysis of the Dirigent Pan-Gene Family in 26 Diverse Inbred Lines Reveals Genomic Diversity in Maize. Genes 2025, 16, 1285. https://doi.org/10.3390/genes16111285
Liu Z, Xue Y, Xie Y, Zhao Y, Yang W, Yang W, Wang F, Ren X. Analysis of the Dirigent Pan-Gene Family in 26 Diverse Inbred Lines Reveals Genomic Diversity in Maize. Genes. 2025; 16(11):1285. https://doi.org/10.3390/genes16111285
Chicago/Turabian StyleLiu, Zhihao, Yingjie Xue, Yuxi Xie, Yikun Zhao, Wei Yang, Weiguang Yang, Fengge Wang, and Xuejiao Ren. 2025. "Analysis of the Dirigent Pan-Gene Family in 26 Diverse Inbred Lines Reveals Genomic Diversity in Maize" Genes 16, no. 11: 1285. https://doi.org/10.3390/genes16111285
APA StyleLiu, Z., Xue, Y., Xie, Y., Zhao, Y., Yang, W., Yang, W., Wang, F., & Ren, X. (2025). Analysis of the Dirigent Pan-Gene Family in 26 Diverse Inbred Lines Reveals Genomic Diversity in Maize. Genes, 16(11), 1285. https://doi.org/10.3390/genes16111285




