Molecular Characteristics, Circularization Mechanism, and Potential Functions of circRNA-0172
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Isolation and Culture
2.2. Total RNA Extraction and Quality Detection
2.3. Genomic DNA Extraction
2.4. Validation of circRNA-0172
2.5. Quantitative Real-Time PCR (qRT-PCR) Analysis
2.6. Subcellular Localization
2.7. RNase Digestion Assay
2.8. Vector Construction
2.9. Cell Transfection
2.10. Cell Viability Assay
2.11. Bioinformatics Analysis
2.12. Statistical Analysis
3. Results
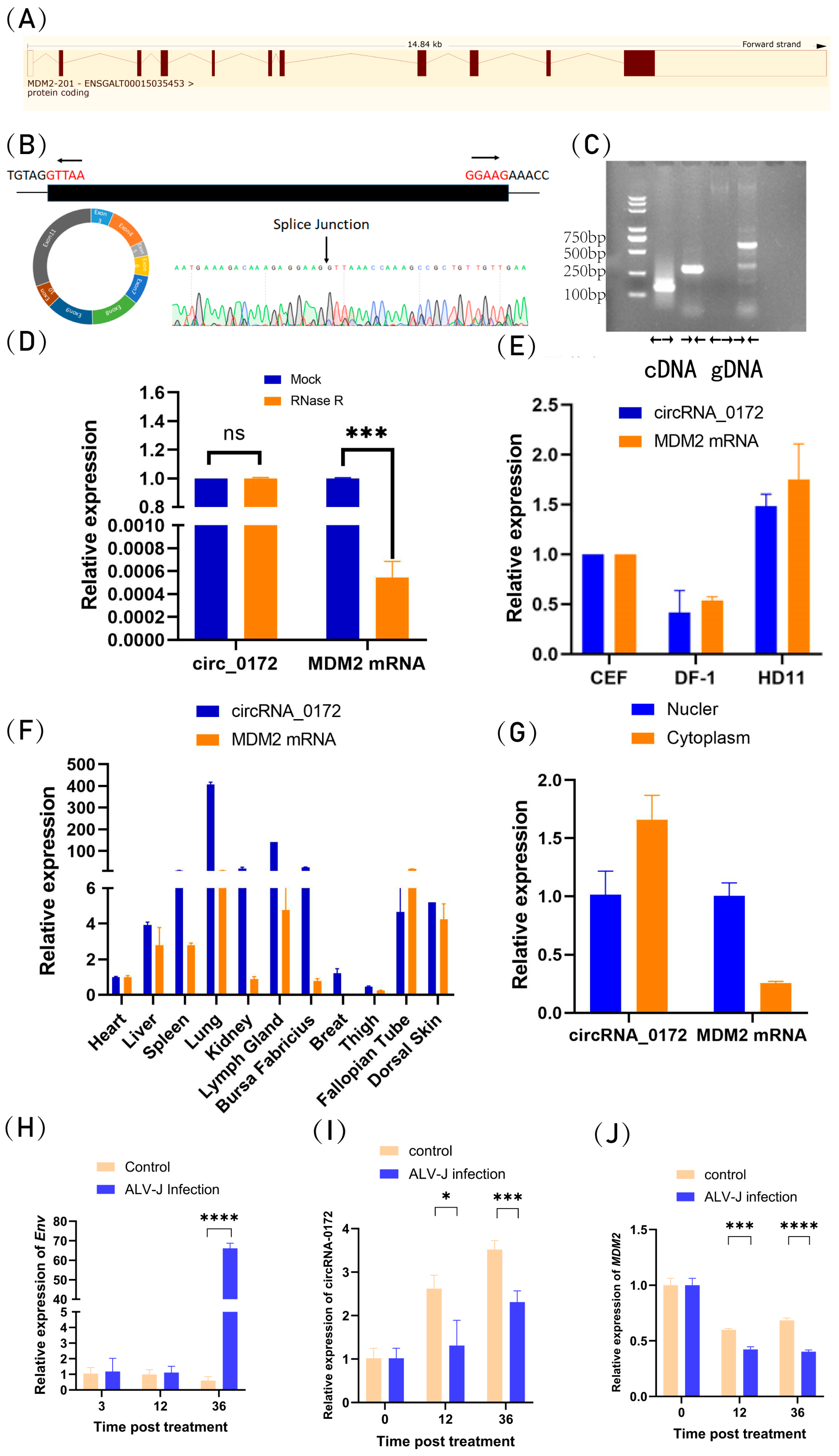
3.1. Characterization of Chicken circRNA-0172
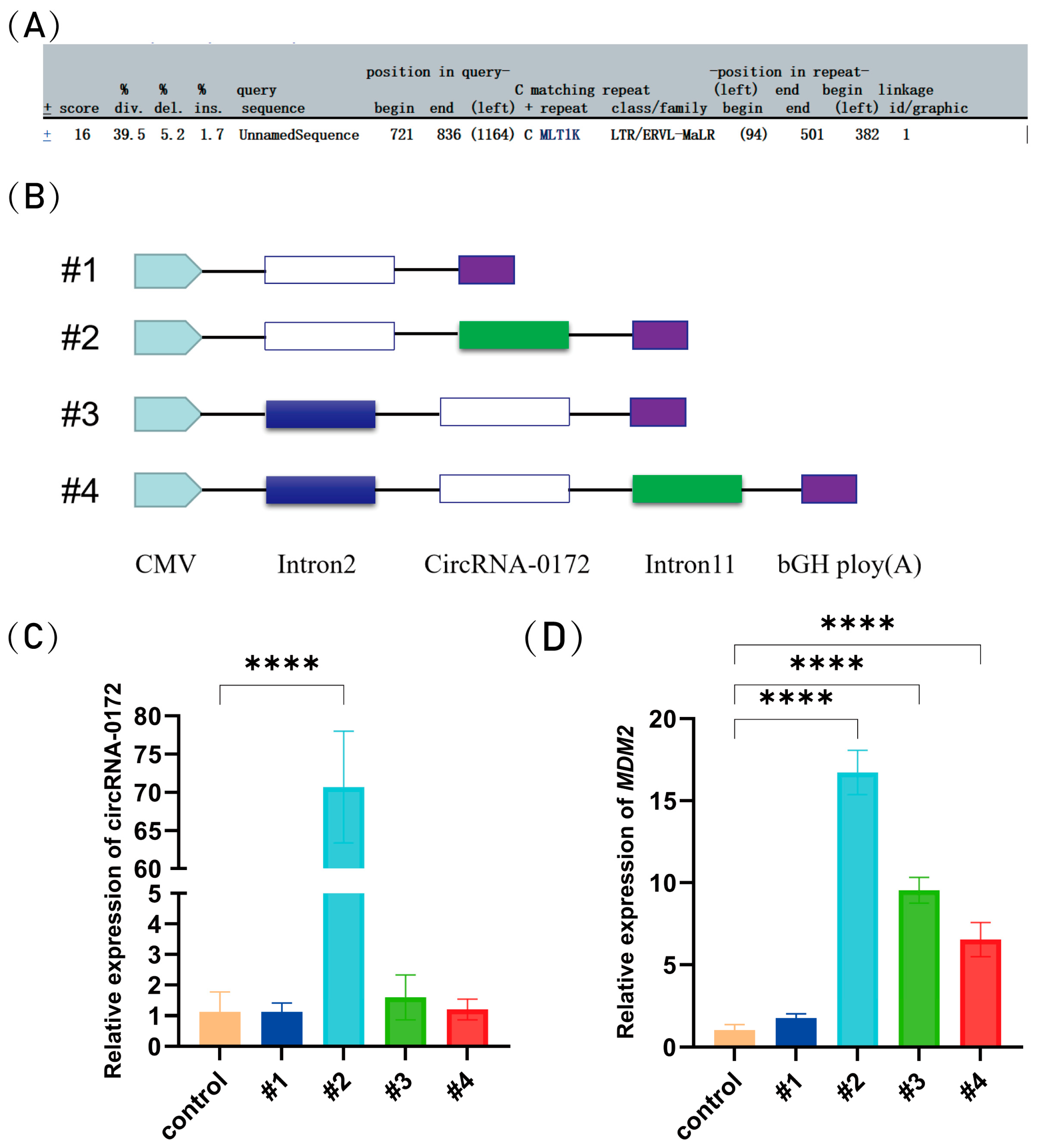
3.2. Circularization of circRNA-0172 Depends on Flanking Sequences
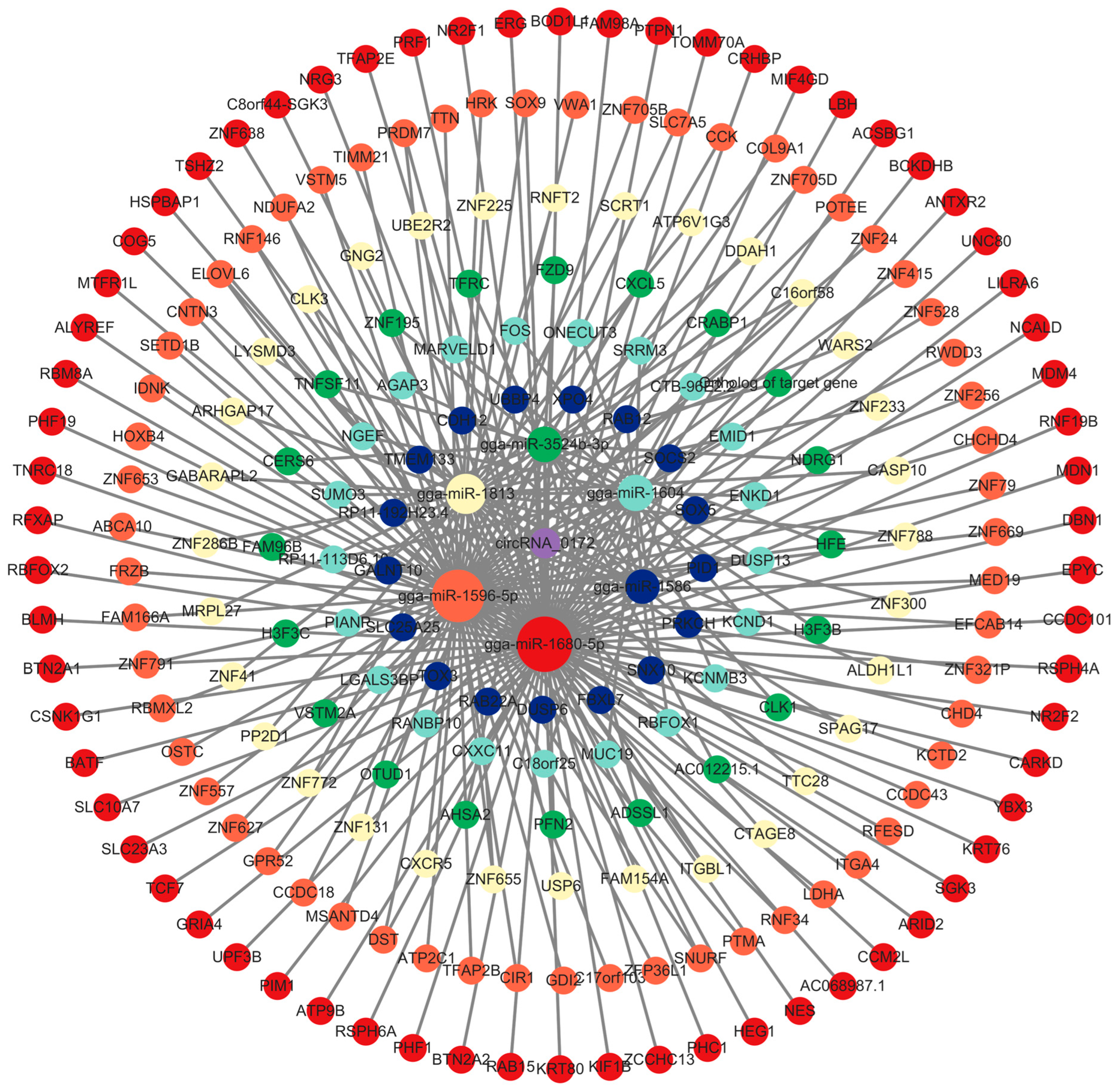
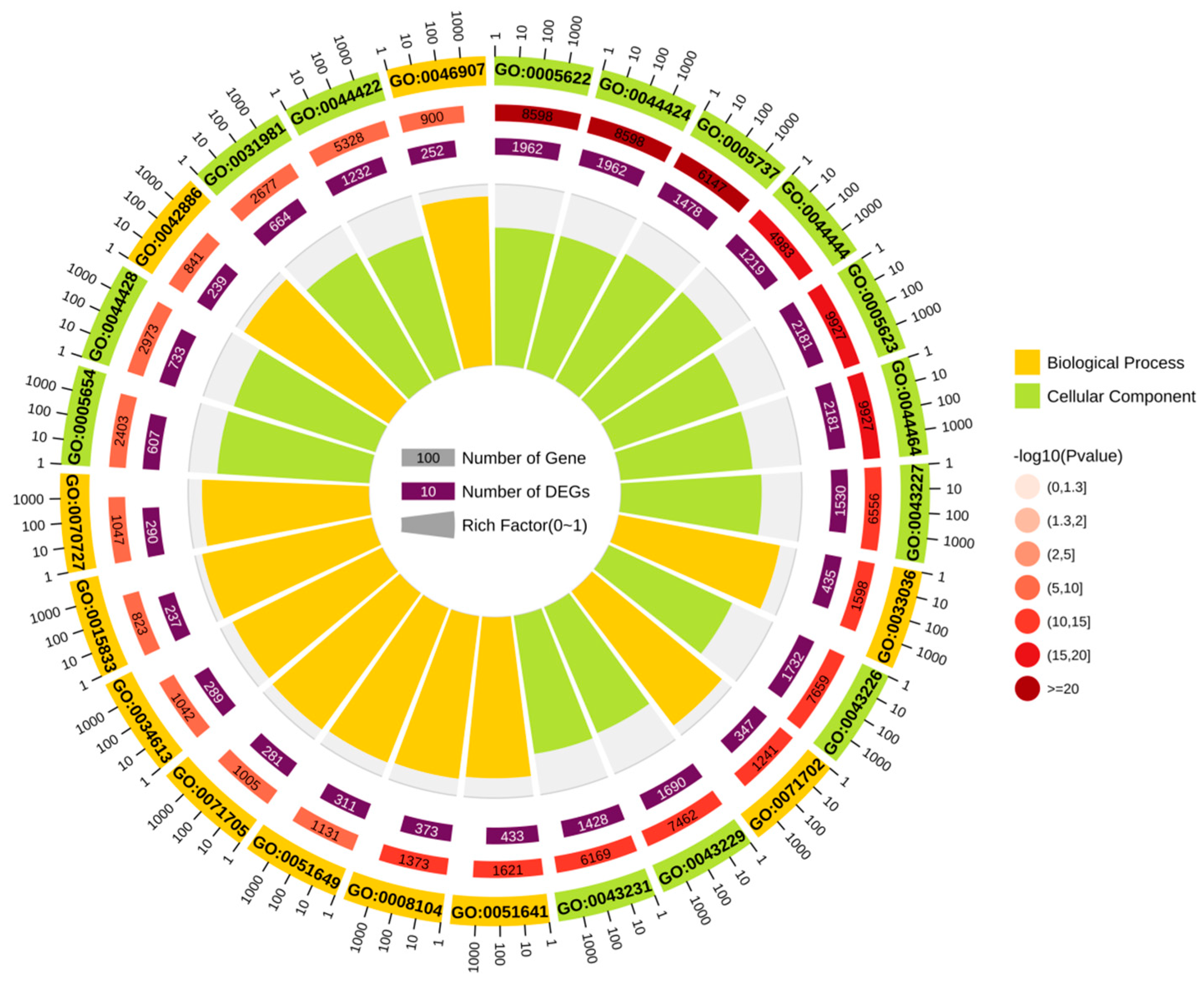
3.3. Prediction of Potential Function of circRNA-0172
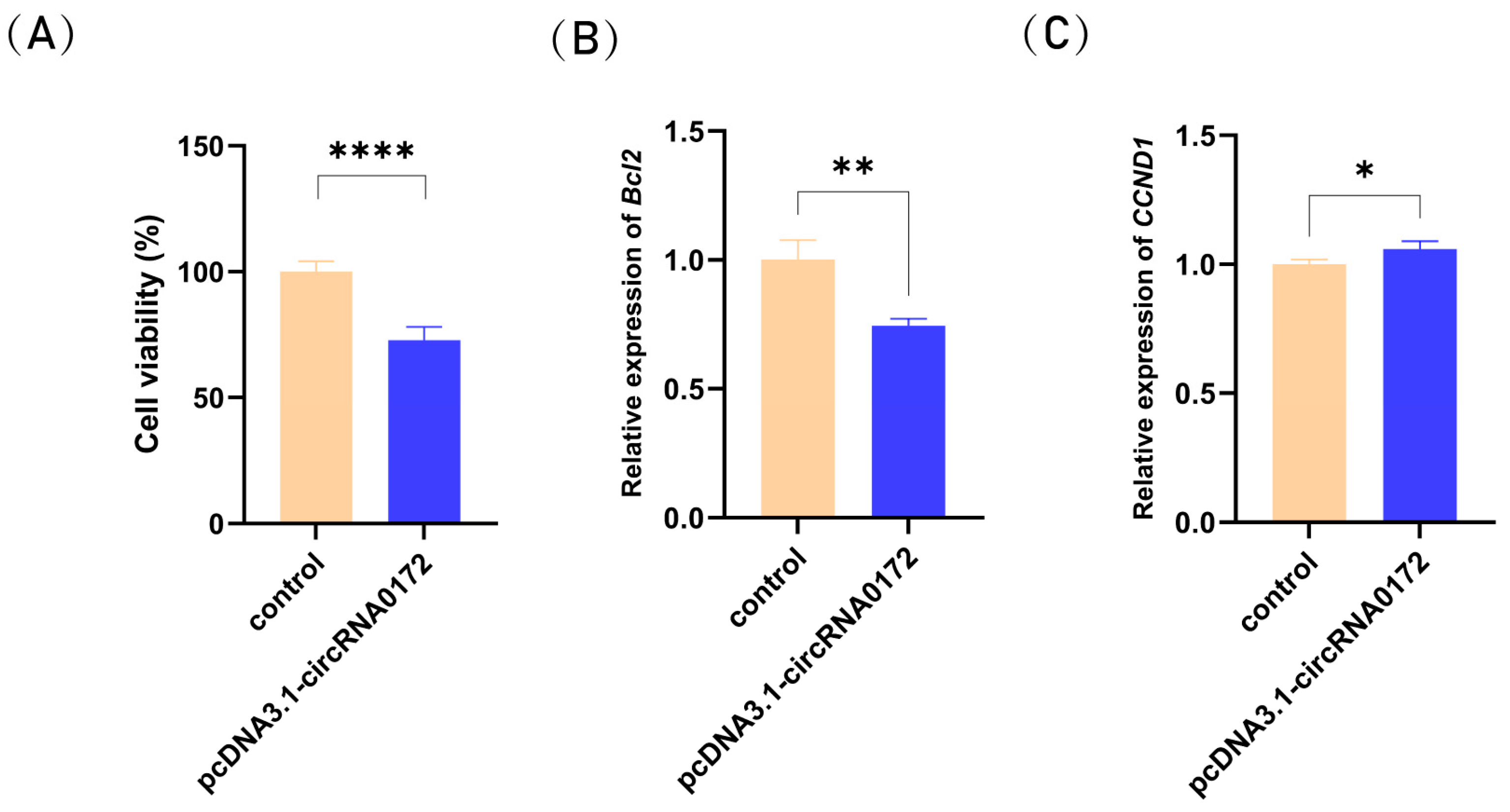
3.4. circRNA-0172 Promotes Apoptosis and Inhibits Proliferation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ALV-J | Avian leukosis virus subgroup J |
| MDM2 | Mouse doubleminute 2 homolog |
| DF-1 | Douglas Foster-1 |
References
- Fan, Y.-J.; Ding, Z.; Zhang, Y.; Su, R.; Yue, J.-L.; Liang, A.-M.; Huang, Q.-W.; Meng, Y.-R.; Li, M.; Xue, Y.; et al. Sex-lethal regulates back-splicing and generation of the sex-differentially expressed circular RNAs. Nucleic Acids Res. 2023, 51, 5228–5241. [Google Scholar] [CrossRef]
- Wang, M.; Yu, F.; Wu, W.; Zhang, Y.; Chang, W.; Ponnusamy, M.; Wang, K.; Li, P. Circular RNAs: A novel type of non-coding RNA and their potential implications in antiviral immunity. Int. J. Biol. Sci. 2017, 13, 1497–1506. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Wang, S.K.; Belk, J.A.; Amaya, L.; Li, Z.; Cardenas, A.; Abe, B.T.; Chen, C.-K.; Wender, P.A.; Chang, H.Y. Engineering circular RNA for enhanced protein production. Nat. Biotechnol. 2023, 41, 293, Erratum in Nat. Biotechnol. 2023, 41, 262–272. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-X.; Li, X.; Nan, F.; Jiang, S.; Gao, X.; Guo, S.-K.; Xue, W.; Cui, Y.; Dong, K.; Ding, H.; et al. Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity. Cell 2019, 177, 865–880. [Google Scholar] [CrossRef] [PubMed]
- Misir, S.; Wu, N.; Yang, B.B. Specific expression and functions of circular RNAs. Cell Death Differ. 2022, 29, 481–491. [Google Scholar] [CrossRef]
- Jiao, J.; Zhang, Z.J. Nuclear circRNA: Impairing genome stability via circR loops. Clin. Transl. Discov. 2023, 3, e229. [Google Scholar] [CrossRef]
- Hoque, P.; Romero, B.; Akins, R.E.; Batish, M. Exploring the Multifaceted Biologically Relevant Roles of circRNAs: From Regulation, Translation to Biomarkers. Cells 2023, 12, 2813. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Chen, G.; Shi, Y.; Liu, M.; Sun, J. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. 2018, 9, 175. [Google Scholar] [CrossRef]
- Abdelmohsen, K.; Panda, A.C.; Munk, R.; Grammatikakis, I.; Dudekula, D.B.; De, S.; Kim, J.; Noh, J.H.; Kim, K.M.; Martindale, J.L.; et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017, 14, 361–369. [Google Scholar] [CrossRef]
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A.; et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016, 7, 12429. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2017, 24, 194, Erratum in Nat. Struct. Mol. Biol. 2017, 24, 194. [Google Scholar] [CrossRef]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37. [Google Scholar] [CrossRef]
- Yang, Y.; Gao, X.; Zhang, M.; Yan, S.; Sun, C.; Xiao, F.; Huang, N.; Yang, X.; Zhao, K.; Zhou, H.; et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J. Natl. Cancer Inst. 2018, 110, 304–315. [Google Scholar] [CrossRef]
- Momand, J.; Zambetti, G.P.; Olson, D.C.; George, D.; Levine, A.J. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 1992, 69, 1237–1245. [Google Scholar] [CrossRef]
- Deb, S.P.; Singh, S.; Deb, S. MDM2 overexpression, activation of signaling networks, and cell proliferation. Sub-Cell. Biochem. 2014, 85, 215–234. [Google Scholar] [CrossRef]
- Wang, W.; Albadari, N.; Du, Y.; Fowler, J.F.; Sang, H.T.; Xian, W.; McKeon, F.; Li, W.; Zhou, J.; Zhang, R. MDM2 Inhibitors for Cancer Therapy: The Past, Present, and Future. Pharmacol. Rev. 2024, 76, 414–453. [Google Scholar] [CrossRef]
- Haupt, Y.; Maya, R.; Kazaz, A.; Oren, M. MDM2 promotes the rapid degradation of p53. Nature 1997, 387, 296–299. [Google Scholar] [CrossRef] [PubMed]
- Kubbutat, M.H.; Jones, S.N.; Vousden, K.H. Regulation of p53 stability by MDM2. Nature 1997, 387, 299–303. [Google Scholar] [CrossRef]
- Chinnam, M.; Xu, C.; Lama, R.; Zhang, X.; Cedeno, C.D.; Wang, Y.; Stablewski, A.B.; Goodrich, D.W.; Wang, X. MDM2 E3 ligase activity is essential for p53 regulation and cell cycle integrity. PLoS Genet. 2022, 18, e1010171. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, K.; Taya, Y.; Tamai, K.; Yamaizumi, M. Requirement of ATM in phosphorylation of the human p53 protein at serine 15 following DNA double-strand breaks. Mol. Cell. Biol. 1999, 19, 2828–2834. [Google Scholar] [CrossRef]
- Saito, S.; Goodarzi, A.A.; Higashimoto, Y.; Noda, Y.; Lees-Miller, S.P.; Appella, E.; Anderson, C.W. ATM mediates phosphorylation at multiple p53 sites, including Ser(46), in response to ionizing radiation. J. Biol. Chem. 2002, 277, 12491–12494. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-H.; Sheetz, M.P.P. Transcription-independent functions of p53 in DNA repair pathway selection. Bioessays 2023, 45, e2200122. [Google Scholar] [CrossRef]
- el-Deiry, W.S.; Tokino, T.; Velculescu, V.E.; Levy, D.B.; Parsons, R.; Trent, J.M.; Lin, D.; Mercer, W.E.; Kinzler, K.W.; Vogelstein, B. WAF1, a potential mediator of p53 tumor suppression. Cell 1993, 75, 817–825. [Google Scholar] [CrossRef]
- Shimada, S.; Ishizawa, T.; Ishizawa, K.; Matsumura, T.; Hasegawa, T.; Hirose, T. The value of MDM2 and CDK4 amplification levels using real-time polymerase chain reaction for the differential diagnosis of liposarcomas and their histologic mimickers. Hum. Pathol. 2006, 37, 1123–1129. [Google Scholar] [CrossRef]
- Mejia-Guerrero, S.; Quejada, M.; Gokgoz, N.; Gill, M.; Parkes, R.K.; Wunder, J.S.; Andrulis, I.L. Characterization of the 12q15 MDM2 and 12q13-14 CDK4 Amplicons and Clinical Correlations in Osteosarcoma. Genes Chromosomes Cancer 2010, 49, 518–525. [Google Scholar] [CrossRef]
- Yoon, Y.J.; Chang, H.Y.; Ahn, S.H.; Kim, J.K.; Park, Y.K.; Kang, D.R.; Park, J.Y.; Myoung, S.M.; Kim, D.Y.; Chon, C.Y.; et al. MDM2 and p53 polymorphisms are associated with the development of hepatocellular carcinoma in patients with chronic hepatitis B virus infection. Carcinogenesis 2008, 29, 1192–1196. [Google Scholar] [CrossRef]
- Michalk, M.; Meinrath, J.; Kuenstlinger, H.; Koitzsch, U.; Drebber, U.; Merkelbach-Bruse, S.; Bollschweiler, E.; Kloth, M.; Hartmann, W.; Hoelscher, A.; et al. MDM2 gene amplification in esophageal carcinoma. Oncol. Rep. 2016, 35, 2223–2227. [Google Scholar] [CrossRef] [PubMed]
- Bond, G.L.; Hu, W.W.; Bond, E.E.; Robins, H.; Lutzker, S.G.; Arva, N.C.; Bargonetti, J.; Bartel, F.; Taubert, H.; Wuerl, P.; et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell 2004, 119, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Vousden, K.H.; Prives, C. Blinded by the Light: The Growing Complexity of p53. Cell 2009, 137, 413–431. [Google Scholar] [CrossRef]
- Shinohara, T.; Uesugi, M. In-vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Tanpakushitsu Kakusan Koso. Protein Nucleic Acid Enzym. 2007, 52, 1816–1817. [Google Scholar]
- Katz, C.; Low-Calle, A.M.; Choe, J.H.; Laptenko, O.; Tong, D.; Joseph-Chowdhury, J.-S.N.; Garofalo, F.; Zhu, Y.; Friedler, A.; Prives, C. Wild-type and cancer-related p53 proteins are preferentially degraded by MDM2 as dimers rather than tetramers. Genes Dev. 2018, 32, 430–447. [Google Scholar] [CrossRef]
- Thomasova, D.; Mulay, S.R.; Bruns, H.; Anders, H.-J. p53-Independent Roles of MDM2 in NF-κB Signaling: Implications for Cancer Therapy, Wound Healing, and Autoimmune Diseases. Neoplasia 2012, 14, 1097–1101. [Google Scholar] [CrossRef]
- Senturk, E.; Manfredi, J.J. MDM2 and tumorigenesis: Evolving theories and unsolved mysteries. Genes Cancer 2012, 3, 192–198. [Google Scholar] [CrossRef]
- Brummer, T.; Zeiser, R. The role of the MDM2/p53 axis in antitumor immune responses. Blood 2024, 143, 2701–2709. [Google Scholar] [CrossRef]
- Koo, N.; Sharma, A.K.; Narayan, S. Therapeutics Targeting p53-MDM2 Interaction to Induce Cancer Cell Death. Int. J. Mol. Sci. 2022, 23, 5005. [Google Scholar] [CrossRef] [PubMed]
- Rayburn, E.; Zhang, R.W.; He, J.; Wang, H. MDM2 and human malignancies: Expression, clinical pathology, prognostic markers, and implications for chemotherapy. Curr. Cancer Drug Targets 2005, 5, 27–41. [Google Scholar] [CrossRef]
- Bueso-Ramos, C.E.; Manshouri, T.; Haidar, M.A.; Yang, Y.; McCown, P.; Ordonez, N.; Glassman, A.; Sneige, N.; Albitar, M. Abnormal expression of MDM-2 in breast carcinomas. Breast Cancer Res. Treat. 1996, 37, 179–188. [Google Scholar] [CrossRef]
- Teoh, G.; Urashima, M.; Ogata, A.; Chauhan, D.; Decaprio, J.A.; Treon, S.P.; Schlossman, R.L.; Anderson, K.C. MDM2 protein overexpression promotes proliferation and survival of multiple myeloma cells. Blood 1997, 90, 1982–1992. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Chang, G.; Li, Z.; Bi, Y.; Liu, X.; Chen, G. Comprehensive Transcriptome Analysis Reveals Competing Endogenous RNA Networks During Avian Leukosis Virus, Subgroup J-Induced Tumorigenesis in Chickens. Front. Physiol. 2018, 9, 996. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Li, Z.; Chen, B.; Zhao, J.; Yu, H.; Hu, J.; Lai, H.; Zhang, H.; Li, Y.; Meng, Z.; et al. RNA splicing junction landscape reveals abundant tumor-specific transcripts in human cancer. Cell Rep. 2024, 43, 114893. [Google Scholar] [CrossRef]
- Diener, C.; Keller, A.; Meese, E. The miRNA-target interactions: An underestimated intricacy. Nucleic Acids Res. 2024, 52, 1544–1557. [Google Scholar] [CrossRef] [PubMed]
- Bisogno, L.S.Z. Post-Transcriptional Regulation of Cancer Traits and Gene Expression in a Genetically Defined, Primary Cell-Derived Model of Breast Tumorigenesis. Ph.D. Thesis, Duke University, Durham, NC, USA, 2017. [Google Scholar]
- Karni-Schmidt, O.; Lokshin, M.; Prives, C. The Roles of MDM2 and MDMX in Cancer. Annu. Rev. Pathol. 2016, 11, 617–644. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Fu, J.; Wu, J.; Ren, J.; Xiang, R.; Kong, C.; Fu, T. CircFBXW4 Suppresses Colorectal Cancer Progression by Regulating the MiR-338-5p/SLC5A7 Axis. Adv. Sci. 2024, 11, e2300129. [Google Scholar] [CrossRef]
- Yao, S.; Jia, X.; Wang, F.; Sheng, L.; Song, P.; Cao, Y.; Shi, H.; Fan, W.; Ding, X.; Gao, S.-J.; et al. CircRNA ARFGEF1 functions as a ceRNA to promote oncogenic KSHV-encoded viral interferon regulatory factor induction of cell invasion and angiogenesis by upregulating glutaredoxin 3. PLoS Pathog. 2021, 17, e1009294. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Zhao, X.; Liu, J.; Yang, W. Epstein-Barr virus circRNAome as host miRNA sponge regulates virus infection, cell cycle, and oncogenesis. Bioengineered 2019, 10, 593–603. [Google Scholar] [CrossRef]


| Primer Name | Primer Sequence (5′–3′) |
|---|---|
| circRNA-0172 (divergent primer) | F: CGAAGATTATTCTCAGCCAT R: CACCAGCTAACTTCAACAG |
| Convergent primer | F: GTTAAACCAAAGCCGCTGTTG R: CTCTTCCTCTTTGTCTTTCATTTCTTT |
| MDM2 | F: CTCTACGCTGGCTGTTCC R: AATCGCTGCTATTGCTCC |
| GAPDH | F: CGATCTGAACTACATGGTTTAC R: TCTGCCCATTTGATGTTGC |
| #4-Intron2 | F: agaGGAAGAGGTATGGAGTCCAGTCTGCCTGT R:aacgggccctctagactcgagAGTTCTTCATTTGTCTTTCCTGTTCACAT |
| #4-0172 | F: CTCCACACAGGTTAAACCAAAGCCGCTGTTGTTG R: GACTCCATACCTCTTCCtctttgtctttcatttctttcttctcag |
| #4-Intron11 | F: tagtccagtgtggtggaattcTGCGCTCCCCACGTGG R: TTGGTTTAACCTGTGTGGAGATGAAAACAGAAGAACAG |
| #3-Intron2 | F: agaGGAAGAGGTATGGAGTCCAGTCTGCCTGT R:aacgggccctctagactcgagAGTTCTTCATTTGTCTTTCCTGTTCACAT |
| #3-0172 | F: CTCCACACAGGTTAAACCAAAGCCGCTGTTGTTG R:AACGGGCCCTCTAGACTCGAGCTTCCtctttgtctttcatttctttcttctcag |
| #2-0172 | F: tagtccagtgtggtggaattcGTTAAACCAAAGCCGCTGTTGTTGA R: GACTCCATACCTCTTCCtctttgtctttcatttctttcttctcag |
| #2-Intron11 | F: tagtccagtgtggtggaattcTGCGCTCCCCACGTGG R: TTGGTTTAACCTGTGTGGAGATGAAAACAGAAGAACAG |
| #1-0172 | F: tagtccagtgtggtggaattcGTTAAACCAAAGCCGCTGTTGTTGA R:AACGGGCCCTCTAGACTCGAGCTTCCtctttgtctttcatttctttcttctcag |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Li, W.; Yang, T.; Bai, H.; Qiu, L.; Chang, G. Molecular Characteristics, Circularization Mechanism, and Potential Functions of circRNA-0172. Genes 2025, 16, 1282. https://doi.org/10.3390/genes16111282
Wang H, Li W, Yang T, Bai H, Qiu L, Chang G. Molecular Characteristics, Circularization Mechanism, and Potential Functions of circRNA-0172. Genes. 2025; 16(11):1282. https://doi.org/10.3390/genes16111282
Chicago/Turabian StyleWang, Haojie, Wenhao Li, Ting Yang, Hao Bai, Lingling Qiu, and Guobin Chang. 2025. "Molecular Characteristics, Circularization Mechanism, and Potential Functions of circRNA-0172" Genes 16, no. 11: 1282. https://doi.org/10.3390/genes16111282
APA StyleWang, H., Li, W., Yang, T., Bai, H., Qiu, L., & Chang, G. (2025). Molecular Characteristics, Circularization Mechanism, and Potential Functions of circRNA-0172. Genes, 16(11), 1282. https://doi.org/10.3390/genes16111282





