1. Introduction
SATB2-associated syndrome (SAS; OMIM #612313), also called Glass syndrome, is a rare multisystemic disorder caused by haploinsufficiency or dysfunction of the
SATB2 gene (OMIM #608148), located on chromosome 2q33.1. Clinically, SAS is primarily characterized by neurodevelopmental impairment with severely limited or absent expressive speech, intellectual disability, and dental anomalies. Additional recurrent features include craniofacial dysmorphisms, palatal defects, variable behavioral anomalies, growth retardation, low bone density, and epilepsy [
1].
The most distinctive hallmark of SAS is the early and persistent deficit in verbal communication, with nearly all affected individuals presenting delayed language milestones, often progressing to absent or extremely limited speech [
1,
2]. Orofacial abnormalities, mostly represented by dental anomalies and cleft palate, are reported in up to 50% of cases and have been associated with more severe language and feeding difficulties [
2]. In addition, a failure to thrive, associated with normal linear growth, is frequently observed in individuals with SAS, often accompanied by scoliosis and low bone density [
2,
3].
The
SATB2 gene encodes a nuclear matrix-associated transcription factor that organizes higher-order chromatin architecture and regulates transcriptional programs essential for neurodevelopment, craniofacial morphogenesis, and osteogenesis [
4]. SATB2 binds matrix-attachment regions (MARs) of DNA and recruits chromatin-modifying enzymes, including histone deacetylases (HDACs) and acetyltransferases (HATs), to control gene activation or repression at key regulatory elements [
5]. During corticogenesis, SATB2 represses Ctip2/BCL11B to specify upper-layer cortical projection neurons, thereby orchestrating callosal connectivity [
6,
7]. In skeletal development, SATB2 functions as a molecular hub by directly binding enhancers of osteogenic genes such as
Hoxa2,
IBSP, and
BGLAP, and synergistically interacting with ATF4 and RUNX2 to potentiate osteoblast differentiation [
8]. Mice with combined Satb2 and Runx2 or Atf4 deficiency exhibit severe skeletal defects, underscoring SATB2 pivotal role in bone formation [
4,
9].
Genotype–phenotype correlation studies suggest that variant position within SATB2 domains influences clinical severity. Missense variants located outside the main functional domains are associated with lower scores in impaired cognition, behavior, sleep, and sialorrhea categories, reflecting milder neurodevelopmental impairment.
To better systematize this phenotypic variability, Zarate et al. [
10] developed a standardized SAS severity scoring system, enabling structured evaluation of neurodevelopmental and systemic involvement across 164 genetically confirmed cases. This tool revealed that null variants and large chromosomal deletions are associated with a more severe phenotype, especially in terms of communication, feeding, and adaptive functioning.
Although knowledge of SAS has expanded, its diagnosis during early childhood remains challenging, especially when craniofacial dysmorphisms are subtle or absent. The present study contributes to the clinical delineation of SAS by reporting a new case series of molecularly confirmed individuals, with detailed phenotypic documentation. Our aim is to enrich current understanding of the clinical spectrum, particularly in relation to neurodevelopment, craniofacial traits, and familial background, and to contribute to future genotype–phenotype stratification efforts.
3. Results
3.1. Patients Descriptions (Table 1)
Patient 1 (
Figure 1A): The patient is a male who was referred to our unit at the age of 56 yrs. for a pervasive NeuroDevelopmental Disorder (NDD) characterized by absent speech, autistic behavior, and moderate intellectual disability. The NDD was related to an episode of infectious encephalitis, although the etiology was never clearly established. This occurred in conjunction with a single febrile seizure. Brain Magnetic Resonance Imaging (MRI) was largely unremarkable, except for an isolated focal area of gliosis of nonspecific significance located in the subcortical white matter of the right superior frontal gyrus. Neuropsychiatric evaluation revealed good receptive verbal comprehension, use of gestural communication, sleep disturbances, and ritualistic, stereotyped, and compulsive behaviors, including self-directed aggression. A significant impairment was observed across all domains of adaptive functioning. In recent years, episodes of fluctuating lower limb hyposthenia have been reported. The patient also exhibits peculiar eating behavior, including hyperphagia with a BMI of 20 (stature of 175 cm and weight of 61 kg). Dental anomalies could not be fully characterized due to multiple prior dental extractions, which have resulted in partial edentulism with only the incisors remaining. The patient also presents with bilateral pes planus and brachydactyly with an enlargement of the distal phalanx of the thumb. Genetic testing included array-CGH, which yielded non-diagnostic results. Singleton exome sequencing (parental samples were unavailable) subsequently identified a heterozygous in-frame duplication of two amino acids in the
SATB2 gene (NM_001172509.2): c.961_966dup, p.(Ile321_Ala322dup).
Patient 2 (
Figure 1B): This patient was a male born at term (40 weeks of gestation). Neonatal auxometric parameters were within normal limits; the Apgar score was 9 at the first minute. In the first days of life, facial dysmorphisms were noted, including a mildly flattened nasal bridge, subtle midface hypoplasia, laterally sparse eyebrows, and syndactyly of the proximal phalanges of the second and third toes. At 19 months of age, the patient was diagnosed with laryngomalacia, which required endoscopic correction. Independent walking was achieved at 6 years of age and was described as markedly delayed and unstable, with a persistently flexed posture. At the most recent clinical evaluation (10 years of age), anthropometric measurements were: weight 25 kg (5th centile), height 127 cm (4th centile), and head circumference 52 cm (27th centile). Brain MRI revealed diffuse white matter demyelination, ventricular enlargement, and global white matter volume reduction. Ophthalmologic evaluation documented reduced visual acuity, likely of neurological (cortical) origin, in the absence of ocular abnormalities. Genetic work-up included array-CGH, which was non-diagnostic. Trio exome sequencing subsequently identified a de novo heterozygous missense variant in the
SATB2 gene (NM_001172509.2): c.1573G>A, resulting in the amino acid substitution p.(Glu525Lys).
Patient 3 (
Figure 1C): A 9-year-old female was referred for global NDD and facial dysmorphisms, with an unremarkable family history. The pregnancy was reported as uneventful, with delivery occurring at 40 + 0 weeks of gestation. Birth parameters included a weight of 3790 g and a length of 51 cm, OFC was not reported. Physical examination revealed multiple craniofacial anomalies, including cleft palate, unruly dentition, short philtrum, small mouth opening, under folded helices, hypertelorism, downward slanting palpebral fissures, blue sclerae, long eyebrows, and prominent ears. Independent walking was achieved at 32 months. A clinical evaluation at 4 years of age confirmed persistent motor developmental delay. At 9 years of age, the patient was able to articulate only a few words, but presented non-verbal communication abilities, eye contact, and understanding spoken language. Cognitive assessment yielded an IQ score of 50, consistent with moderate intellectual disability. Brain MRI was unremarkable apart from the presence of arachnoid cysts. No seizures have been observed to date, although electroencephalogram (EEG) recordings demonstrated generalized immaturity of background activity. Array-CGH was normal, while a trio exome sequencing identified a de novo heterozygous variant in the
SATB2 gene (NM_001172509.2): c.1196G>A, resulting in the missense change p.(Arg399His).
Patient 4: This patient was referred for global NDD in the context of an unremarkable family history. No complications were reported during pregnancy or the perinatal period. Physical examination revealed nonspecific facial dysmorphisms and an abnormal bilateral morphology of the distal phalanges of the thumbs. Neuropsychiatric evaluation highlighted global neurodevelopmental and language immaturity, with findings consistent with intellectual disability and a clinical diagnosis of autism spectrum disorder (ASD). Behavioral assessment identified obsessive–compulsive traits and episodes of aggressive behavior. The patient experienced transient epileptic events, and EEG recordings confirmed abnormal cerebral activity. The array-CGH yielded inconclusive results. Subsequent trio exome sequencing revealed a de novo heterozygous missense variant in the SATB2 gene (NM_001172509.2): c.587C>T, resulting in the amino acid substitution p.(Pro196Leu).
Patient 5: This female patient was referred at 2 years old for a significant speech delay and absent social interaction. Family history was negative, and she had no perinatal complications. Upon physical examination, she presented with a rounded facial shape, prominent nasal bridge, and micrognathia. Her developmental milestones were substantially delayed: independent walking was achieved at 24 months, and her first spoken words were not reported before 48 months, indicating a significant delay in expressive language development. A neuropsychiatric evaluation conducted at 9 years of age revealed an unstable gait and marked deficits in both gross and fine motor skills. Her expressive speech remained severely limited, and she continued to exhibit absent social engagement alongside persistent sleep disturbances. Cognitive assessment documented an IQ score of 60, consistent with moderate intellectual disability. An EEG showed an abnormal pattern characterized by alpha-theta rhythm with spikes and slow waves, as well as a degenerated spike–wave complex in the frontal regions. After a normal array-CGH, a trio exome sequencing identified a de novo heterozygous pathogenic variant in the SATB2 gene (NM_001172509.2): c.857dup, resulting in a frameshift and premature termination variant p.(Pro287AlafsTer17).
Patient 6: This 11-year-old female was born after an uneventful pregnancy and delivery. At birth, she presented with a cleft palate, lingualized lower incisors, and congenital clubfoot. At 11 years old, growth assessment showed she was underweight (25 kg, −2.2 SD), with height and head circumference at the lower limits of normal. Neurodevelopmental evaluation revealed intellectual disability and language delay, evident from early childhood. An ophthalmological assessment identified strabismus and astigmatism. Brain MRI showed hypoplasia of the corpus callosum. EEG revealed abnormalities in the absence of clinical seizures. Karyotype and array-CGH were normal. Trio exome sequencing identified a de novo in-frame deletion in the SATB2 gene (NM_001172509.2): c.1979_1981del, resulting in the deletion of one isoleucine residue p.(Ile660del). Family history was notable for a maternal uncle with a cleft lip and palate.
Table 1.
Clinical summary of six patients with Glass syndrome.
Table 1.
Clinical summary of six patients with Glass syndrome.
| Case | NDD | ASD | Language Delay | MRI Anomalies | Behav. Anom. | Motor Delay | Sleep Difficult. | Facial Dysmorphism | Cleft Palate | Dental Anomalies | Growth Retardation | Clinical Seizures |
|---|
| 1 | + | + | + | − | + | + | + | + | − | + | − | − |
| 2 | + | + | + | + | + | + | − | + | − | − | + | − |
| 3 | + | − | + | − | − | + | − | + | + | + | − | − |
| 4 | + | + | + | NA | + | + | − | + | − | − | − | + |
| 5 | + | + | + | NA | + | + | + | + | − | − | − | − |
| 6 | + | − | + | + | − | + | − | + | + | + | + | − |
3.2. Variants Analysis
The
SATB2 gene (OMIM #608148), located on chromosome 2q33, spans approximately 191 kb and comprises 12 exons. It encodes a 733-amino-acid DNA-binding protein that selectively binds AT-rich sequences and is highly conserved across species [
17,
18]. According to the Genome Aggregation Database (gnomAD v4.1;
https://gnomad.broadinstitute.org/; accessed on 1 August 2025),
SATB2 exhibits a high probability of loss-of-function intolerance (pLI = 1.0) and a Z-score of 5.53, supporting its classification as a haploinsufficient gene with strong constraint against variation.
Table 2 provides a detailed summary of the identified variants in our cases, which are also schematically summarized on the protein in
Figure 2A. Extended data on ACMG criteria and variant classification, together with Clinvar variant information, are reported in
Table S4.
3.2.1. Missense Variants
Cases 2–4 harbor de novo heterozygous missense variants: c.1573G>A p.(Glu525Lys) within the CUT2 domain, c.1196G>A p.(Arg399His) within the CUT1 domain and c.587C>T p.(Pro196Leu) within the CUTL domain, respectively, as illustrated in
Figure 2A. All variants were absent from the GnomAD population database. Scores derived from MetaRNN (
http://www.liulab.science/metarnn.html; accessed on 1 October 2025) demonstrate a supportive but non-conclusive pathogenicity assessment of all three variants.
MetaDome analysis (
https://stuart.radboudumc.nl/metadome/; accessed on 1 October 2025) confirms they are in regions intolerant to variation, further supporting their pathogenicity (
Figure 2B). Based on ACMG (
Table 2) criteria, all three variants were classified as likely pathogenic.
The c.1196G>A p.(Arg399His) variant has been previously reported in ClinVar (ID 373069), the LOVD database (ID SATB2_000007), and described by [
19]. The c.1573G>A p.(Glu525Lys) variant has also been reported in ClinVar (ID 2577951). To the best of our knowledge, the c.587C>T p.(Pro196Leu) variant has not been previously reported (
Table 2).
3.2.2. Indel Variants
The remaining three cases carry two in-frame variants (one duplication and one deletion) and one frameshift duplication, all of which are absent in the control population (gnomAD v4.1). Based on ACMG criteria [
11], all variants were classified as likely pathogenic or pathogenic (
Table 2).
MetaDome analysis (
https://stuart.radboudumc.nl/metadome/; accessed on 1 October 2025) indicates that the regions affected by the in-frame indel variants fall within protein segments that are intolerant to variation, further supporting the potential pathogenicity of these alterations (
Figure 2B).
The c.857dup p.(Pro287AlafsTer17) variant introduces a frameshift, predicted to result in nonsense mediated mRNA decay, and it has not been previously described.
The c.1979_1981del p.(Ile660del) variant results in an in-frame deletion affecting a conserved residue and has been previously reported in ClinVar as pathogenic (ID 3062110). Exploiting in silico 3D protein stability prediction analysis, we generated and compared the WT and mutated (p.(Ile660del)) form of the SATB2 HOX domain (
Figure 3). The prediction indicates a potential increase in protein stability (ΔΔG = −41.02 ± 18 kcal/mol;
Table S1). Nevertheless, these results were considered only as qualitative evidence and will require further functional assays to substantiate the interpretation of the variant’s effect (
Figure 3).
The c.961_966dup p.(Ile321_Ala322dup) variant causes an in-frame insertion involving two conserved residues and has not been previously reported. In this case, parental segregation analysis was not performed, so the de novo status could not be confirmed (
Table 2).
4. Discussion
SATB2-associated syndrome (SAS) is primarily characterized by severe speech impairment, intellectual disability, behavioral issues, palatal defects, and dental anomalies. Despite these features, a definitive clinical diagnosis is often difficult to obtain because the variable and nonspecific signs overlap with other NDDs. The diagnostic challenge is further compounded by the fact that nearly all cases result from de novo variants [
1], rendering family history uninformative. Consequently, diagnosis is frequently delayed, and it is most often confirmed through next-generation sequencing (NGS).
This manuscript reports on six novel cases of SAS, highlighting key clinical and genetic insights. We present the oldest described case of SAS in a 56-year-old male (patient 1), who demonstrates comparable age-dependent non-progressive clinical features previously described in the literature [
20]. This patient’s phenotype has been stable and non-progressive throughout his life. The only new feature to emerge was a late-onset, mild hyposthenia of the lower limbs at age 45, which appeared to be related to episodes of compulsive activity. This patient also exhibited the notable combination of hyperphagia and a normal-to-low BMI, a feature that aligns with recent in vitro evidence suggesting SATB2 dysregulation may impact the metabolism of carbohydrates and amino acids [
21,
22]. The patient’s delayed diagnosis at age 56 yr., previously misattributed to an unclarified post-infective event, underscores the critical importance of updated clinical evaluations to avoid misdiagnosis and improve patient management. Genetically, this patient carries a heterozygous in-frame duplication of two amino acids p.(Ile321_Ala322dup), affecting a highly conserved linker domain, which is classified as likely pathogenic (LP). This variant is not found in the GnomAD database, but its de novo status could not be confirmed due to parental DNA unavailability.
Our findings from two additional cases, patient 2 and patient 3, corroborate existing literature on the impact of missense variants in critical functional domains of SATB2. These patients carry de novo missense variants, p.(Arg399His) and p.(Glu525Lys), affecting the CUT1 and CUT2 domains, respectively. The CUT1 and CUT2 domains of SATB2 play a central role in its ability to function as a global chromatin organizer. These two highly conserved DNA-binding motifs act in coordination with the C-terminal homeodomain to recognize and anchor SATB2 to matrix-attachment regions (MARs), thereby establishing the chromatin loops necessary for long-range transcriptional regulation [
1]. Functional studies suggest that the CUT1 domain is particularly important for initiating the interaction with chromatin, whereas CUT2 facilitates the release of SATB2 once transcriptional remodeling has been executed, ensuring the dynamic nature of chromatin binding [
23]. Through this cycle of attachment and dissociation, SATB2 maintains a finely tuned balance between chromatin stability and plasticity, which is essential for the regulation of gene networks involved in cortical neuron specification, craniofacial patterning, and odontogenesis [
4,
24].
The clinical presentation of these two patients is remarkably severe, with a complete absence of expressive language accompanied by global motor and neurodevelopmental delay, together with variable craniofacial, skeletal, and dental anomalies. Such features reinforce the concept that specific
SATB2 variants can profoundly disrupt protein function. Previous genotype–phenotype studies have already emphasized the severe presentation in patients with CUT1 and CUT2 domains to pathogenic changes [
10].
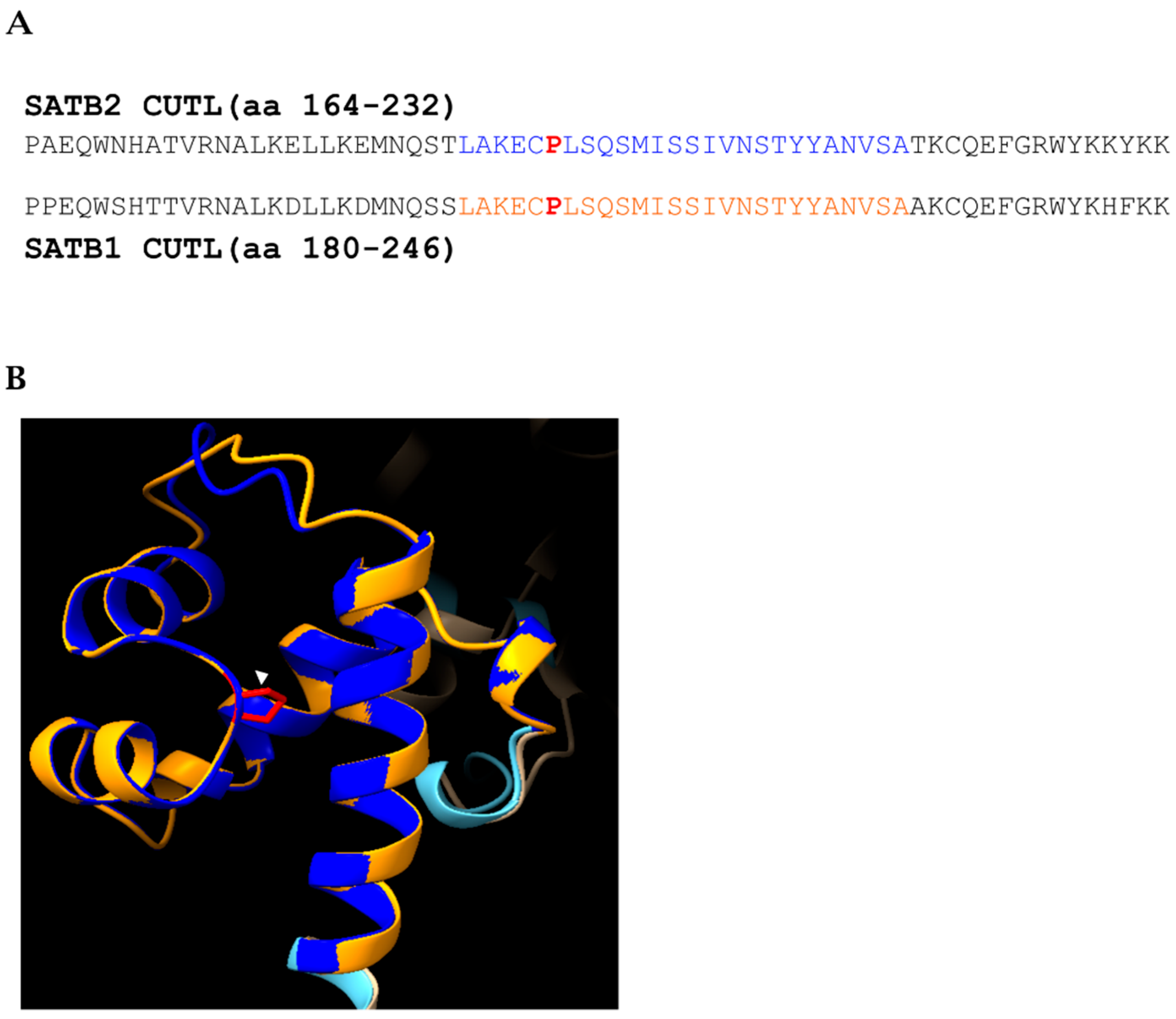
We also identified novel variants that expand our understanding of the gene’s functional domains. Patient 4 carries the p.(Pro196Leu) variant, a novel missense change in the CUT-Like (CUTL) domain. While this domain is highly conserved, its functional role in SATB2, particularly in DNA binding, remains largely unexplored [
25]. However, structural and biochemical work on SATB1 demonstrated that the ULD–CUTL tandem is essential for the multiple-domain–coordinated mechanism of DNA recognition, with CUTL contributing, beyond CUT1, CUT2 and HOX, to the DNA binding capacity of full-length protein and enabling simultaneous engagement of two DNA targets, a plausible basis for MAR-anchored loop formation. The high degree of homology (86%) between the CUTL domains of SATB1 and SATB2, reaching complete conservation at the position of the missense variant p.(Pro196Leu) (
Figure 4A,B), suggests a potential effect on ULD–CUTL coupling and oligomerization, ultimately leading to reduced multi-site DNA engagement and defective chromatin-loop organization as a potential pathogenic mechanism [
25].
Variant p.(Ile660del) was found in patient 6, affecting the HOX domain. Recently, functional assays have shown that missense variants within this domain impair transcriptional repression and disrupt normal nuclear localization, resembling the effects observed in HOX-domain-lacking truncating variants and supporting its critical role in MAR (Matrix Attachment Region) binding and chromatin remodeling activity. It is therefore conceivable that the variant p.(Ile660del) has a similar role in the loss of functional integrity of the HOX domain [
26]. Our 3D folding stability predictions suggest increased stability of the mutant protein compared with the wild type. While this finding may imply an effect on protein function, it should be interpreted with caution and confirmed through dedicated functional assays to elucidate the underlying pathogenic mechanism (
Figure 3). This represents the first reported in-frame deletion in this domain and indeed in any region of the
SATB2 gene.
Furthermore, in patients 2 and 6, neuroradiological anomalies were reported as diffuse white matter hypomyelination and volume loss for patient 2 and hypoplasia of the corpus callosum for patient 6. Those findings are consistent with previous reports that describe abnormality of white matter, delay of myelination, ventriculomegaly and small corpus callosum as the four most frequently reported neuroimaging anomalies [
19].
Interestingly, patient 2 presented with a unique visual phenotype of severely reduced visual acuity without apparent ocular abnormalities combined with neuroradiological features of diffuse white matter hypomyelination and volume loss, suggesting a cortical origin of the vision loss. It is the first time, to our knowledge, that a central vision impairment is reported in a SAS patient. According to the Human Protein Atlas, SATB2 shows strong nuclear expression in pyramidal neurons of the cerebral cortex, including the occipital lobe and visual areas, supporting its potential role in the development and function of cortical circuits essential for the processing of visual information, combined with the assessed role of SATB2 in the regulation of the callosal projection during cerebral development [
6].
A consistent finding across our cohort is a trend toward lower-end body weight in patients 1, 2, and 6. Detailed longitudinal growth curves were not available for our patients; however, the most recent clinical measurements and parental reports consistently indicated a history of chronic low weight. This finding is in line with previous observations in individuals with
SATB2-associated syndrome (SAS), where growth delay predominantly manifests as reduced weight-for-length or BMI, reflecting difficulties in sustaining adequate weight gain over time [
3]. Interestingly, the role of SATB2 as a key regulator for gene networks involved in growth and energy metabolism was suggested by recent in vitro studies that highlight a defective use of several energy substrates of cellular metabolism [
21,
22]. Further investigation using patient-derived cellular models will be essential to determine whether this metabolic dysregulation is a direct contributor to the SAS phenotype.
5. Conclusions
This case series expands the clinical and mutational spectrum of SATB2-associated syndrome (SAS) by describing six new patients carrying rare or novel SATB2 variants, including missense, in-frame, and frameshift changes. Our findings confirm the marked phenotypic variability of SAS and emphasize the importance of comprehensive genetic testing for individuals with unexplained neurodevelopmental delay, particularly when accompanied by speech impairment and craniofacial anomalies. Notably, we report the oldest, molecularly confirmed case of SAS to date, as well as a unique visual phenotype likely of cortical origin, further broadening the phenotypic landscape. In addition, the recurrent observation of growth parameters at the lower limits of normal and recent evidence of metabolic dysregulation invite future studies to explore SATB2’s role in systemic homeostasis. Altogether, our results underscore the need for integrated clinical, neuroimaging, and molecular approaches to ensure timely and accurate diagnosis, as well as to inform long-term management strategies in affected individuals.