Integrative Single-Cell and Bulk Transcriptomic Analysis Identifies Macrophage-Related Gene Signatures Predictive of Hepatocellular Carcinoma in Cirrhosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Data Preprocessing and Analysis for Single-Cell RNA Sequencing
2.3. Differential Expression Analysis and Gene Set Enrichment Analysis
2.4. Cell Interaction Analysis
2.5. High-Dimensional Weighted Gene Co-Expression Network Analysis (hdWGCNA)
2.6. Transcriptomic Data Analysis and Gene Selection
2.7. Model Construction and Performance Evaluation
3. Results
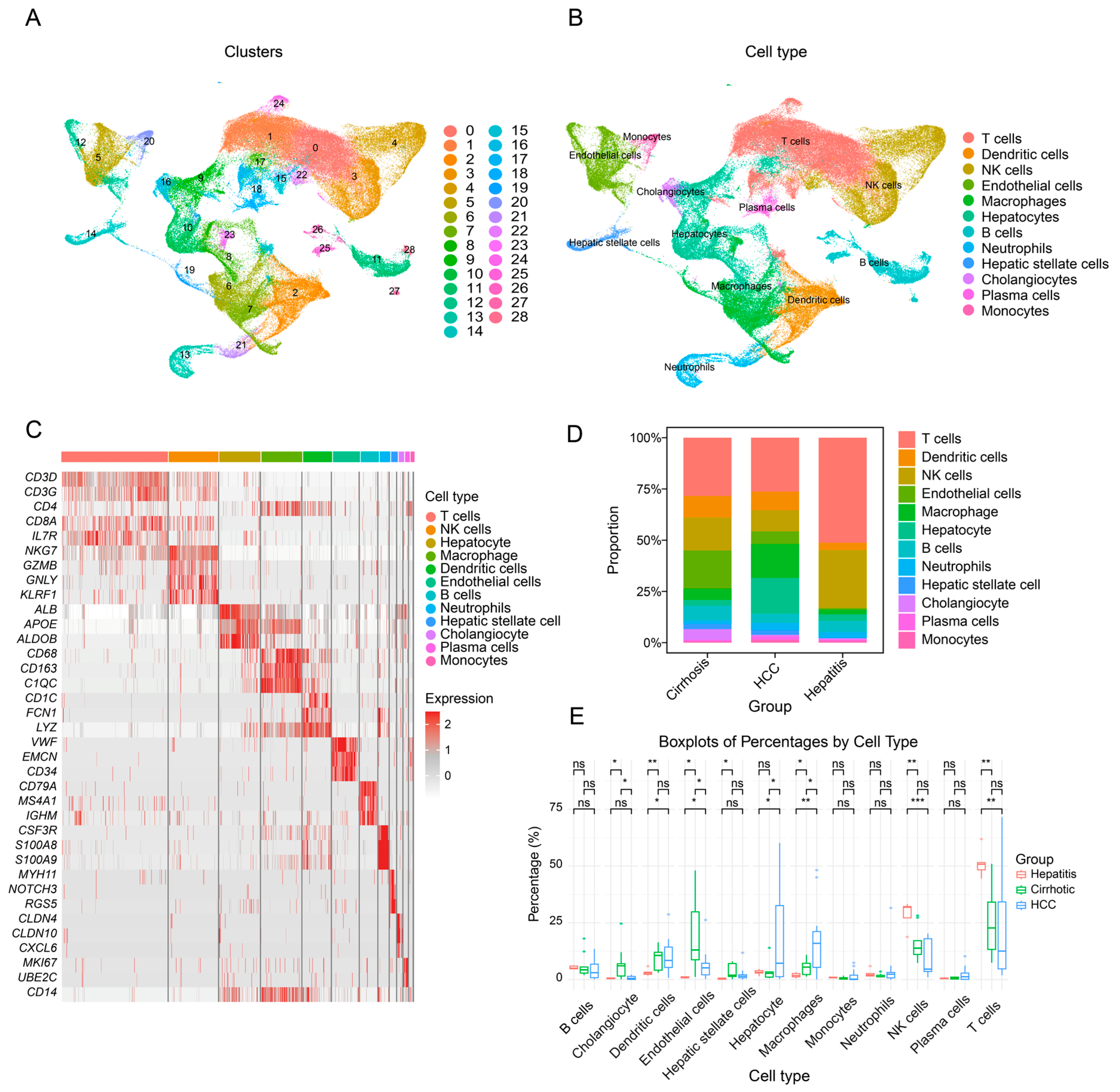
3.1. Single-Cell Transcriptomic Landscape of Hepatitis, Cirrhosis, Hepatocellular Carcinoma, and Adjacent Non-Tumorous Liver Tissue
3.2. Macrophage Profiling in Hepatic Diseases: Gene Expression, Pathways, and Polarization Dynamics
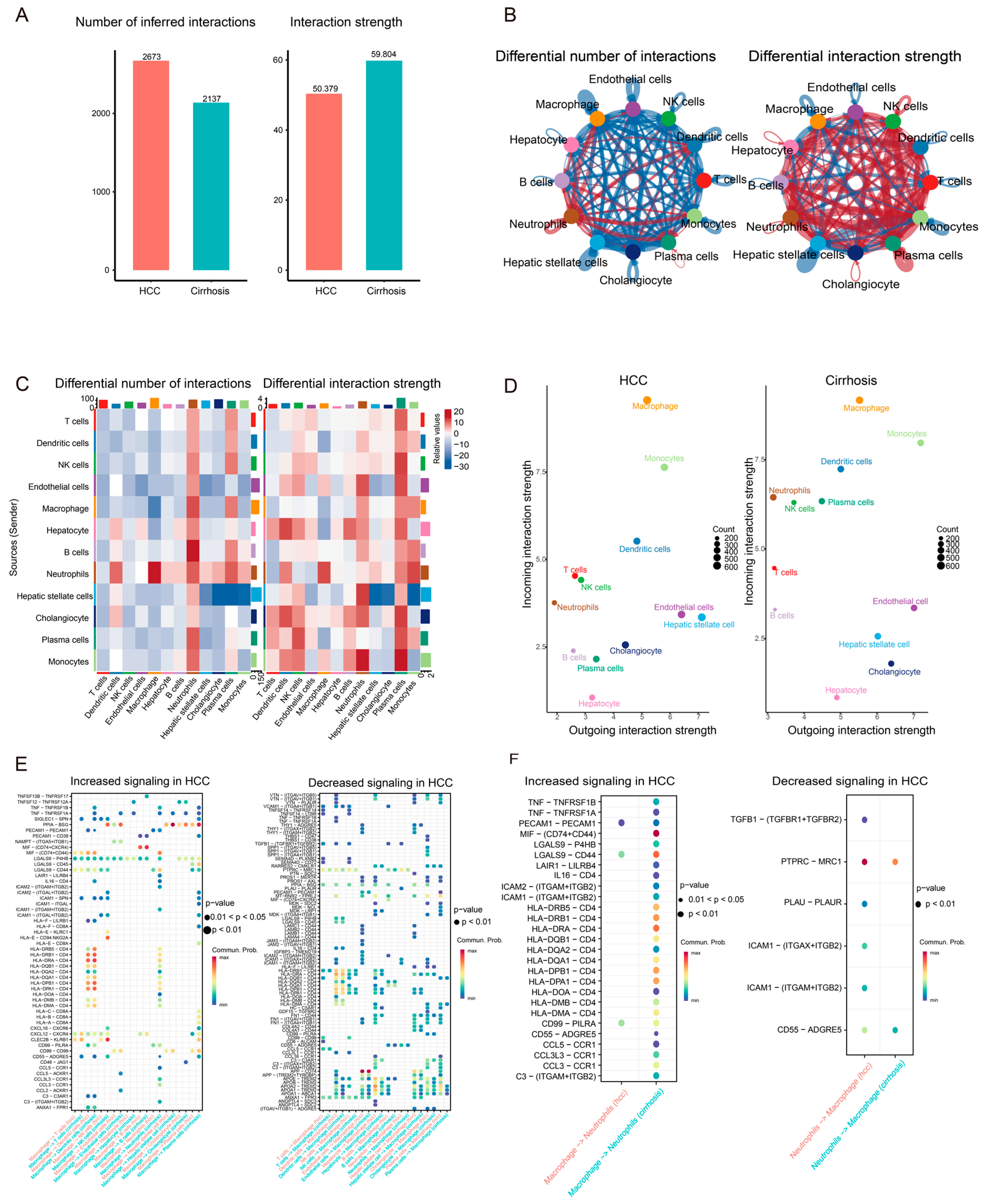
3.3. Communication Network Differences Between Cirrhosis and HCC
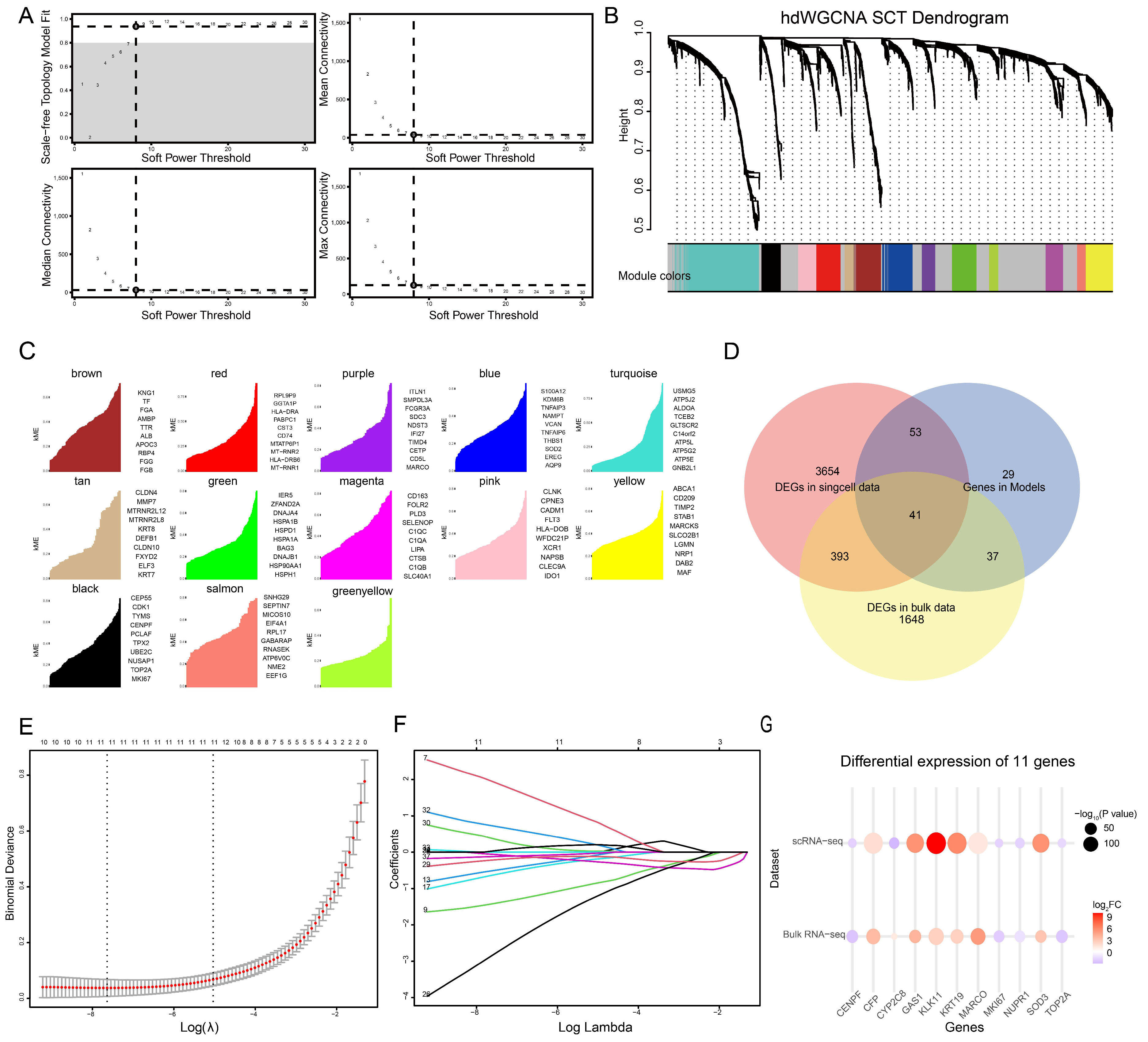
3.4. Gene Selection by High-Dimensional Weighted Gene Co-Expression Network Analysis (hdWGCNA) and Gene Selection by Bulk RNA Sequencing and Lasso Regression
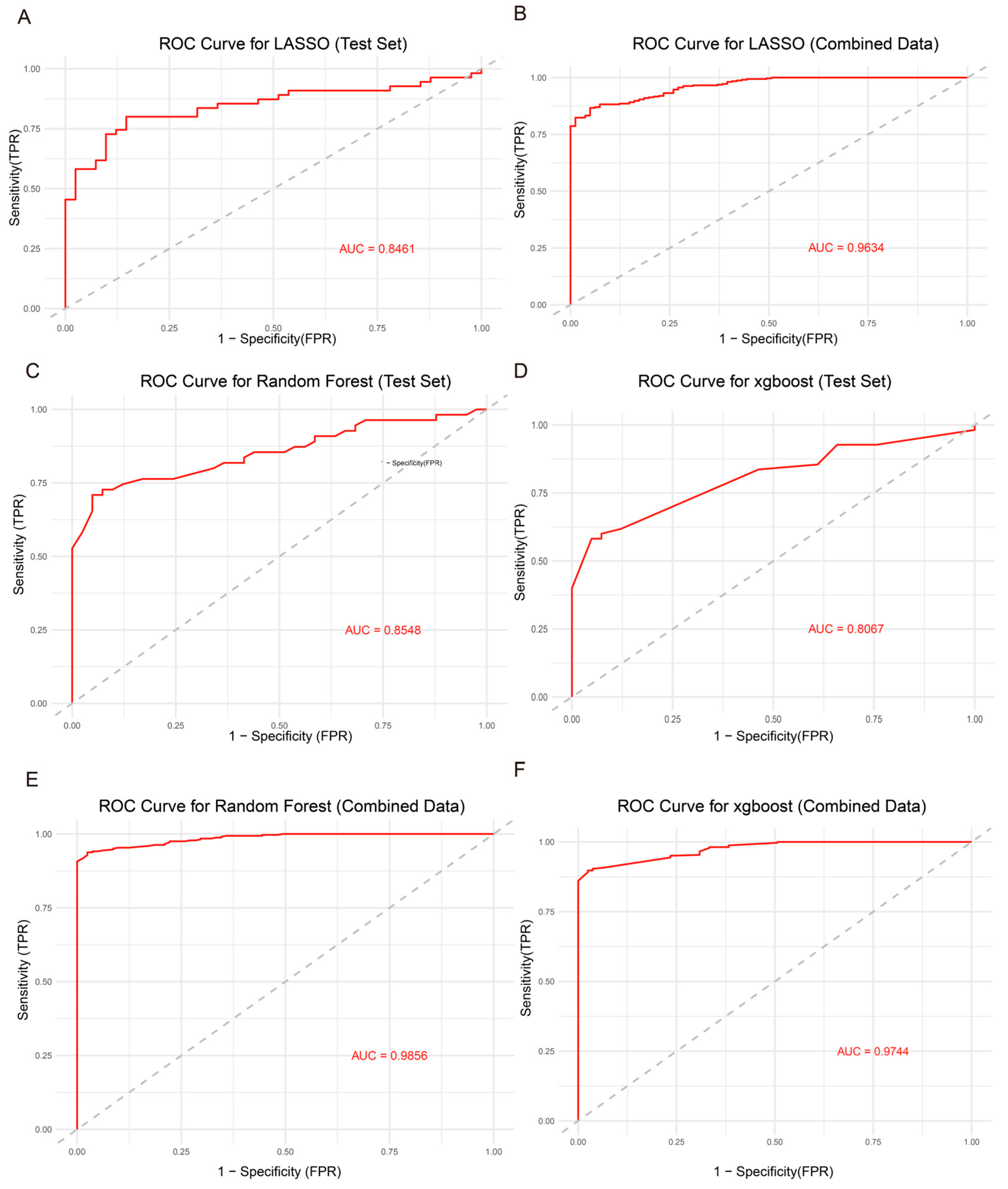
3.5. Model Construction and Performance Evaluation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A Global View of Hepatocellular Carcinoma: Trends, Risk, Prevention and Management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of Hepatocellular Carcinoma. J. Hepatol. 2018, 69, 182–236. [Google Scholar] [CrossRef]
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular Carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef]
- Llovet, J.M.; Zucman-Rossi, J.; Pikarsky, E.; Sangro, B.; Schwartz, M.; Sherman, M.; Gores, G. Hepatocellular Carcinoma. Nat. Rev. Dis. Primer 2016, 2, 16018. [Google Scholar] [CrossRef]
- Jia, W.; Liang, S.; Cheng, B.; Ling, C. The Role of Cancer-Associated Fibroblasts in Hepatocellular Carcinoma and the Value of Traditional Chinese Medicine Treatment. Front. Oncol. 2021, 11, 763519. [Google Scholar] [CrossRef]
- Yoshiji, H.; Nagoshi, S.; Akahane, T.; Asaoka, Y.; Ueno, Y.; Ogawa, K.; Kawaguchi, T.; Kurosaki, M.; Sakaida, I.; Shimizu, M.; et al. Evidence-Based Clinical Practice Guidelines for Liver Cirrhosis 2020. J. Gastroenterol. 2021, 56, 593–619. [Google Scholar] [CrossRef]
- Morgan, R.L.; Baack, B.; Smith, B.D.; Yartel, A.; Pitasi, M.; Falck-Ytter, Y. Eradication of Hepatitis C Virus Infection and the Development of Hepatocellular Carcinoma: A Meta-Analysis of Observational Studies. Ann. Intern. Med. 2013, 158, 329–337. [Google Scholar] [CrossRef] [PubMed]
- van der Meer, A.J.; Veldt, B.J.; Feld, J.J.; Wedemeyer, H.; Dufour, J.-F.; Lammert, F.; Duarte-Rojo, A.; Heathcote, E.J.; Manns, M.P.; Kuske, L.; et al. Association between Sustained Virological Response and All-Cause Mortality among Patients with Chronic Hepatitis C and Advanced Hepatic Fibrosis. JAMA 2012, 308, 2584–2593. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Lambrecht, J.; Ju, C.; Tacke, F. Hepatic Macrophages in Liver Homeostasis and Diseases-Diversity, Plasticity and Therapeutic Opportunities. Cell. Mol. Immunol. 2021, 18, 45–56. [Google Scholar] [CrossRef]
- Casanova-Acebes, M.; Dalla, E.; Leader, A.M.; LeBerichel, J.; Nikolic, J.; Morales, B.M.; Brown, M.; Chang, C.; Troncoso, L.; Chen, S.T.; et al. Tissue-Resident Macrophages Provide a pro-Tumorigenic Niche to Early NSCLC Cells. Nature 2021, 595, 578–584. [Google Scholar] [CrossRef] [PubMed]
- Laviron, M.; Boissonnas, A. Ontogeny of Tumor-Associated Macrophages. Front. Immunol. 2019, 10, 1799. [Google Scholar] [CrossRef] [PubMed]
- Mass, E.; Ballesteros, I.; Farlik, M.; Halbritter, F.; Günther, P.; Crozet, L.; Jacome-Galarza, C.E.; Händler, K.; Klughammer, J.; Kobayashi, Y.; et al. Specification of Tissue-Resident Macrophages during Organogenesis. Science 2016, 353, aaf4238. [Google Scholar] [CrossRef] [PubMed]
- MacParland, S.A.; Liu, J.C.; Ma, X.-Z.; Innes, B.T.; Bartczak, A.M.; Gage, B.K.; Manuel, J.; Khuu, N.; Echeverri, J.; Linares, I.; et al. Single Cell RNA Sequencing of Human Liver Reveals Distinct Intrahepatic Macrophage Populations. Nat. Commun. 2018, 9, 4383. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhang, S.; Liu, Y.; He, X.; Qu, M.; Xu, G.; Wang, H.; Huang, M.; Pan, J.; Liu, Z.; et al. Single-Cell RNA Sequencing Reveals the Heterogeneity of Liver-Resident Immune Cells in Human. Cell Discov. 2020, 6, 22. [Google Scholar] [CrossRef]
- Kourou, K.; Exarchos, T.P.; Exarchos, K.P.; Karamouzis, M.V.; Fotiadis, D.I. Machine Learning Applications in Cancer Prognosis and Prediction. Comput. Struct. Biotechnol. J. 2015, 13, 8–17. [Google Scholar] [CrossRef]
- Swanson, K.; Wu, E.; Zhang, A.; Alizadeh, A.A.; Zou, J. From Patterns to Patients: Advances in Clinical Machine Learning for Cancer Diagnosis, Prognosis, and Treatment. Cell 2023, 186, 1772–1791. [Google Scholar] [CrossRef]
- Boehm, K.M.; Khosravi, P.; Vanguri, R.; Gao, J.; Shah, S.P. Harnessing Multimodal Data Integration to Advance Precision Oncology. Nat. Rev. Cancer 2022, 22, 114–126. [Google Scholar] [CrossRef]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H.J.W.L. Artificial Intelligence in Radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef]
- López-Bueno, R.; Andersen, L.L.; Koyanagi, A.; Núñez-Cortés, R.; Calatayud, J.; Casaña, J.; del Pozo Cruz, B. Thresholds of Handgrip Strength for All-Cause, Cancer, and Cardiovascular Mortality: A Systematic Review with Dose-Response Meta-Analysis. Ageing Res. Rev. 2022, 82, 101778. [Google Scholar] [CrossRef]
- Mokoatle, M.; Marivate, V.; Mapiye, D.; Bornman, R.; Hayes, V.M. A Review and Comparative Study of Cancer Detection Using Machine Learning: SBERT and SimCSE Application. BMC Bioinform. 2023, 24, 112. [Google Scholar] [CrossRef]
- Hao, Y.; Stuart, T.; Kowalski, M.H.; Choudhary, S.; Hoffman, P.; Hartman, A.; Srivastava, A.; Molla, G.; Madad, S.; Fernandez-Granda, C.; et al. Dictionary Learning for Integrative, Multimodal and Scalable Single-Cell Analysis. Nat. Biotechnol. 2024, 42, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Hafemeister, C.; Satija, R. Normalization and Variance Stabilization of Single-Cell RNA-Seq Data Using Regularized Negative Binomial Regression. Genome Biol. 2019, 20, 296. [Google Scholar] [CrossRef]
- Becht, E.; McInnes, L.; Healy, J.; Dutertre, C.-A.; Kwok, I.W.H.; Ng, L.G.; Ginhoux, F.; Newell, E.W. Dimensionality Reduction for Visualizing Single-Cell Data Using UMAP. Nat. Biotechnol. 2018, 37, 38–44. [Google Scholar] [CrossRef]
- Hu, C.; Li, T.; Xu, Y.; Zhang, X.; Li, F.; Bai, J.; Chen, J.; Jiang, W.; Yang, K.; Ou, Q.; et al. CellMarker 2.0: An Updated Database of Manually Curated Cell Markers in Human/Mouse and Web Tools Based on scRNA-Seq Data. Nucleic Acids Res. 2023, 51, D870–D876. [Google Scholar] [CrossRef]
- Jiang, S.; Qian, Q.; Zhu, T.; Zong, W.; Shang, Y.; Jin, T.; Zhang, Y.; Chen, M.; Wu, Z.; Chu, Y.; et al. Cell Taxonomy: A Curated Repository of Cell Types with Multifaceted Characterization. Nucleic Acids Res. 2023, 51, D853–D860. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284. [Google Scholar] [CrossRef]
- Jin, S.; Guerrero-Juarez, C.F.; Zhang, L.; Chang, I.; Ramos, R.; Kuan, C.-H.; Myung, P.; Plikus, M.V.; Nie, Q. Inference and Analysis of Cell-Cell Communication Using CellChat. Nat. Commun. 2021, 12, 1088. [Google Scholar] [CrossRef]
- Xu, K.; Dong, M.; Wu, Z.; Luo, L.; Xie, F.; Li, F.; Huang, H.; Wang, F.; Xiong, X.; Wen, Z. Single-Cell RNA Sequencing Identifies Crucial Genes Influencing the Polarization of Tumor-Associated Macrophages in Liver Cancer. Int. J. Genomics 2024, 2024, 7263358. [Google Scholar] [CrossRef] [PubMed]
- Ke, H.; Yuan, R.; Liu, H.; Luo, M.; Hu, H.; Zhang, E.; Zhuang, K.; Yang, Y.; Yang, R. Serum Protein Biomarkers for HCC Risk Prediction in HIV/HBV Co-Infected People: A Clinical Proteomic Study Using Mass Spectrometry. Front. Immunol. 2023, 14, 1282469. [Google Scholar] [CrossRef]
- Crouchet, E.; Bandiera, S.; Fujiwara, N.; Li, S.; El Saghire, H.; Fernández-Vaquero, M.; Riedl, T.; Sun, X.; Hirschfield, H.; Jühling, F.; et al. A Human Liver Cell-Based System Modeling a Clinical Prognostic Liver Signature for Therapeutic Discovery. Nat. Commun. 2021, 12, 5525. [Google Scholar] [CrossRef]
- Chaudhary, K.; Poirion, O.B.; Lu, L.; Garmire, L.X. Deep Learning-Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin. Cancer Res. 2018, 24, 1248–1259. [Google Scholar] [CrossRef]
- Sun, J.; Mao, F.; Liu, C.; Zhang, F.; Jiang, D.; Guo, W.; Huo, L.; Zhou, L.; Lau, W.Y.; Shi, J.; et al. Combined FOLFOX4 with All-Trans Retinoic Acid versus FOLFOX4 with Placebo in Treatment of Advanced Hepatocellular Carcinoma with Extrahepatic Metastasis: A Randomized, Double-Blind Comparative Study. Signal Transduct. Target. Ther. 2023, 8, 368. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, T.-M.; Luo, J.-Z.; Lan, C.-L.; Wei, Z.-L.; Fu, T.-H.; Liao, X.-W.; Zhu, G.-Z.; Ye, X.-P.; Peng, T. CYP2C8 Suppress Proliferation, Migration, Invasion and Sorafenib Resistance of Hepatocellular Carcinoma via PI3K/Akt/P27kip1 Axis. J. Hepatocell. Carcinoma 2021, 8, 1323–1338. [Google Scholar] [CrossRef]
- Wang, T.; Lu, J.; Wang, R.; Cao, W.; Xu, J. TOP2A Promotes Proliferation and Metastasis of Hepatocellular Carcinoma Regulated by miR-144-3p. J. Cancer 2022, 13, 589–601. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, X.; Wang, L.; Wang, T.; Tang, X.; Su, X. Centromere Protein F (CENPF) Serves as a Potential Prognostic Biomarker and Target for Human Hepatocellular Carcinoma. J. Cancer 2021, 12, 2933–2951. [Google Scholar] [CrossRef]
- Saeed, M.M.; Ma, X.; Fu, X.; Ullah, I.; Ali, T.; Bai, C.; Liu, Y.; Dong, C.; Cui, X. RACGAP1 and MKI67 Are Potential Prognostic Biomarker in Hepatocellular Carcinoma Caused by HBV/HCV via Lactylation. Front. Oncol. 2025, 15, 1537084. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Zhang, Z.; Liu, Y.; Fang, Y.; Xie, Y.; Zheng, Y.; Li, G.; Liang, L.; Ding, Y. NUPR1 Contributes to Radiation Resistance by Maintaining ROS Homeostasis via AhR/CYP Signal Axis in Hepatocellular Carcinoma. BMC Med. 2022, 20, 365. [Google Scholar] [CrossRef] [PubMed]
- Toh, M.R.; Wong, E.Y.T.; Wong, S.H.; Ng, A.W.T.; Loo, L.-H.; Chow, P.K.-H.; Ngeow, J. Global Epidemiology and Genetics of Hepatocellular Carcinoma. Gastroenterology 2023, 164, 766–782. [Google Scholar] [CrossRef]
- Brar, G.; Greten, T.F.; Graubard, B.I.; McNeel, T.S.; Petrick, J.L.; McGlynn, K.A.; Altekruse, S.F. Hepatocellular Carcinoma Survival by Etiology: A SEER-Medicare Database Analysis. Hepatol. Commun. 2020, 4, 1541–1551. [Google Scholar] [CrossRef]
- Zhou, W.-C.; Zhang, Q.-B.; Qiao, L. Pathogenesis of Liver Cirrhosis. World J. Gastroenterol. 2014, 20, 7312–7324. [Google Scholar] [CrossRef]
- Smith, A.; Baumgartner, K.; Bositis, C. Cirrhosis: Diagnosis and Management. Am. Fam. Physician 2019, 100, 759–770. [Google Scholar]
- Yunna, C.; Mengru, H.; Lei, W.; Weidong, C. Macrophage M1/M2 Polarization. Eur. J. Pharmacol. 2020, 877, 173090. [Google Scholar] [CrossRef]
- Aydın, M.M.; Akçalı, K.C. Liver Fibrosis. Turk. J. Gastroenterol. 2018, 29, 14–21. [Google Scholar] [CrossRef]
- Elpek, G.Ö. Cellular and Molecular Mechanisms in the Pathogenesis of Liver Fibrosis: An Update. World J. Gastroenterol. 2014, 20, 7260–7276. [Google Scholar] [CrossRef]
- Krenkel, O.; Tacke, F. Liver Macrophages in Tissue Homeostasis and Disease. Nat. Rev. Immunol. 2017, 17, 306–321. [Google Scholar] [CrossRef]
- Barron, L.; Wynn, T.A. Fibrosis Is Regulated by Th2 and Th17 Responses and by Dynamic Interactions between Fibroblasts and Macrophages. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 300, G723–G728. [Google Scholar] [CrossRef]
- Kisseleva, T.; Cong, M.; Paik, Y.; Scholten, D.; Jiang, C.; Benner, C.; Iwaisako, K.; Moore-Morris, T.; Scott, B.; Tsukamoto, H.; et al. Myofibroblasts Revert to an Inactive Phenotype during Regression of Liver Fibrosis. Proc. Natl. Acad. Sci. USA 2012, 109, 9448–9453. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Hilakivi-Clarke, L.; Shaha, A.; Wang, Y.; Wang, X.; Deng, Y.; Lai, J.; Kang, N. Metabolic Reprogramming and Its Clinical Implication for Liver Cancer. Hepatology 2023, 78, 1602–1624. [Google Scholar] [CrossRef] [PubMed]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does It Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Tan, Y.; Yin, P.; Ye, G.; Gao, P.; Lu, X.; Wang, H.; Xu, G. Metabolic Characterization of Hepatocellular Carcinoma Using Nontargeted Tissue Metabolomics. Cancer Res. 2013, 73, 4992–5002. [Google Scholar] [CrossRef] [PubMed]
- Komohara, Y.; Fujiwara, Y.; Ohnishi, K.; Takeya, M. Tumor-Associated Macrophages: Potential Therapeutic Targets for Anti-Cancer Therapy. Adv. Drug Deliv. Rev. 2016, 99, 180–185. [Google Scholar] [CrossRef]
- Parker, K.H.; Sinha, P.; Horn, L.A.; Clements, V.K.; Yang, H.; Li, J.; Tracey, K.J.; Ostrand-Rosenberg, S. HMGB1 Enhances Immune Suppression by Facilitating the Differentiation and Suppressive Activity of Myeloid-Derived Suppressor Cells. Cancer Res. 2014, 74, 5723–5733. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, T.; Saito, H.; Ikeguchi, M. Increased B7-H1 and B7-H4 Expressions on Circulating Monocytes and Tumor-Associated Macrophages Are Involved in Immune Evasion in Patients with Gastric Cancer. Yonago Acta Med. 2011, 54, 1–10. [Google Scholar] [PubMed]
- Wang, L.; Zhang, H.; Sun, L.; Gao, W.; Xiong, Y.; Ma, A.; Liu, X.; Shen, L.; Li, Q.; Yang, H. Manipulation of Macrophage Polarization by Peptide-Coated Gold Nanoparticles and Its Protective Effects on Acute Lung Injury. J. Nanobiotechnol. 2020, 18, 38. [Google Scholar] [CrossRef]
- Xu, F.; Guo, M.; Huang, W.; Feng, L.; Zhu, J.; Luo, K.; Gao, J.; Zheng, B.; Kong, L.-D.; Pang, T.; et al. Annexin A5 Regulates Hepatic Macrophage Polarization via Directly Targeting PKM2 and Ameliorates NASH. Redox Biol. 2020, 36, 101634. [Google Scholar] [CrossRef]
- Ma, P.-F.; Gao, C.-C.; Yi, J.; Zhao, J.-L.; Liang, S.-Q.; Zhao, Y.; Ye, Y.-C.; Bai, J.; Zheng, Q.-J.; Dou, K.-F.; et al. Cytotherapy with M1-Polarized Macrophages Ameliorates Liver Fibrosis by Modulating Immune Microenvironment in Mice. J. Hepatol. 2017, 67, 770–779. [Google Scholar] [CrossRef]
- Yang, Y.; Ye, Y.-C.; Chen, Y.; Zhao, J.-L.; Gao, C.-C.; Han, H.; Liu, W.-C.; Qin, H.-Y. Crosstalk between Hepatic Tumor Cells and Macrophages via Wnt/β-Catenin Signaling Promotes M2-Like Macrophage Polarization and Reinforces Tumor Malignant Behaviors. Cell Death Dis. 2018, 9, 793. [Google Scholar] [CrossRef]
- Mantovani, A.; Biswas, S.K.; Galdiero, M.R.; Sica, A.; Locati, M. Macrophage Plasticity and Polarization in Tissue Repair and Remodelling. J. Pathol. 2013, 229, 176–185. [Google Scholar] [CrossRef]
- Zhao, X.; Sun, L.; Mu, T.; Yi, J.; Ma, C.; Xie, H.; Liu, M.; Tang, H. An HBV-Encoded miRNA Activates Innate Immunity to Restrict HBV Replication. J. Mol. Cell Biol. 2020, 12, 263–276. [Google Scholar] [CrossRef]
- Yi, H.; Zhang, Y.; Yang, X.; Li, M.; Hu, H.; Xiong, J.; Wang, N.; Jin, J.; Zhang, Y.; Song, Y.; et al. Hepatitis B Core Antigen Impairs the Polarization While Promoting the Production of Inflammatory Cytokines of M2 Macrophages via the TLR2 Pathway. Front. Immunol. 2020, 11, 535. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, Y.; Zhai, N.; Song, H.; Li, H.; Yang, Y.; Li, T.; Guo, X.; Chi, B.; Niu, J.; et al. HCV Core Protein Inhibits Polarization and Activity of Both M1 and M2 Macrophages through the TLR2 Signaling Pathway. Sci. Rep. 2016, 6, 36160. [Google Scholar] [CrossRef]
- Ho, D.W.-H.; Lam, W.-L.M.; Chan, L.-K.; Ng, I.O.-L. Investigation of Functional Synergism of CENPF and FOXM1 Identifies POLD1 as Downstream Target in Hepatocellular Carcinoma. Front. Med. 2022, 9, 860395. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Song, Z.; Tan, B.; Zhang, J.; Zhang, J.; Liu, S. Identification and Validation of Hub Genes Associated with Hepatocellular Carcinoma Via Integrated Bioinformatics Analysis. Front. Oncol. 2021, 11, 614531. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Ji, Y.; Jiang, X.; Qi, X. Downregulation of CYP2A6 and CYP2C8 in Tumor Tissues Is Linked to Worse Overall Survival and Recurrence-Free Survival from Hepatocellular Carcinoma. BioMed Res. Int. 2018, 2018, 5859415. [Google Scholar] [CrossRef]
- Gao, X.; Ren, X.; Wang, F.; Ren, X.; Liu, M.; Cui, G.; Liu, X. Immunotherapy and Drug Sensitivity Predictive Roles of a Novel Prognostic Model in Hepatocellular Carcinoma. Sci. Rep. 2024, 14, 9509. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xu, X.; Zhang, Z.; Xue, C.; Kong, Z.; Wu, S.; Yun, X.; Fu, Y.; Zhu, C.; Qin, X. Linc-KILH Potentiates Notch1 Signaling through Inhibiting KRT19 Phosphorylation and Promotes the Malignancy of Hepatocellular Carcinoma. Int. J. Biol. Sci. 2021, 17, 768–780. [Google Scholar] [CrossRef]
- Tang, J.; Zhuo, H.; Zhang, X.; Jiang, R.; Ji, J.; Deng, L.; Qian, X.; Zhang, F.; Sun, B. A Novel Biomarker Linc00974 Interacting with KRT19 Promotes Proliferation and Metastasis in Hepatocellular Carcinoma. Cell Death Dis. 2014, 5, e1549. [Google Scholar] [CrossRef]
- Zhang, Q.; Wei, Y.; Li, Y.; Jiao, X. Low MARCO Expression Is Associated with Poor Survival in Patients with Hepatocellular Carcinoma Following Liver Transplantation. Cancer Manag. Res. 2022, 14, 1935–1944. [Google Scholar] [CrossRef]
- Li, K.; Yang, Y.; Ma, M.; Lu, S.; Li, J. Hypoxia-Based Classification and Prognostic Signature for Clinical Management of Hepatocellular Carcinoma. World J. Surg. Oncol. 2023, 21, 216. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Zhang, P.; Cai, Y.; Zhu, G.; Chen, S.; Song, L.; Du, J.; Wang, B.; Dai, W.; Zhou, J.; et al. Lactylation-Driven NUPR1 Promotes Immunosuppression of Tumor-Infiltrating Macrophages in Hepatocellular Carcinoma. Adv. Sci. 2025, 12, e2413095. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.-L.; Bai, T.; Zhou, L.; Zhu, R.-T.; Wang, W.-J.; Liang, R.-P.; Li, J.; Zhang, C.-X.; Gou, J.-J. SOD3 Deficiency Induces Liver Fibrosis by Promoting Hepatic Stellate Cell Activation and Epithelial-Mesenchymal Transition. J. Cell. Physiol. 2021, 236, 4313–4329. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Shao, B.; Zhou, Y.; Chen, Z. High Expression of TOP2A in Hepatocellular Carcinoma Is Associated with Disease Progression and Poor Prognosis. Oncol. Lett. 2020, 20, 232. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Zeng, C.; Yong, X.; Zhou, W.; Xie, Y.; Shu, J. Integrative Single-Cell and Bulk Transcriptomic Analysis Identifies Macrophage-Related Gene Signatures Predictive of Hepatocellular Carcinoma in Cirrhosis . Genes 2025, 16, 1213. https://doi.org/10.3390/genes16101213
Zhang Z, Zeng C, Yong X, Zhou W, Xie Y, Shu J. Integrative Single-Cell and Bulk Transcriptomic Analysis Identifies Macrophage-Related Gene Signatures Predictive of Hepatocellular Carcinoma in Cirrhosis . Genes. 2025; 16(10):1213. https://doi.org/10.3390/genes16101213
Chicago/Turabian StyleZhang, Zhongyuan, Chuisheng Zeng, Xuetong Yong, Wenping Zhou, Yongfang Xie, and Jianzhong Shu. 2025. "Integrative Single-Cell and Bulk Transcriptomic Analysis Identifies Macrophage-Related Gene Signatures Predictive of Hepatocellular Carcinoma in Cirrhosis " Genes 16, no. 10: 1213. https://doi.org/10.3390/genes16101213
APA StyleZhang, Z., Zeng, C., Yong, X., Zhou, W., Xie, Y., & Shu, J. (2025). Integrative Single-Cell and Bulk Transcriptomic Analysis Identifies Macrophage-Related Gene Signatures Predictive of Hepatocellular Carcinoma in Cirrhosis . Genes, 16(10), 1213. https://doi.org/10.3390/genes16101213





