Comprehensive Identification of miRNAs and circRNAs in the Regulation of Intramuscular and Subcutaneous Fat Deposition in Meat Ducks
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Samples Preparation
2.2. RNA Sequencing
2.3. circRNA Identification and Differential Analysis
2.4. miRNA Identification
2.5. Target Gene Prediction of DE miRNAs
2.6. GO and KEGG Functional Enrichment Analysis
2.7. Interaction Network Construction
2.8. Validation of Quantitative Real−Time PCR (qRT−PCR)
2.9. Statistical Analyses
3. Results
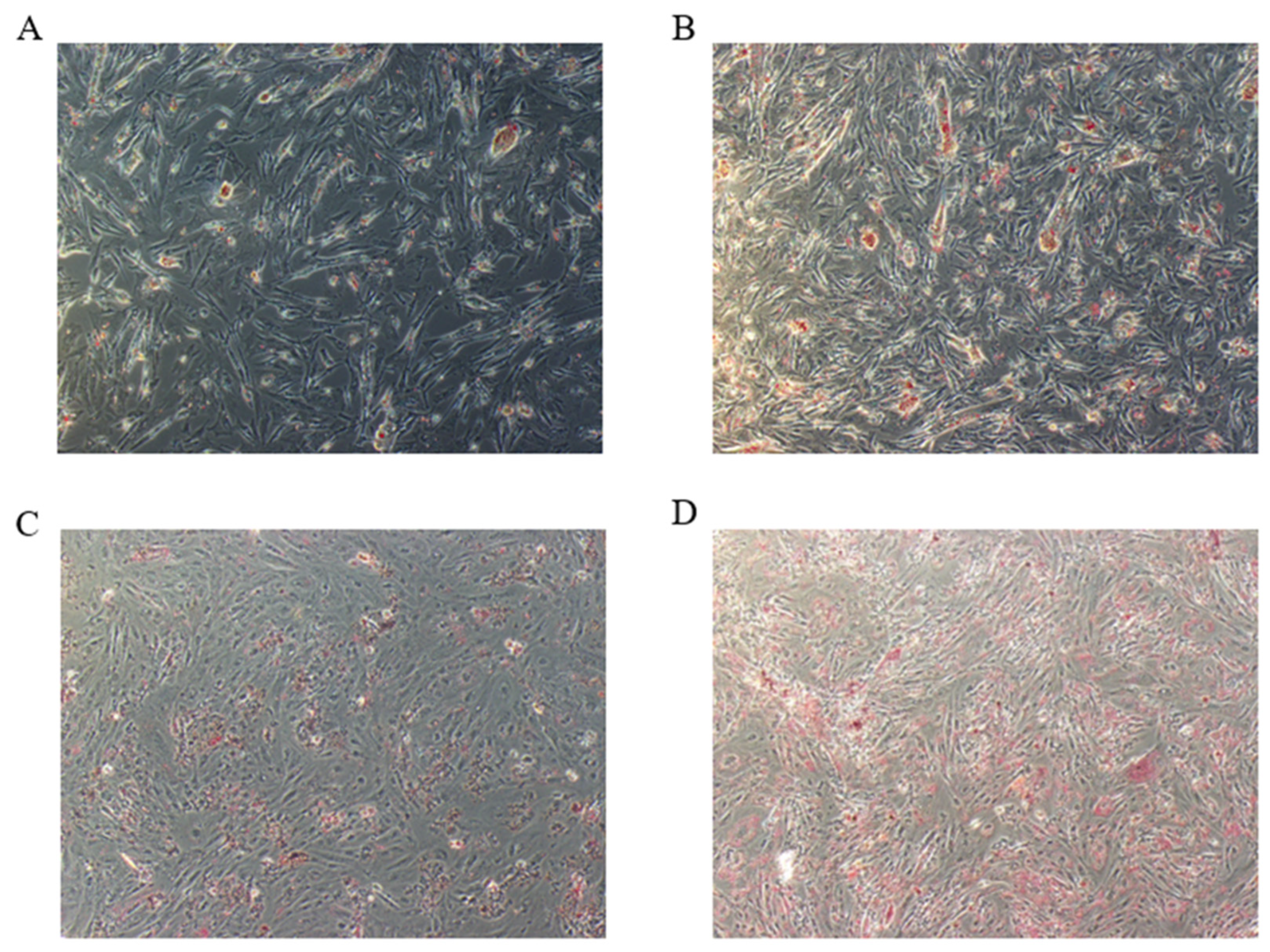
3.1. In Vitro Cell Culture and Results of Oil Red O Staining
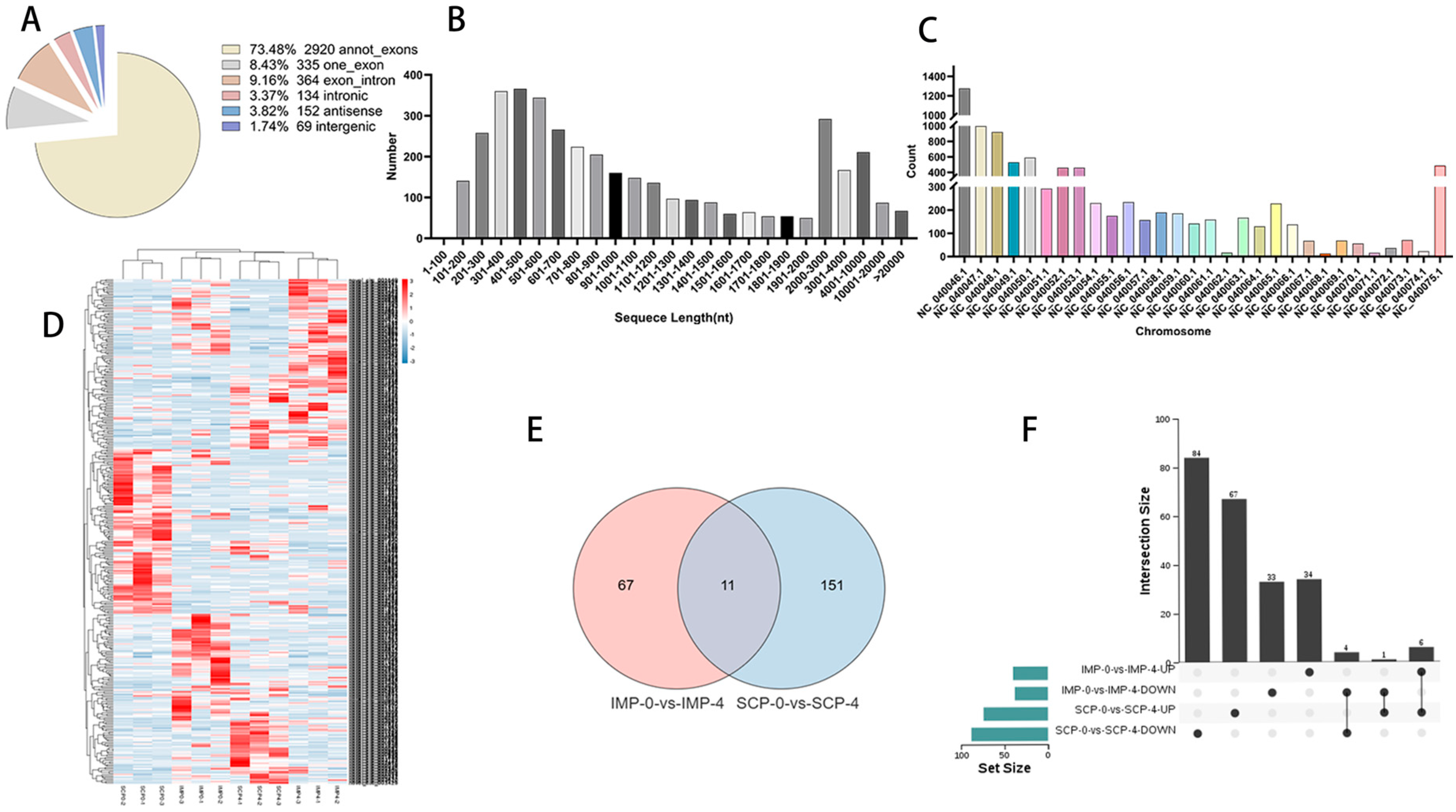
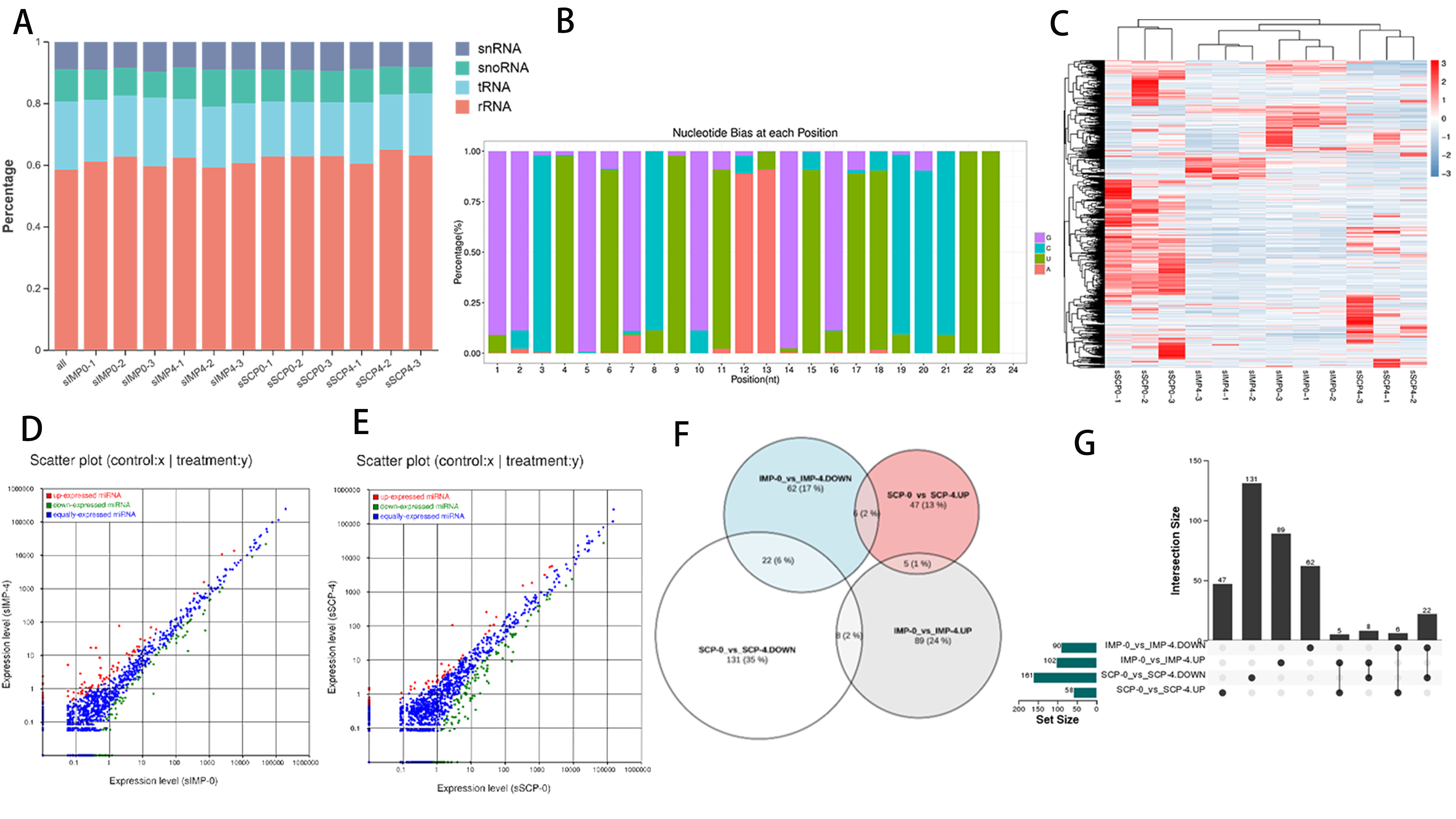
3.2. Global Responses of circRNAs to Fat Deposition
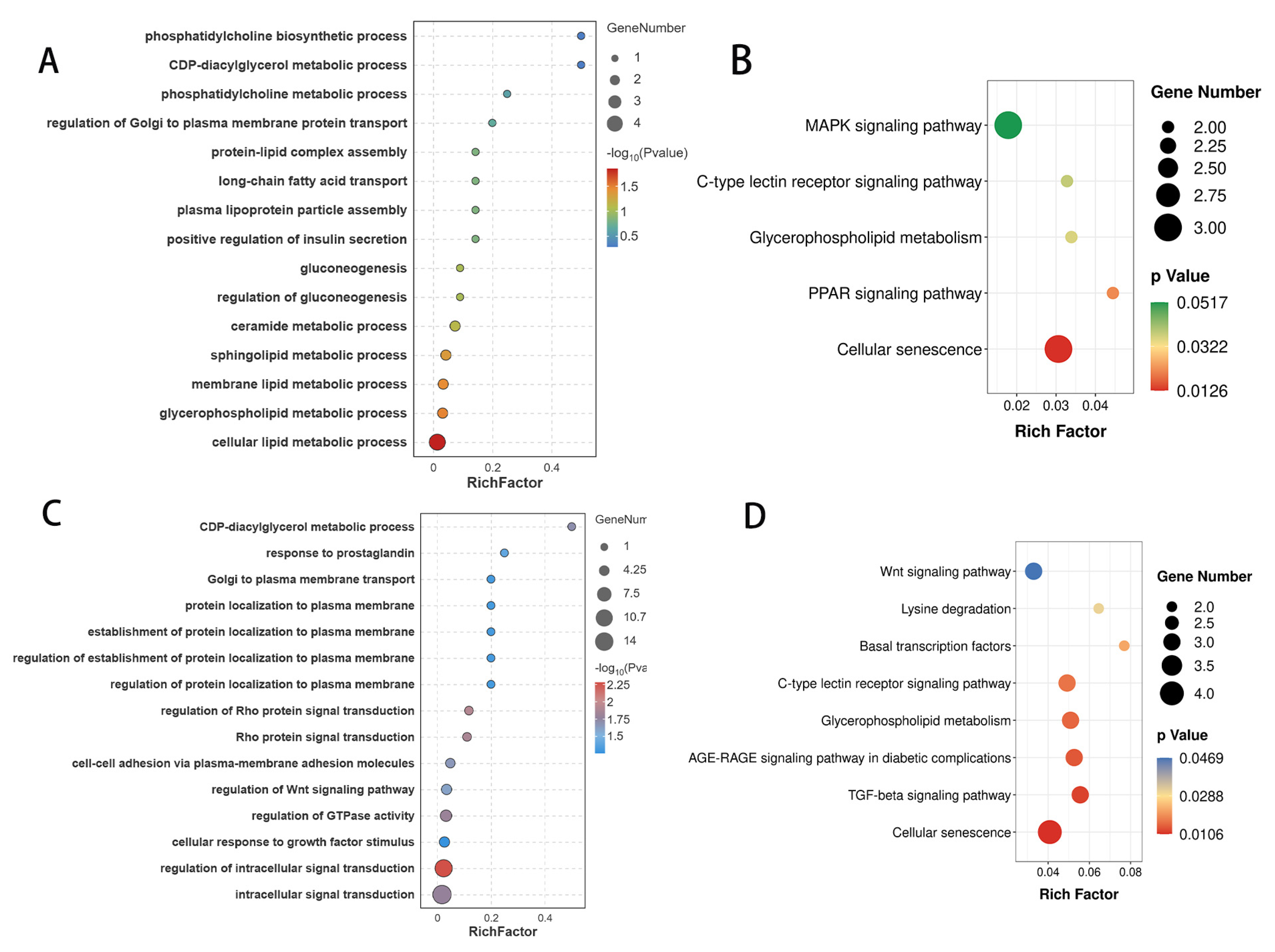
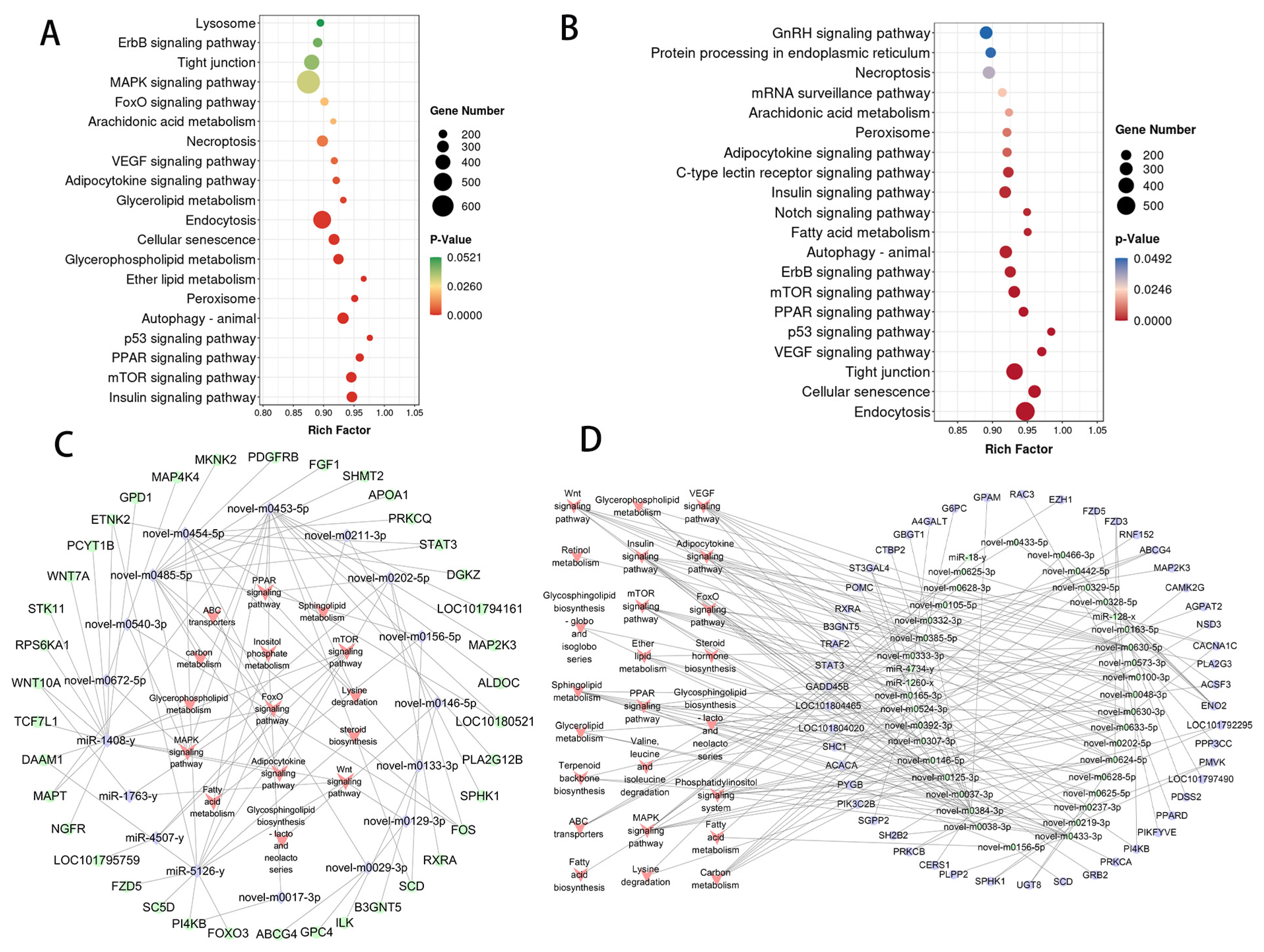
3.3. Functional Annotation of circRNA Host Gene
3.4. Global Responses of miRNAs to Fat Deposition
3.5. Functional Annotation of miRNA Target Genes
3.6. Integrative Analysis of miRNA and mRNA Expression Profiles
3.7. Construction of the DE circRNA–miRNA–mRNA ceRNA Regulatory Network
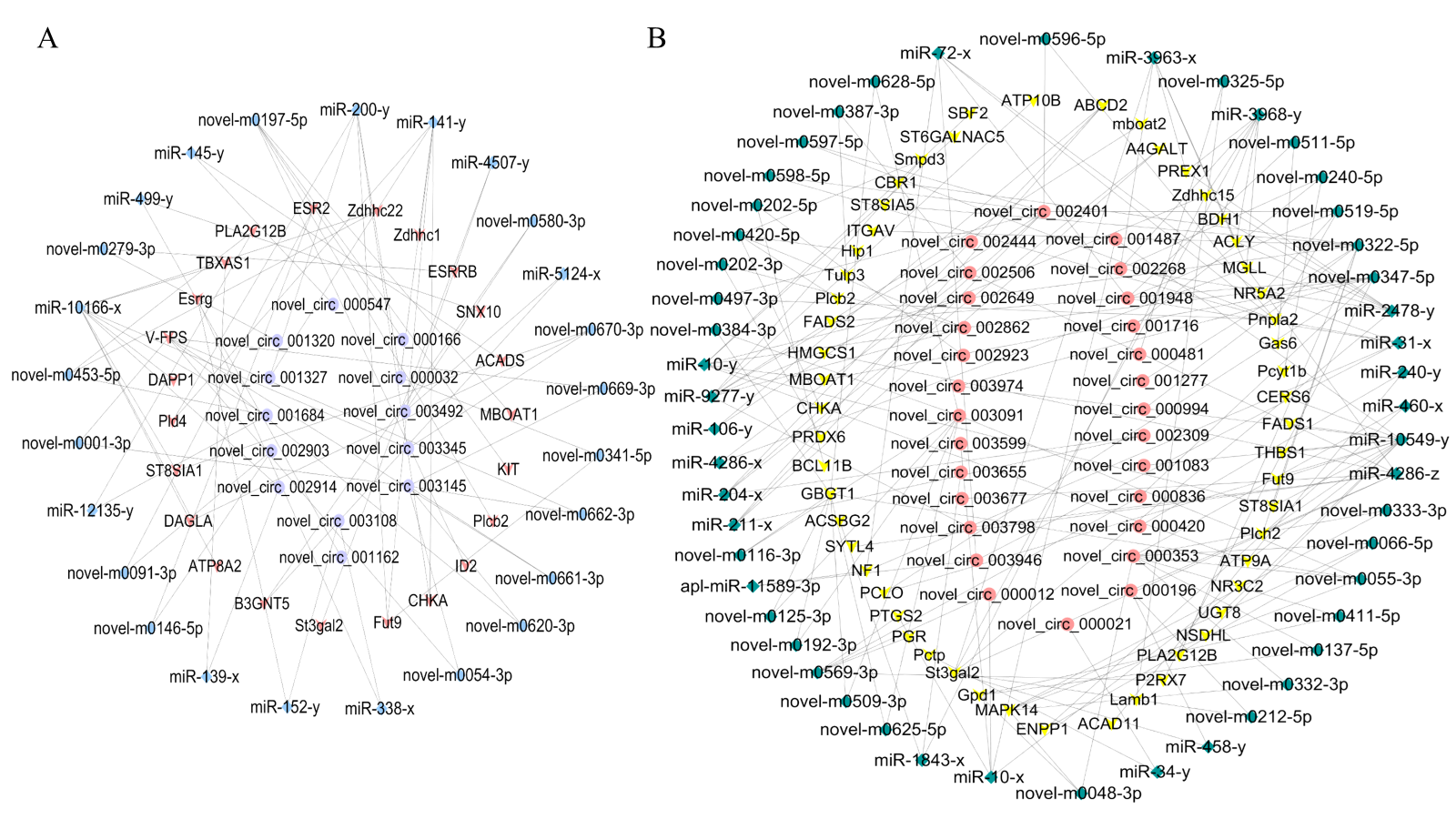
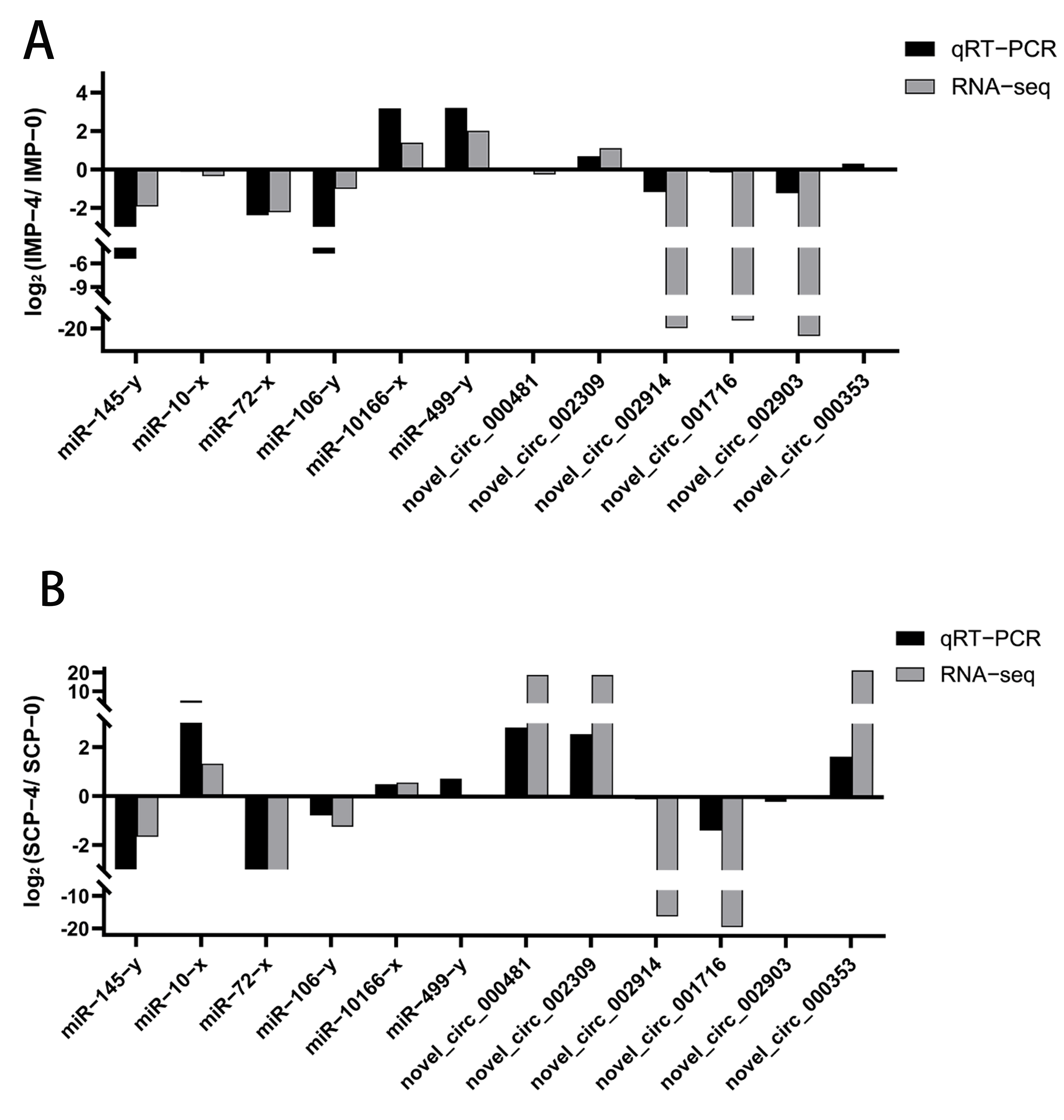
3.8. qRT-PCR Validation of DEcircRNAs and DEmiRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, H.; Bai, W.; Xu, W.; Li, B.S.; Ruan, Z. Research on integrated preservation technology for braised duck meat. Chin. Poult. 2007, 20, 2. [Google Scholar]
- Huang, Y.; He, Z.; Li, H.; Zhou, G.H.; Xu, X.L. Effects of Subcutaneous Fat and Intramuscular Fat on Pork Flavor. Chin. J. Agric. Sci. 2011, 44, 13. [Google Scholar]
- Moody, W.; Cassens, R.G. A Quantitative and Morphological Study of Bovine Longissimus Fat Cells. J. Food Sci. 1968, 33, 47–52. [Google Scholar] [CrossRef]
- Bagchi, D.P.; Forss, I.; Mandrup, S.; MacDougald, O.A. SnapShot: Niche Determines Adipocyte Character I. Cell Metab. 2018, 27, 264. [Google Scholar] [CrossRef]
- Shi, B.; Zhang, Z.; Lv, X.; An, K.; Li, L.; Xia, Z. Screening of Genes Related to Fat Deposition of Pekin Ducks Based on Transcriptome Analysis. Animals 2024, 14, 268. [Google Scholar] [CrossRef]
- Li, X.; Huang, Q.; Meng, F.; Hong, C.; Li, B.; Yang, Y.; Qu, Z.; Wu, J.; Li, F.; Xin, H.; et al. Analysis of Transcriptome Differences Between Subcutaneous and Intramuscular Adipose Tissue of Tibetan Pigs. Genes 2025, 16, 246. [Google Scholar] [CrossRef]
- Siersbæk, R.; Nielsen, R.; Mandrup, S. Transcriptional networks and chromatin remodeling controlling adipogenesis. Trends Endocrinol. Metab. 2012, 23, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Huang, H. Screening of Genes and miRNAs Related to Chicken Abdominal Fat Deposition and Functional Identification of the BNP Gene. Ph.D. Dissertation, Chinese Academy of Agricultural Sciences, Beijing, China, 2014. [Google Scholar]
- Tian, W.; Hao, X.; Nie, R.; Ling, Y.; Zhang, B.; Zhang, H.; Wu, C. Integrative analysis of miRNA and mRNA profiles reveals that gga-miR-106-5p inhibits adipogenesis by targeting the KLF15 gene in chickens. J. Anim. Sci. Biotechnol. 2022, 13, 81. [Google Scholar] [CrossRef]
- Wang, L.; Hu, X.; Wang, S.; Yuan, C.; Wang, Z.; Chang, G.; Chen, G. MicroRNA analysis reveals the role of miR-214 in duck adipocyte differentiation. Anim. Biosci. 2022, 35, 1327–1339. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhao, Y.; Jin, W.; Li, Y.; Zhang, Y.; Ma, X.; Sun, G.; Han, R.; Tian, Y.; Li, H.; et al. MicroRNAs and their regulatory networks in Chinese Gushi chicken abdominal adipose tissue during postnatal late development. BMC Genom. 2019, 20, 778. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, J.; Zheng, Q.; Wang, S.; Wei, X.; Li, F.; Shang, J.; Lei, C.; Ma, Y. Characterization of Circular RNAs in Chinese Buffalo (Bubalus bubalis) Adipose Tissue: A Focus on Circular RNAs Involved in Fat Deposition. Animals 2019, 9, 403. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yang, C.; Zhuang, Z.; Mao, J.; Chen, A.; Zhou, T.; Bai, H.; Jiang, Y.; Chang, G.; Wang, Z. RNA-Seq Analysis Revealed circRNAs and Genes Associated with Abdominal Fat Deposition in Ducks. Animals 2024, 14, 260. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, S.; Yue, B.; Zhang, S.; Jiang, E.; Chen, H.; Lan, X. CircRNA Profiling Reveals CircPPARγ Modulates Adipogenic Differentiation via Sponging miR-92a-3p. J. Agric. Food. Chem. 2022, 70, 6698–6708. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, H.; Ding, L.; Chen, Y.; Yin, C.; Zhou, P.; Yan, H.; Gou, Q. ceRNA Profiling Reveals circSAMD4A Promoted Porcine Adipocytes Differentiation via Targeting miR-127/PRKAR2B. Anim. Sci. J. 2025, 96, e70072. [Google Scholar] [CrossRef]
- Liu, S.; Yang, Y.; Luo, H.; Pang, W.; Martin, G. Fat deposition and partitioning for meat production in cattle and sheep. Anim. Nutr. 2024, 17, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Liu, T.; Yousuf, S.; Zhang, X.; Huang, W.; Li, A.; Xie, L.; Miao, X. Identification of potential miRNA-mRNA regulatory network and the key miRNAs in intramuscular and subcutaneous adipose. Front. Vet. Sci. 2022, 9, 976603. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, S.; Yang, J.; Zhao, F. Accurate quantification of circular RNAs identifies extensive circular isoform switching events. Nat. Commun. 2020, 11, 90. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Li, B.; Sun, M. Experimental Report on Compound Feed for Cherry Valley Ducks. Feed. Ind. 1987, 4, 8–9. [Google Scholar]
- Dodson, M.V.; Du, M.; Wang, S.; Bergen, W.G.; Fernyhough-Culver, M.; Basu, U.; Poulos, S.P.; Hausman, G.J. Adipose depots differ in cellularity, adipokines produced, gene expression, and cell systems. Adipocyte 2014, 3, 236–241. [Google Scholar] [CrossRef]
- Gardan, D.; Gondret, F.; Louveau, I. Lipid metabolism and secretory function of porcine intramuscular adipocytes compared with subcutaneous and perirenal adipocytes. Am. J. Physiol.-Endocrinol. Metab. 2006, 291, E372–E380. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhuang, Z.; Jia, W.; Xie, M.; Zhou, Z.; Tang, J.; Bai, H.; Chang, G.; Chen, G.; Hou, S. Comparative transcriptome analysis reveals the key genes involved in lipid deposition in pekin ducks (Anas platyrhynchos domesticus). Agriculture 2022, 12, 1775. [Google Scholar] [CrossRef]
- Gao, Q. Study on Histological and Molecular Biological Characteristics of Intramuscular Fat in Growing Erhualian Pigs. Ph.D. Dissertation, Nanjing Agricultural University, Nanjing, China, 2003. [Google Scholar]
- Kouba, M.; Bonneau, M.; Noblet, J. Relative development of subcutaneous, intermuscular, and kidney fat in growing pigs with different body compositions. J. Anim. Sci. 1999, 77, 622–629. [Google Scholar] [CrossRef]
- Chen, J.; Dodson, M.V.; Jiang, Z. Cellular and molecular comparison of redifferentiation of intramuscular- and visceral-adipocyte derived progeny cells. Int. J. Biol. Sci. 2010, 6, 80–88. [Google Scholar] [CrossRef]
- Wang, L.; Liang, W.; Wang, S.; Wang, Z.; Bai, H.; Jiang, Y.; Bi, Y.; Chen, G.; Chang, G. Circular RNA expression profiling reveals that circ-PLXNA1 functions in duck adipocyte differentiation. PLoS ONE 2020, 15, e0236069. [Google Scholar] [CrossRef]
- Tian, W.; Liu, Y.; Zhang, W.; Nie, R.; Ling, Y.; Zhang, B.; Zhang, H.; Wu, C. CircDOCK7 facilitates the proliferation and adipogenic differentiation of chicken abdominal preadipocytes through the gga-miR-301b-3p/ACSL1 axis. J. Anim. Sci. Biotechno. 2023, 14, 91. [Google Scholar] [CrossRef]
- Sun, W.X.; Nan, W.T.; Gu, S.S.; Yang, Z.P.; Mao, Y.J. Differential Expression Analysis of miR-27b and Its Target Gene PPARγ in Intramuscular and Subcutaneous Adipocytes. J. Nanjing Agric. Univ. 2016, 39, 6. [Google Scholar]
- Xing, B.; Wang, J.; Chen, J.; Zhao, R.Q.; Yang, X.J. Effects of porcine miR-199a-5p on lipid biogenesis and prediction/analysis of its target genes. Chin. J. Anim. Sci. Vet. Med. 2021, 48, 55–63. [Google Scholar]
- Liu, H. Mechanism Study of microRNA-32-5p Regulating Differential Deposition of Intramuscular and Subcutaneous Fat in Pigs by Targeting KLF3. Ph.D. Dissertation, Nanjing Agricultural University, Nanjing, China, 2020. [Google Scholar]
- Li, B.; He, Y.; Wu, W.; Zhang, H.; Li, X.W.; Zhu, L. Circular RNA Profiling Identifies Novel circPPARA that Promotes Intramuscular Fat Deposition in Pigs. J. Agric. Food. Chem. 2022, 70, 4123–4137. [Google Scholar] [CrossRef] [PubMed]
| Gene | Primer Sequence (5′–3′) |
|---|---|
| U6 | F: GGAACGATACAGAGAAGATTAGC R: TGGAACGCTTCACGAATTTGCG |
| GAPDH | F: AGATGCTGGTGCTGAATACG R: CGGAGATGATGACACGCTTA |
| novel_circ_000481 | F: ATGTGCTTGCACCTATGGCT R: AGCTACCTCCGTTCATGCAC |
| novel_circ_002309 | F: CGCTTTGTCCGAGGAGACAT R: TTGTATGCCACTGTCAACGC |
| novel_circ_000353 | F: CAGCCTCAGTCCGAATGGTT R: AGCCAGGAATGTCAGATGCC |
| novel_circ_002903 | F: CGCTTTGTCCGAGGAGACAT R: TTGTATGCCACTGTCAACGC |
| novel_circ_002914 | F: TGCCAGCACTAACAATTTGCC R: ATACAGCCATCCCCATTATCCG |
| novel_circ_001716 | F: GGTCTGCTTTTCCATTGGGC R: TTCAAGCCAGCTAACACCGT |
| miR-145-y | GCGCGATTCCTGGAAATACT |
| miR-10166-x | GCGCGTGCAGCTGATGA |
| miR-72-x | AGGCAAGATGTTGGCATAGCTG |
| miR-106-y | CGCGACTGCAGTATAAGCACT |
| miR-10-x | TACCCTGTAGATCCGAATTTGT |
| miR-499-y | CGCGCGCGAACACACA |
| circRNA−ID | log2(FC) | p-Value | Source_Gene |
|---|---|---|---|
| novel_circ_002903 | −21.98876634 | 1.64 × 10−6 | ncbi_101798557 |
| novel_circ_001112 | −21.06765464 | 1.84 × 10−4 | ncbi_101796118 |
| novel_circ_001327 | −20.75001229 | 5.53 × 10−5 | ncbi_101798557 |
| novel_circ_003145 | −20.4450971 | 4.46 × 10−4 | ncbi_101797443 |
| novel_circ_000955 | −20.09971448 | 9.18 × 10−3 | ncbi_101790241 |
| novel_circ_000347 | −20.02493364 | 1.45 × 10−2 | ncbi_101799066 |
| novel_circ_002914 | −19.95022905 | 5.97 × 10−3 | ncbi_101799066 |
| novel_circ_000683 | 20.40296203 | 2.61 × 10−3 | ncbi_101792777 |
| novel_circ_003292 | 20.40454937 | 4.34 × 10−3 | ncbi_101802634 |
| novel_circ_002224 | 20.44693427 | 4.67 × 10−4 | ncbi_101789710 |
| novel_circ_003570 | 20.45954433 | 1.80 × 10−3 | ncbi_101792634 |
| circRNA−ID | log2(FC) | p-Value | Source_Gene |
|---|---|---|---|
| novel_circ_002525 | −20.33703397 | 2.16 × 10−5 | ncbi_101790166 |
| novel_circ_002703 | −20.32357405 | 3.65 × 10−5 | ncbi_106015233 |
| novel_circ_002444 | −20.28053857 | 2.43 × 10−5 | ncbi_101792223 |
| novel_circ_003697 | −20.0386573 | 4.15 × 10−4 | ncbi_101790695 |
| novel_circ_001312 | −20.00303916 | 1.75 × 10−4 | ncbi_101801709 |
| novel_circ_002176 | −19.95397623 | 5.67 × 10−4 | ncbi_101805096 |
| novel_circ_001016 | −19.89744 | 2.99 × 10−4 | ncbi_101803584 |
| novel_circ_002829 | −19.79543522 | 3.50 × 10−3 | ncbi_101795905 |
| novel_circ_003429 | 20.69082673 | 2.60 × 10−4 | ncbi_101800418 |
| novel_circ_000353 | 21.82125304 | 4.26 × 10−14 | ncbi_101798353 |
| IMP−miRNA−ID | IMP−log2(FC) | IMP-p-Value | SCP−miRNA−ID | SCP−log2(FC) | SCP-p-Value |
|---|---|---|---|---|---|
| novel-m0001-5p | −6.912210731 | 9.15 × 10−6 | novel-m0355-5p | −9.925366519 | 5.55 × 10−16 |
| novel-m0054-5p | −6.594946589 | 6.73 × 10−5 | novel-m0336-5p | −8.617798026 | 1.62 × 10−8 |
| novel-m0620-5p | −6.5360529 | 1.52 × 10−4 | novel-m0630-3p | −8.035935434 | 9.30 × 10−6 |
| novel-m0661-5p | 6.503235273 | 2.54 × 10−14 | novel-m0338-5p | −7.726513368 | 7.56 × 10−5 |
| novel-m0662-5p | 6.503235273 | 2.42 × 10−14 | miR-240-y | −7.700023496 | 7.31 × 10−5 |
| novel-m0669-5p | 6.503235273 | 2.11 × 10−14 | novel-m0301-3p | −7.680793175 | 7.41 × 10−5 |
| novel-m0298-5p | 7.070961712 | 5.20 × 10−8 | novel-m0241-5p | −7.364776683 | 1.40 × 10−3 |
| novel-m0212-5p | 7.203266423 | 8.60 × 10−8 | novel-m0267-5p | −7.276341569 | 6.78 × 10−4 |
| novel-m0589-5p | 7.852935186 | 5.05 × 10−11 | novel-m0618-5p | −7.276341569 | 7.08 × 10−4 |
| novel-m0670-5p | 7.898833127 | 4.13 × 10−10 | miR-10549-y | 7.256664742 | 1.28 × 10−4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Zhou, T.; Liang, W.; Song, Q.; Jiang, Y.; Bai, H.; Chen, G.; Chang, G. Comprehensive Identification of miRNAs and circRNAs in the Regulation of Intramuscular and Subcutaneous Fat Deposition in Meat Ducks. Genes 2025, 16, 1208. https://doi.org/10.3390/genes16101208
Wang Z, Zhou T, Liang W, Song Q, Jiang Y, Bai H, Chen G, Chang G. Comprehensive Identification of miRNAs and circRNAs in the Regulation of Intramuscular and Subcutaneous Fat Deposition in Meat Ducks. Genes. 2025; 16(10):1208. https://doi.org/10.3390/genes16101208
Chicago/Turabian StyleWang, Zhixiu, Tingting Zhou, Wenshuang Liang, Qianqian Song, Yong Jiang, Hao Bai, Guohong Chen, and Guobin Chang. 2025. "Comprehensive Identification of miRNAs and circRNAs in the Regulation of Intramuscular and Subcutaneous Fat Deposition in Meat Ducks" Genes 16, no. 10: 1208. https://doi.org/10.3390/genes16101208
APA StyleWang, Z., Zhou, T., Liang, W., Song, Q., Jiang, Y., Bai, H., Chen, G., & Chang, G. (2025). Comprehensive Identification of miRNAs and circRNAs in the Regulation of Intramuscular and Subcutaneous Fat Deposition in Meat Ducks. Genes, 16(10), 1208. https://doi.org/10.3390/genes16101208





