A Genome-Wide Association Study Identifying Novel Genetic Markers of Response to Treatment with Interleukin-23 Inhibitors in Psoriasis
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Treatment Response
2.2. Genomic DNA Isolation and Genotyping
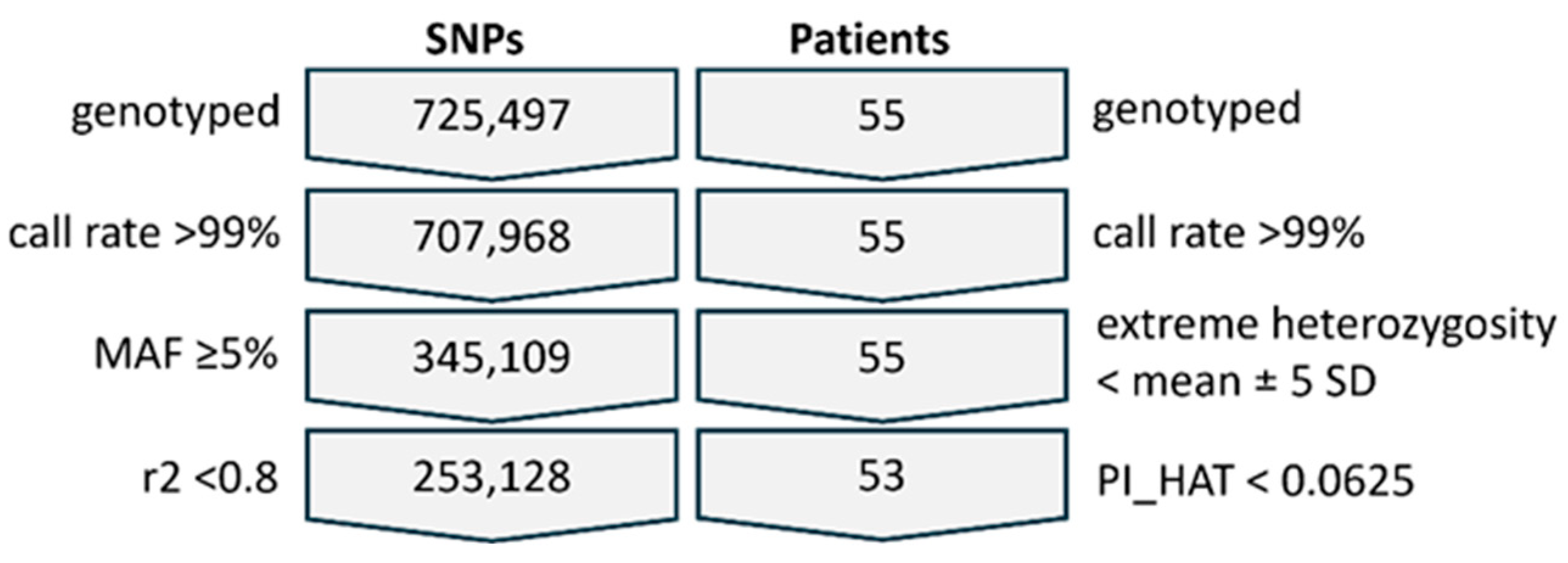
2.3. Quality Control, Filtering, and Association Analysis
2.4. Databases
3. Results
3.1. Patient Characteristics and Treatment Outcomes
3.2. GWAS of Treatment Response
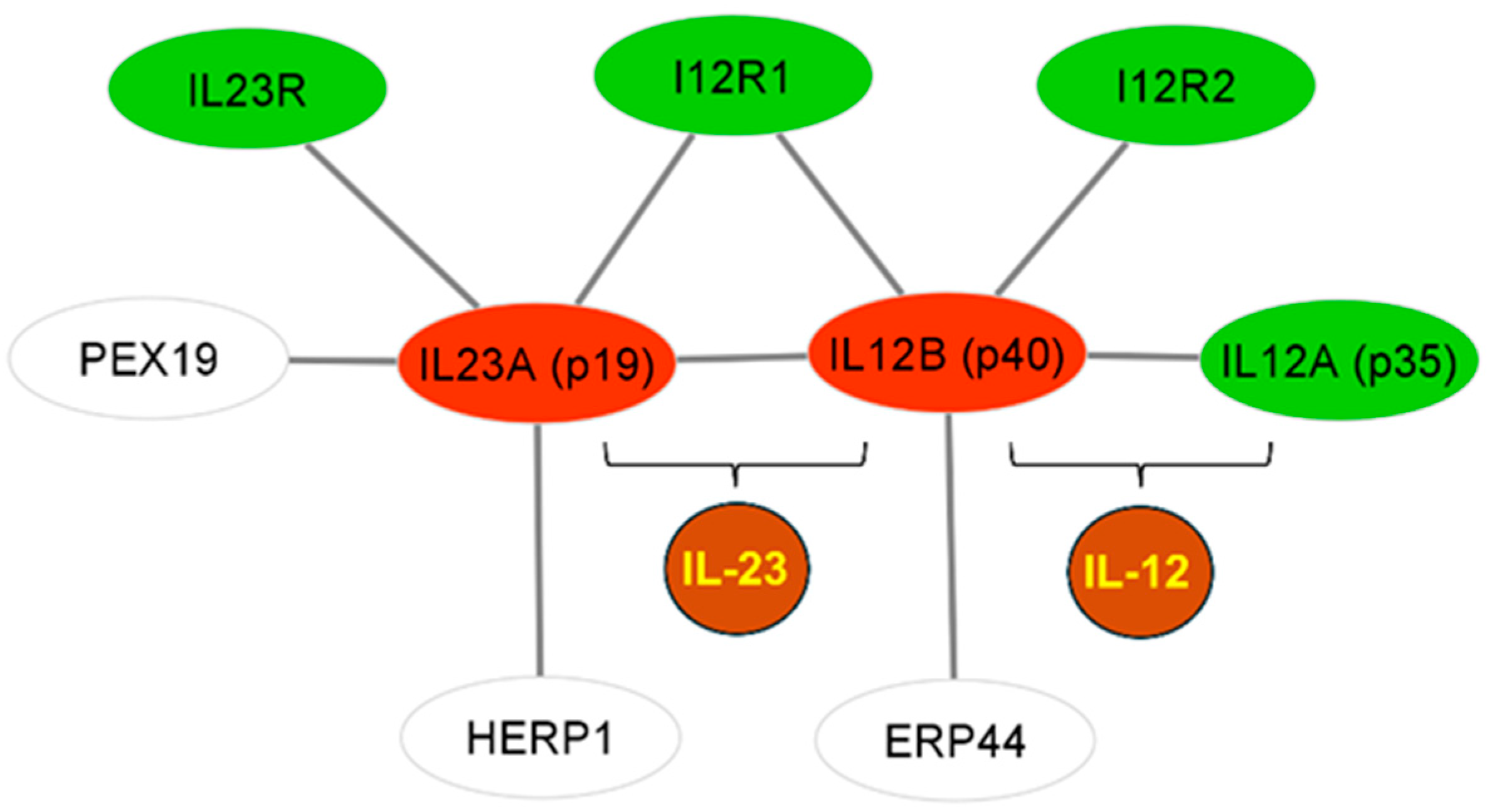
3.3. Association Analysis for Candidate Genes
3.4. Replication Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TNF | Tumor necrosis factor |
| IL | Interleukin |
| PASI | Psoriasis Area and Severity Index |
| PPI | Protein–protein interaction |
| GWAS | Genome-wide association study |
| eQTL | Expression quantitative trait locus |
| SNP | Single-nucleotide polymorphism |
References
- Armstrong, A.W.; Read, C. Pathophysiology, Clinical Presentation, and Treatment of Psoriasis: A Review. JAMA 2020, 323, 1945–1960. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.C.; Akbik, M.; Smith, L.R.; Klionsky, Y.; Feldman, S.R. Therapeutic Advancements in Psoriasis and Psoriatic Arthritis. J. Clin. Med. 2025, 14, 1312. [Google Scholar] [CrossRef] [PubMed]
- Menter, A.; Krueger, G.G.; Paek, S.Y.; Kivelevitch, D.; Adamopoulos, I.E.; Langley, R.G. Interleukin-17 and Interleukin-23: A Narrative Review of Mechanisms of Action in Psoriasis and Associated Comorbidities. Dermatol. Ther. 2021, 11, 385–400. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, C.E.M.; Armstrong, A.W.; Gudjonsson, J.E.; Barker, J.N.W.N. Psoriasis. Lancet 2021, 397, 1301–1315. [Google Scholar] [CrossRef]
- Pang, Y.; D’Cunha, R.; Winzenborg, I.; Veldman, G.; Pivorunas, V.; Wallace, K. Risankizumab: Mechanism of action, clinical and translational science. Clin. Transl. Sci. 2024, 17, e13706. [Google Scholar] [CrossRef]
- Krueger, J.G.; Eyerich, K.; Kuchroo, V.K.; Ritchlin, C.T.; Abreu, M.T.; Elloso, M.M.; Fourie, A.; Fakharzadeh, S.; Sherlock, J.P.; Yang, Y.-W.; et al. IL-23 past, present, and future: A roadmap to advancing IL-23 science and therapy. Front. Immunol. 2024, 15, 1331217. [Google Scholar] [CrossRef]
- Aggarwal, P.; Fleischer, A.B., Jr. IL-17 and IL-23 Inhibitors Have the Fastest Time to Meaningful Clinical Response for Plaque Psoriasis: A Network Meta-Analysis. J. Clin. Med. 2024, 13, 5139. [Google Scholar] [CrossRef]
- Jairath, V.; Acosta Felquer, M.L.; Cho, R.J. IL-23 inhibition for chronic inflammatory disease. Lancet 2024, 404, 1679–1692. [Google Scholar] [CrossRef]
- Thomas, S.E.; Barenbrug, L.; Hannink, G.; Seyger, M.M.B.; de Jong, E.M.G.J.; van den Reek, J.M.P.A. Drug Survival of IL-17 and IL-23 Inhibitors for Psoriasis: A Systematic Review and Meta-Analysis. Drugs 2024, 84, 565–578. [Google Scholar] [CrossRef]
- Reich, K.; Armstrong, A.W.; Foley, P.; Song, M.; Wasfi, Y.; Randazzo, B.; Li, S.; Shen, Y.K.; Gordon, K.B. Efficacy and safety of guselkumab, an anti-interleukin-23 monoclonal antibody, compared with adalimumab for the treatment of patients with moderate to severe psoriasis with randomized withdrawal and retreatment: Results from the phase III, double-blind, placebo- and active comparator-controlled VOYAGE 2 trial. J. Am. Acad. Dermatol. 2017, 76, 418–431. [Google Scholar] [CrossRef]
- Reich, K.; Papp, K.A.; Blauvelt, A.; Tyring, S.K.; Sinclair, R.; Thaçi, D.; Nograles, K.; Mehta, A.; Cichanowitz, N.; Li, Q.; et al. Tildrakizumab versus placebo or etanercept for chronic plaque psoriasis (reSURFACE 1 and reSURFACE 2): Results from two randomised controlled, phase 3 trials. Lancet 2017, 390, 276–288. [Google Scholar] [CrossRef]
- Reich, K.; Griffiths, C.E.M.; Gordon, K.B.; Papp, K.A.; Song, M.; Randazzo, B.; Li, S.; Shen, Y.K.; Han, C.; Kimball, A.B.; et al. Maintenance of clinical response and consistent safety profile with up to 3 years of continuous treatment with guselkumab: Results from the VOYAGE 1 and VOYAGE 2 trials. J. Am. Acad. Dermatol. 2020, 82, 936–945. [Google Scholar] [CrossRef]
- Papp, K.; Thaçi, D.; Reich, K.; Riedl, E.; Langley, R.G.; Krueger, J.G.; Gottlieb, A.B.; Nakagawa, H.; Bowman, E.P.; Mehta, A.; et al. Tildrakizumab (MK-3222), an antiinterleukin-23p19 monoclonal antibody, improves psoriasis in a phase IIb randomized placebo-controlled trial. Br. J. Dermatol. 2015, 173, 930–939. [Google Scholar] [CrossRef] [PubMed]
- Papp, K.A.; Blauvelt, A.; Bukhalo, M.; Gooderham, M.; Krueger, J.G.; Lacour, J.P.; Menter, A.; Padula, S.J. Risankizumab versus ustekinumab for moderate-to-severe plaque psoriasis. N. Engl. J. Med. 2017, 376, 1551–1560. [Google Scholar] [CrossRef] [PubMed]
- Kopp, T.; Riedl, E.; Bangert, C.; Bowman, E.P.; Greisenegger, E.; Horowitz, A.; Kittler, H.; Blumenschein, W.M.; McClanahan, T.K.; Marbury, T.; et al. Clinical improvement in psoriasis with specific targeting of interleukin-23. Nature 2015, 521, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Ständer, S.; Thaçi, D. Interleukin-23p19 inhibitors for the treatment of moderate-to-severe psoriasis: An expert opinion of real-world evidence studies in Europe. J. Dermatol. Treat. 2025, 36, 2438803. [Google Scholar] [CrossRef]
- Ingelman-Sundberg, M.; Nebert, D.W.; Lauschke, V.M. Emerging trends in pharmacogenomics: From common variant associations toward comprehensive genomic profiling. Hum. Genom. 2023, 17, 105. [Google Scholar] [CrossRef]
- Loras, A.; Gil-Barrachina, M.; Hernando, B.; Perez-Pastor, G.; Martinez-Domenech, A.; Mahiques, L.; Pitarch, G.; Valcuende-Cavero, F.; Ballester-Sanchez, R.; Marques-Torrejon, M.A.; et al. Association between several immune response-related genes and the effectiveness of biological treatments in patients with moderate-to-severe psoriasis. Exp. Dermatol. 2024, 33, e15003. [Google Scholar] [CrossRef]
- Manchanda, Y.; De, A.; Das, S.; Chakraborty, D. Disease Assessment in Psoriasis. Indian J. Dermatol. 2023, 68, 278–281. [Google Scholar] [CrossRef]
- Chaplin, Μ.; Kirkham, J.J.; Dwan, K.; Sloan, D.J.; Davies, G.; Jorgensen, A.L. STrengthening the Reporting Of Pharmacogenetic Studies: Development of the STROPS guideline. PLoS Med. 2020, 17, e1003344. [Google Scholar] [CrossRef]
- Liadaki, K.; Zafiriou, E.; Giannoulis, T.; Alexouda, S.; Chaidaki, K.; Gidarokosta, P.; Roussaki-Schulze, A.-V.; Tsiogkas, S.G.; Daponte, A.; Mamuris, Z.; et al. PDE4 Gene Family Variants Are Associated with Response to Apremilast Treatment in Psoriasis. Genes 2024, 15, 369. [Google Scholar] [CrossRef] [PubMed]
- Kirsten, N.; Rustenbach, S.; von Kiedrowski, R.; Sorbe, C.; Reich, K.; Augustin, M. Which PASI Outcome Is Most Relevant to the Patients in Real-World Care? Life 2021, 11, 1151. [Google Scholar] [CrossRef] [PubMed]
- Yangorcid, H.J.; Leeorcid, H.S. Common statistical methods used in medical research. Kosin Med. J. 2025, 40, 21–30. [Google Scholar] [CrossRef]
- RStudio Team. RStudio: Integrated Development Environment for R. RStudio; PBC: Boston, MA, USA, 2023. [Google Scholar]
- Chang, C.C. Data Management and Summary Statistics with PLINK. Methods Mol. Biol. 2020, 2090, 49–65. [Google Scholar] [CrossRef]
- Sherry, S.T.; Ward, M.H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef]
- Martin, F.J.; Amode, M.R.; Aneja, A.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Becker, A.; Bennett, R.; Berry, A.; Bhai, J.; et al. Ensembl 2023. Nucleic Acids Res. 2023, 51, D933–D941. [Google Scholar] [CrossRef]
- Klapa, M.I.; Tsafou, K.; Theodoridis, E.; Tsakalidis, A.; Moschonas, N.K. Reconstruction of the experimentally supported human protein interactome: What can we learn? BMC Syst. Biol. 2013, 7, 96. [Google Scholar] [CrossRef]
- Dimitrakopoulos, G.N.; Klapa, M.I.; Moschonas, N.K. PICKLE 3.0: Enriching the human meta-database with the mouse protein interactome extended via mouse-human orthology. Bioinformatics 2020, 37, 145–146. [Google Scholar] [CrossRef]
- Ward, L.D.; Kellis, M. HaploReg v4: Systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016, 44, D877–D881. [Google Scholar] [CrossRef]
- Franz, M.; Lopes, C.T.; Fong, D.; Kucera, M.; Cheung, M.; Siper, M.C.; Huck, G.; Dong, Y.; Sumer, O.; Bader, G.D. Cytoscape.js 2023 update: A graph theory library for visualization and analysis. Bioinformatics 2023, 39, btad031. [Google Scholar] [CrossRef]
- Aguet, F.; Barbeira, A.N.; Bonazzola, R.; Jo, B.; Kasela, S.; Liang, Y.; Parsana, P.; Aguet, F.; Battle, A.; Brown, A.; et al. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020, 369, 1318–1330. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- García-Domínguez, M. The Role of IL-23 in the Development of Inflammatory Diseases. Biology 2025, 14, 347. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, K.A.-M.; Schulze, L.L.; Paap, E.-M.; Müller, T.M.; Neurath, M.F.; Zundler, S. Immunology of IL-12: An update on functional activities and implications for disease. EXCLI J. 2020, 19, 1563–1589. [Google Scholar] [CrossRef]
- Lyschik, S.; Lauer, A.A.; Roth, T.; Janitschke, D.; Hollander, M.; Will, T.; Hartmann, T.; Kopito, R.R.; Helms, V.; Grimm, M.O.W.; et al. PEX19 Coordinates Neutral Lipid Storage in Cells in a Peroxisome-Independent Fashion. Front. Cell Dev. Biol. 2022, 26, 859052. [Google Scholar] [CrossRef]
- Alayoubi, A.M.; Ijaz, A.; Wali, A.; Hashmi, J.A.; Alharbi, A.; Basit, S. Zellweger syndrome; identification of mutations in PEX19 and PEX26 gene in Saudi families. Ann. Med. 2025, 57, 2447400. [Google Scholar] [CrossRef]
- Schulz, J.; Avci, D.; Queisser, M.A.; Gutschmidt, A.; Dreher, L.-S.; Fenech, E.J.; Volkmar, N.; Hayashi, Y.; Hoppe, T.; Christianson, J.C. Conserved cytoplasmic domains promote Hrd1 ubiquitin ligase complex formation for ER-associated degradation (ERAD). J. Cell Sci. 2017, 130, 3322–3335. [Google Scholar] [CrossRef]
- Tempio, T.; Anelli, T. The pivotal role of ERp44 in patrolling protein secretion. J. Cell Sci. 2020, 133, jcs240366. [Google Scholar] [CrossRef]
- Ko, B.S.; Lee, S.B.; Kim, T.-K. A brief guide to analyzing expression quantitative trait loci. Mol. Cells 2024, 47, 100139. [Google Scholar] [CrossRef]
- Perumal, N.; Gopalakrishnan, P.; Burkovetskaya, M.; Doss, D.; Dukkipati, S.S.; Kanchan, R.K.; Mahapatra, S. Nuclear factor I/B: Duality in action in cancer pathophysiology. Cancer Lett. 2025, 609, 217349. [Google Scholar] [CrossRef]
- Sun, H.; Li, N.; Tan, J.; Li, H.; Zhang, J.; Qu, L.; Lamont, S.J. Transcriptional Regulation of RIP2 Gene by NFIB Is Associated with Cellular Immune and Inflammatory Response to APEC Infection. Int. J. Mol. Sci. 2022, 30, 3814. [Google Scholar] [CrossRef]
- Wu, W.; Wang, X.; Yu, X.; Lan, H.Y. Smad3 Signatures in Renal Inflammation and Fibrosis. Int. J. Biol. Sci. 2022, 18, 2795–2806. [Google Scholar] [CrossRef]
- Liu, S.; Li, Z.; Lan, S.; Hao, H.; Baz, A.A.; Yan, X.; Gao, P.; Chen, S.; Chu, Y. The Dual Roles of Activating Transcription Factor 3 (ATF3) in Inflammation, Apoptosis, Ferroptosis, and Pathogen Infection Responses. Int. J. Mol. Sci. 2024, 25, 824. [Google Scholar] [CrossRef]
- Kamarck, M.L.; Trimmer, C.; Murphy, N.R.; Gregory, K.M.; Manoel, D.; Logan, D.W.; Saraiva, L.R.; Mainland, J.D. Identifying Candidate Genes Underlying Isolated Congenital Anosmia. Clin. Genet. 2023, 105, 376–385. [Google Scholar] [CrossRef]
| Characteristics | Patients |
|---|---|
| Gender (males/females) | 32/21 |
| Age of disease onset (mean ± SD) years | 36.7 (±16.4) |
| Age of treatment onset (mean ± SD) years | 56.1 (±13.3) |
| Biologic-naïve patients * (%) | 26% |
| Baseline PASI (mean ± SD) | 10.4 (±9.1) |
| Baseline weight (kg) (mean ± SD) | 89.2 (±20.4) |
| Baseline Body Mass Index (BMI) (mean ± SD) | 30.9 (±7.3) |
| Comorbidities (%) | 65% |
| rs73641950; Chr. 9: 14.499.258 bp Intronic, NFIB Gene | rs6627462; Chr. X: 151.758.565 bp Intergenic | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| F_R | F_NR | p Adj | OR | 95% CI | F_R | F_NR | p Adj | OR | 95% CI | |
| 3 m.: R (n = 43) > PASI 75 NR (n = 7) < PASI 50 | 0.012 | 0.429 | 0.0036 | 63.75 | 7–597 | 0.100 | 0.889 | 0.0103 | 72 | 7.6–679 |
| 3 m.: R (n = 38) > PASI 90 NR (n = 7) < PASI75 | 0.013 | 0.429 | 0.0244 | 56.18 | 6–528 | 0.096 | 0.889 | 0.0208 | 75 | 7.7–731 |
| 6 m.: R (n = 47) > PASI 75 NR (n = 6) < PASI 50 | 0.021 | 0.500 | 0.0009 | 46.08 | 7.6–278 | |||||
| 6 m.: R (n = 46) > PASI 90 NR (n = 6) < PASI 75 | 0.022 | 0.500 | 0.0013 | 45.05 | 7.4–273 | |||||
| F_R | F_NR | Raw p Value | p Adj | OR | 95% CI | |
|---|---|---|---|---|---|---|
| 3 m.: R (n = 43) > PASI 75 NR (n = 7) < PASI 50 | 0.023 | 0.357 | 5.6 × 10−6 | 0.0073 | 23.3 | 3.9–138 |
| 3 m.: R (n = 38) > PASI 90 NR (n = 7) < PASI75 | 0.026 | 0.357 | 2.2 × 10−5 | 0.0282 | 20.6 | 3.5–122 |
| 6 m.: R (n = 47) > PASI 75 NR (n = 6) < PASI 50 | 0.032 | 0.417 | 2.0 × 10−6 | 0.0026 | 21.7 | 4.3–110 |
| 6 m.: R (n = 46) > PASI 90 NR (n = 6) < PASI 75 | 0.033 | 0.417 | 2.7 × 10−6 | 0.0035 | 21.2 | 4.2–108 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zachari, S.; Liadaki, K.; Planaki, A.; Zafiriou, E.; Kouvarou, O.; Gerogianni, K.; Giannoulis, T.; Mamuris, Z.; Bogdanos, D.P.; Moschonas, N.K.; et al. A Genome-Wide Association Study Identifying Novel Genetic Markers of Response to Treatment with Interleukin-23 Inhibitors in Psoriasis. Genes 2025, 16, 1195. https://doi.org/10.3390/genes16101195
Zachari S, Liadaki K, Planaki A, Zafiriou E, Kouvarou O, Gerogianni K, Giannoulis T, Mamuris Z, Bogdanos DP, Moschonas NK, et al. A Genome-Wide Association Study Identifying Novel Genetic Markers of Response to Treatment with Interleukin-23 Inhibitors in Psoriasis. Genes. 2025; 16(10):1195. https://doi.org/10.3390/genes16101195
Chicago/Turabian StyleZachari, Sophia, Kalliopi Liadaki, Angeliki Planaki, Efterpi Zafiriou, Olga Kouvarou, Kalliopi Gerogianni, Themistoklis Giannoulis, Zissis Mamuris, Dimitrios P. Bogdanos, Nicholas K. Moschonas, and et al. 2025. "A Genome-Wide Association Study Identifying Novel Genetic Markers of Response to Treatment with Interleukin-23 Inhibitors in Psoriasis" Genes 16, no. 10: 1195. https://doi.org/10.3390/genes16101195
APA StyleZachari, S., Liadaki, K., Planaki, A., Zafiriou, E., Kouvarou, O., Gerogianni, K., Giannoulis, T., Mamuris, Z., Bogdanos, D. P., Moschonas, N. K., & Sarafidou, T. (2025). A Genome-Wide Association Study Identifying Novel Genetic Markers of Response to Treatment with Interleukin-23 Inhibitors in Psoriasis. Genes, 16(10), 1195. https://doi.org/10.3390/genes16101195







