The p.Ile202Thr Substitution in TUBB2B Can Be Associated with Syndromic Presentation of Congenital Fibrosis of the Extraocular Muscles
Abstract
1. Introduction
2. Materials and Methods
2.1. Exome Sequencing Data Analysis
2.2. DNA Methylation Profiling Analysis
2.3. RNA Analysis
2.4. WGS Data Analysis
2.5. Structural and Docking Analysis
3. Results
3.1. Clinical Findings
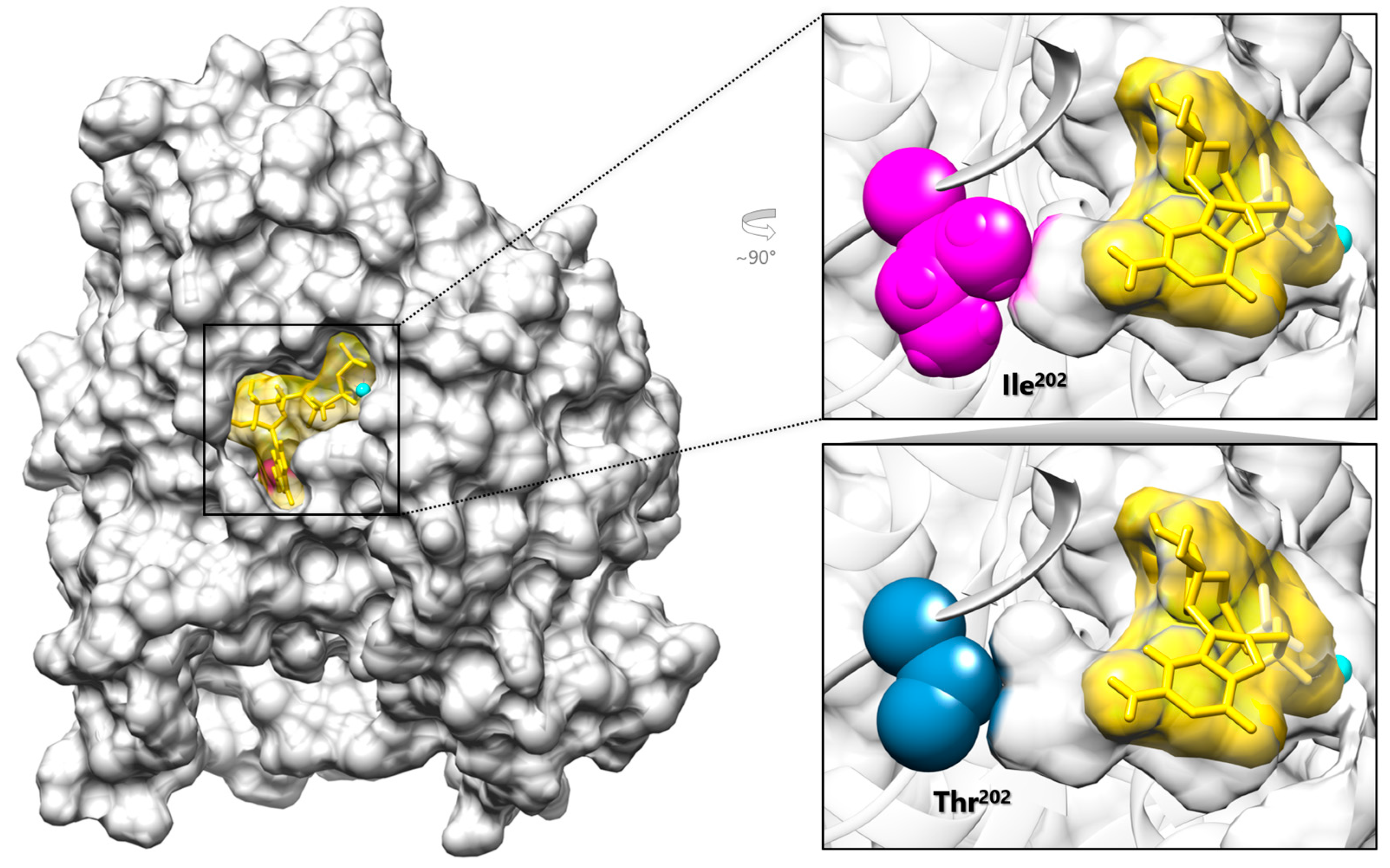
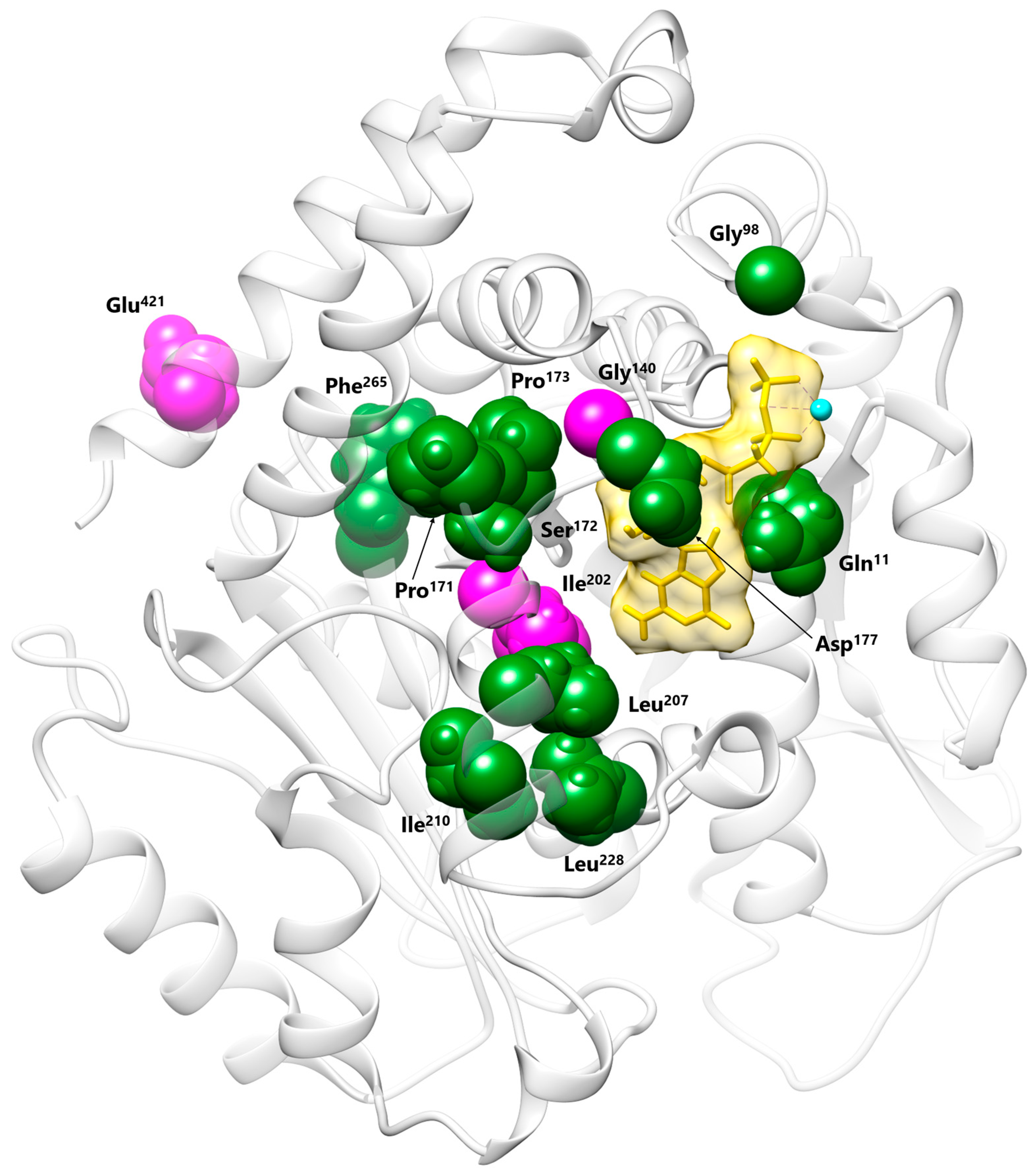
3.2. Molecular Findings and Structural Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bahi-Buisson, N.; Maillard, C. Tubulinopathies Overview. In GeneReviews; Adam, M.P., Feldman, J., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Maillard, C.; Roux, C.J.; Charbit-Henrion, F.; Steffann, J.; Laquerriere, A.; Quazza, F.; Buisson, N.B. Tubulin Mutations in Human Neurodevelopmental Disorders. Semin. Cell Dev. Biol. 2023, 137, 87–95. [Google Scholar] [CrossRef]
- Mutch, C.A.; Poduri, A.; Sahin, M.; Barry, B.; Walsh, C.A.; Barkovich, A.J. Disorders of Microtubule Function in Neurons: Imaging Correlates. AJNR Am. J. Neuroradiol. 2016, 37, 528–535. [Google Scholar] [CrossRef]
- Wu, Q.; Liu, J.; Fang, A.; Li, R.; Bai, Y.; Kriegstein, A.R.; Wang, X. The Dynamics of Neuronal Migration. Adv. Exp. Med. Biol. 2014, 800, 25–36. [Google Scholar] [CrossRef]
- Desai, A.; Mitchison, T.J. Microtubule Polymerization Dynamics. Annu. Rev. Cell Dev. Biol. 1997, 13, 83–117. [Google Scholar] [CrossRef] [PubMed]
- Knossow, M.; Campanacci, V.; Khodja, L.A.; Gigant, B. The Mechanism of Tubulin Assembly into Microtubules: Insights from Structural Studies. iScience 2020, 23, 101511. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Walsh, C.A. Protein-Protein Interactions, Cytoskeletal Regulation and Neuronal Migration. Nat. Rev. Neurosci. 2001, 2, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Guzik, B.W.; Goldstein, L.S.B. Microtubule-Dependent Transport in Neurons: Steps towards an Understanding of Regulation, Function and Dysfunction. Curr. Opin. Cell Biol. 2004, 16, 443–450. [Google Scholar] [CrossRef]
- Ayala, R.; Shu, T.; Tsai, L.-H. Trekking across the Brain: The Journey of Neuronal Migration. Cell 2007, 128, 29–43. [Google Scholar] [CrossRef]
- Tantry, M.S.A.; Santhakumar, K. Insights on the Role of α- and β-Tubulin Isotypes in Early Brain Development. Mol. Neurobiol. 2023, 60, 3803–3823. [Google Scholar] [CrossRef]
- Jaglin, X.H.; Chelly, J. Tubulin-Related Cortical Dysgeneses: Microtubule Dysfunction Underlying Neuronal Migration Defects. Trends Genet. 2009, 25, 555–566. [Google Scholar] [CrossRef]
- Romaniello, R.; Arrigoni, F.; Bassi, M.T.; Borgatti, R. Mutations in α- and β-Tubulin Encoding Genes: Implications in Brain Malformations. Brain Dev. 2015, 37, 273–280. [Google Scholar] [CrossRef]
- Bahi-Buisson, N.; Poirier, K.; Fourniol, F.; Saillour, Y.; Valence, S.; Lebrun, N.; Hully, M.; Bianco, C.F.; Boddaert, N.; Elie, C.; et al. The Wide Spectrum of Tubulinopathies: What Are the Key Features for the Diagnosis? Brain 2014, 137, 1676–1700. [Google Scholar] [CrossRef]
- Dekker, J.; Diderich, K.E.M.; Schot, R.; Husen, S.C.; Dremmen, M.H.G.; Go, A.T.J.I.; Weerts, M.J.A.; van Slegtenhorst, M.A.; Mancini, G.M.S. A Novel Family Illustrating the Mild Phenotypic Spectrum of TUBB2B Variants. Eur. J. Paediatr. Neurol. 2021, 35, 35–39. [Google Scholar] [CrossRef]
- Yamada, K.; Andrews, C.; Chan, W.-M.; McKeown, C.A.; Magli, A.; de Berardinis, T.; Loewenstein, A.; Lazar, M.; O’Keefe, M.; Letson, R.; et al. Heterozygous Mutations of the Kinesin KIF21A in Congenital Fibrosis of the Extraocular Muscles Type 1 (CFEOM1). Nat. Genet. 2003, 35, 318–321. [Google Scholar] [CrossRef]
- Tischfield, M.A.; Baris, H.N.; Wu, C.; Rudolph, G.; Van Maldergem, L.; He, W.; Chan, W.-M.; Andrews, C.; Demer, J.L.; Robertson, R.L.; et al. Human TUBB3 Mutations Perturb Microtubule Dynamics, Kinesin Interactions, and Axon Guidance. Cell 2010, 140, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Cederquist, G.Y.; Luchniak, A.; Tischfield, M.A.; Peeva, M.; Song, Y.; Menezes, M.P.; Chan, W.-M.; Andrews, C.; Chew, S.; Jamieson, R.V.; et al. An Inherited TUBB2B Mutation Alters a Kinesin-Binding Site and Causes Polymicrogyria, CFEOM and Axon Dysinnervation. Hum. Mol. Genet. 2012, 21, 5484–5499. [Google Scholar] [CrossRef]
- Cheng, L.; Desai, J.; Miranda, C.J.; Duncan, J.S.; Qiu, W.; Nugent, A.A.; Kolpak, A.L.; Wu, C.C.; Drokhlyansky, E.; Delisle, M.M.; et al. Human CFEOM1 Mutations Attenuate KIF21A Autoinhibition and Cause Oculomotor Axon Stalling. Neuron 2014, 82, 334–349. [Google Scholar] [CrossRef]
- Balasubramanian, R.; Chew, S.; MacKinnon, S.E.; Kang, P.B.; Andrews, C.; Chan, W.-M.; Engle, E.C. Expanding the Phenotypic Spectrum and Variability of Endocrine Abnormalities Associated with TUBB3 E410K Syndrome. J. Clin. Endocrinol. Metab. 2015, 100, E473–E477. [Google Scholar] [CrossRef]
- Jurgens, J.A.; Barry, B.J.; Lemire, G.; Chan, W.-M.; Whitman, M.C.; Shaaban, S.; Robson, C.D.; MacKinnon, S.; England, E.M.; McMillan, H.J.; et al. Novel Variants in TUBA1A Cause Congenital Fibrosis of the Extraocular Muscles with or without Malformations of Cortical Brain Development. Eur. J. Hum. Genet. 2021, 29, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Luchniak, A.; Roy, P.S.; Kumar, A.; Schneider, I.C.; Gelfand, V.I.; Jernigan, R.L.; Gupta, M.L. Tubulin CFEOM Mutations Both Inhibit or Activate Kinesin Motor Activity. Mol. Biol. Cell 2024, 35, ar32. [Google Scholar] [CrossRef] [PubMed]
- Whitman, M.C.; Jurgens, J.A.; Hunter, D.G.; Engle, E.C. Congenital Fibrosis of the Extraocular Muscles Overview. In GeneReviews; Adam, M.P., Feldman, J., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Kaçar Bayram, A.; Per, H.; Quon, J.; Canpolat, M.; Ülgen, E.; Doğan, H.; Gumus, H.; Kumandas, S.; Bayram, N.; Bilguvar, K.; et al. A Rare Case of Congenital Fibrosis of Extraocular Muscle Type 1A Due to KIF21A Mutation with Marcus Gunn Jaw-Winking Phenomenon. Eur. J. Paediatr. Neurol. 2015, 19, 743–746. [Google Scholar] [CrossRef]
- Motta, M.; Fasano, G.; Gredy, S.; Brinkmann, J.; Bonnard, A.A.; Simsek-Kiper, P.O.; Gulec, E.Y.; Essaddam, L.; Utine, G.E.; Guarnetti Prandi, I.; et al. SPRED2 Loss-of-Function Causes a Recessive Noonan Syndrome-like Phenotype. Am. J. Hum. Genet. 2021, 108, 2112–2129. [Google Scholar] [CrossRef]
- Dentici, M.L.; Niceta, M.; Lepri, F.R.; Mancini, C.; Priolo, M.; Bonnard, A.A.; Cappelletti, C.; Leoni, C.; Ciolfi, A.; Pizzi, S.; et al. Loss-of-Function Variants in ERF Are Associated with a Noonan Syndrome-like Phenotype with or without Craniosynostosis. Eur. J. Hum. Genet. 2024, 32, 954–963. [Google Scholar] [CrossRef]
- Fasano, G.; Muto, V.; Radio, F.C.; Venditti, M.; Mosaddeghzadeh, N.; Coppola, S.; Paradisi, G.; Zara, E.; Bazgir, F.; Ziegler, A.; et al. Dominant ARF3 Variants Disrupt Golgi Integrity and Cause a Neurodevelopmental Disorder Recapitulated in Zebrafish. Nat. Commun. 2022, 13, 6841. [Google Scholar] [CrossRef]
- Bonfiglio, F.; Legati, A.; Lasorsa, V.A.; Palombo, F.; De Riso, G.; Isidori, F.; Russo, S.; Furini, S.; Merla, G.; Coppedè, F.; et al. Best Practices for Germline Variant and DNA Methylation Analysis of Second- and Third-Generation Sequencing Data. Hum. Genom. 2024, 18, 120. [Google Scholar] [CrossRef] [PubMed]
- Yauy, K.; Lecoquierre, F.; Baert-Desurmont, S.; Trost, D.; Boughalem, A.; Luscan, A.; Costa, J.-M.; Geromel, V.; Raymond, L.; Richard, P.; et al. Genome Alert!: A Standardized Procedure for Genomic Variant Reinterpretation and Automated Gene-Phenotype Reassessment in Clinical Routine. Genet. Med. 2022, 24, 1316–1327. [Google Scholar] [CrossRef] [PubMed]
- Birgmeier, J.; Haeussler, M.; Deisseroth, C.A.; Steinberg, E.H.; Jagadeesh, K.A.; Ratner, A.J.; Guturu, H.; Wenger, A.M.; Diekhans, M.E.; Stenson, P.D.; et al. AMELIE Speeds Mendelian Diagnosis by Matching Patient Phenotype and Genotype to Primary Literature. Sci. Transl. Med. 2020, 12, eaau9113. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ Data to High Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Liu, X.; Li, C.; Mou, C.; Dong, Y.; Tu, Y. dbNSFP v4: A Comprehensive Database of Transcript-Specific Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs. Genome Med. 2020, 12, 103. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A Program for Annotating and Predicting the Effects of Single Nucleotide Polymorphisms, SnpEff: SNPs in the Genome of Drosophila Melanogaster Strain W1118; Iso-2; Iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Jagadeesh, K.A.; Wenger, A.M.; Berger, M.J.; Guturu, H.; Stenson, P.D.; Cooper, D.N.; Bernstein, J.A.; Bejerano, G. M-CAP Eliminates a Majority of Variants of Uncertain Significance in Clinical Exomes at High Sensitivity. Nat. Genet. 2016, 48, 1581–1586. [Google Scholar] [CrossRef]
- Kircher, M.; Witten, D.M.; Jain, P.; O’Roak, B.J.; Cooper, G.M.; Shendure, J. A General Framework for Estimating the Relative Pathogenicity of Human Genetic Variants. Nat. Genet. 2014, 46, 310–315. [Google Scholar] [CrossRef]
- Li, Q.; Wang, K. InterVar: Clinical Interpretation of Genetic Variants by the 2015 ACMG-AMP Guidelines. Am. J. Hum. Genet. 2017, 100, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Niceta, M.; Ciolfi, A.; Ferilli, M.; Pedace, L.; Cappelletti, C.; Nardini, C.; Hildonen, M.; Chiriatti, L.; Miele, E.; Dentici, M.L.; et al. DNA Methylation Profiling in Kabuki Syndrome: Reclassification of Germline KMT2D VUS and Sensitivity in Validating Postzygotic Mosaicism. Eur. J. Hum. Genet. 2024, 32, 819–826. [Google Scholar] [CrossRef]
- Ciolfi, A.; Foroutan, A.; Capuano, A.; Pedace, L.; Travaglini, L.; Pizzi, S.; Andreani, M.; Miele, E.; Invernizzi, F.; Reale, C.; et al. Childhood-Onset Dystonia-Causing KMT2B Variants Result in a Distinctive Genomic Hypermethylation Profile. Clin. Epigenetics 2021, 13, 157. [Google Scholar] [CrossRef]
- Butcher, D.T.; Cytrynbaum, C.; Turinsky, A.L.; Siu, M.T.; Inbar-Feigenberg, M.; Mendoza-Londono, R.; Chitayat, D.; Walker, S.; Machado, J.; Caluseriu, O.; et al. CHARGE and Kabuki Syndromes: Gene-Specific DNA Methylation Signatures Identify Epigenetic Mechanisms Linking These Clinically Overlapping Conditions. Am. J. Hum. Genet. 2017, 100, 773–788. [Google Scholar] [CrossRef]
- Aref-Eshghi, E.; Kerkhof, J.; Pedro, V.P.; Groupe, D.I.F.; Barat-Houari, M.; Ruiz-Pallares, N.; Andrau, J.-C.; Lacombe, D.; Van-Gils, J.; Fergelot, P.; et al. Evaluation of DNA Methylation Episignatures for Diagnosis and Phenotype Correlations in 42 Mendelian Neurodevelopmental Disorders. Am. J. Hum. Genet. 2020, 106, 356–370. [Google Scholar] [CrossRef]
- Ferilli, M.; Ciolfi, A.; Pedace, L.; Niceta, M.; Radio, F.C.; Pizzi, S.; Miele, E.; Cappelletti, C.; Mancini, C.; Galluccio, T.; et al. Genome-Wide DNA Methylation Profiling Solves Uncertainty in Classifying NSD1 Variants. Genes 2022, 13, 2163. [Google Scholar] [CrossRef]
- Smedley, D.; Schubach, M.; Jacobsen, J.O.B.; Köhler, S.; Zemojtel, T.; Spielmann, M.; Jäger, M.; Hochheiser, H.; Washington, N.L.; McMurry, J.A.; et al. A Whole-Genome Analysis Framework for Effective Identification of Pathogenic Regulatory Variants in Mendelian Disease. Am. J. Hum. Genet. 2016, 99, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Rausch, T.; Zichner, T.; Schlattl, A.; Stütz, A.M.; Benes, V.; Korbel, J.O. DELLY: Structural Variant Discovery by Integrated Paired-End and Split-Read Analysis. Bioinformatics 2012, 28, i333–i339. [Google Scholar] [CrossRef] [PubMed]
- Geoffroy, V.; Herenger, Y.; Kress, A.; Stoetzel, C.; Piton, A.; Dollfus, H.; Muller, J. AnnotSV: An Integrated Tool for Structural Variations Annotation. Bioinformatics 2018, 34, 3572–3574. [Google Scholar] [CrossRef] [PubMed]
- Ti, S.-C.; Alushin, G.M.; Kapoor, T.M. Human β-Tubulin Isotypes Can Regulate Microtubule Protofilament Number and Stability. Dev. Cell 2018, 47, 175–190.e5. [Google Scholar] [CrossRef]
- Rosignoli, S.; Paiardini, A. DockingPie: A Consensus Docking Plugin for PyMOL. Bioinformatics 2022, 38, 4233–4234. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Adam, M.P.; Banka, S.; Bjornsson, H.T.; Bodamer, O.; Chudley, A.E.; Harris, J.; Kawame, H.; Lanpher, B.C.; Lindsley, A.W.; Merla, G.; et al. Kabuki Syndrome: International Consensus Diagnostic Criteria. J. Med. Genet. 2019, 56, 89–95. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Jaganathan, K.; Kyriazopoulou Panagiotopoulou, S.; McRae, J.F.; Darbandi, S.F.; Knowles, D.; Li, Y.I.; Kosmicki, J.A.; Arbelaez, J.; Cui, W.; Schwartz, G.B.; et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell 2019, 176, 535–548.e24. [Google Scholar] [CrossRef] [PubMed]
- Romaniello, R.; Tonelli, A.; Arrigoni, F.; Baschirotto, C.; Triulzi, F.; Bresolin, N.; Bassi, M.T.; Borgatti, R. A Novel Mutation in the β-Tubulin Gene TUBB2B Associated with Complex Malformation of Cortical Development and Deficits in Axonal Guidance. Dev. Med. Child. Neurol. 2012, 54, 765–769. [Google Scholar] [CrossRef]
- Romaniello, R.; Arrigoni, F.; Panzeri, E.; Poretti, A.; Micalizzi, A.; Citterio, A.; Bedeschi, M.F.; Berardinelli, A.; Cusmai, R.; D’Arrigo, S.; et al. Tubulin-Related Cerebellar Dysplasia: Definition of a Distinct Pattern of Cerebellar Malformation. Eur. Radiol. 2017, 27, 5080–5092. [Google Scholar] [CrossRef]
- Flaherty, M.P.; Grattan-Smith, P.; Steinberg, A.; Jamieson, R.; Engle, E.C. Congenital Fibrosis of the Extraocular Muscles Associated with Cortical Dysplasia and Maldevelopment of the Basal Ganglia. Ophthalmology 2001, 108, 1313–1322. [Google Scholar] [CrossRef] [PubMed]
- Whitman, M.C.; Andrews, C.; Chan, W.-M.; Tischfield, M.A.; Stasheff, S.F.; Brancati, F.; Ortiz-Gonzalez, X.; Nuovo, S.; Garaci, F.; MacKinnon, S.E.; et al. Two Unique TUBB3 Mutations Cause Both CFEOM3 and Malformations of Cortical Development. Am. J. Med. Genet. A 2016, 170A, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.C.; Olney, A.H.; Beavers, A.; Spaulding, J.; Nelson, M.; Nielsen, S.; Sanmann, J.N. The Recurrent TUBB3 Gly98Ser Substitution Is the First Described to Inconsistently Result in CFEOM3. Am. J. Med. Genet. A 2020, 182, 2161–2167. [Google Scholar] [CrossRef]
- Geiger, J.T.; Schindler, A.B.; Blauwendraat, C.; Singer, H.S.; Scholz, S.W. TUBB2B Mutation in an Adult Patient with Myoclonus-Dystonia. Case Rep. Neurol. 2017, 9, 216–221. [Google Scholar] [CrossRef]
- Çitli, Ş.; Serdaroglu, E. Maternal Germline Mosaicism of a de Novo TUBB2B Mutation Leads to Complex Cortical Dysplasia in Two Siblings. Fetal Pediatr. Pathol. 2022, 41, 155–165. [Google Scholar] [CrossRef]
- Breuss, M.W.; Nguyen, T.; Srivatsan, A.; Leca, I.; Tian, G.; Fritz, T.; Hansen, A.H.; Musaev, D.; McEvoy-Venneri, J.; James, K.N.; et al. Uner Tan Syndrome Caused by a Homozygous TUBB2B Mutation Affecting Microtubule Stability. Hum. Mol. Genet. 2017, 26, 258–269. [Google Scholar] [CrossRef] [PubMed]
- Demer, J.L.; Clark, R.A.; Engle, E.C. Magnetic Resonance Imaging Evidence for Widespread Orbital Dysinnervation in Congenital Fibrosis of Extraocular Muscles Due to Mutations in KIF21A. Investig. Ophthalmol. Vis. Sci. 2005, 46, 530–539. [Google Scholar] [CrossRef]
- Demer, J.L.; Clark, R.A.; Tischfield, M.A.; Engle, E.C. Evidence of an Asymmetrical Endophenotype in Congenital Fibrosis of Extraocular Muscles Type 3 Resulting from TUBB3 Mutations. Investig. Ophthalmol. Vis. Sci. 2010, 51, 4600–4611. [Google Scholar] [CrossRef]
- Thomas, M.G.; Maconachie, G.D.E.; Kuht, H.J.; Chan, W.-M.; Sheth, V.; Hisaund, M.; McLean, R.J.; Barry, B.; Al-Diri, B.; Proudlock, F.A.; et al. Optic Nerve Head and Retinal Abnormalities Associated with Congenital Fibrosis of the Extraocular Muscles. Int. J. Mol. Sci. 2021, 22, 2575. [Google Scholar] [CrossRef]
- Cushion, T.D.; Dobyns, W.B.; Mullins, J.G.L.; Stoodley, N.; Chung, S.-K.; Fry, A.E.; Hehr, U.; Gunny, R.; Aylsworth, A.S.; Prabhakar, P.; et al. Overlapping Cortical Malformations and Mutations in TUBB2B and TUBA1A. Brain 2013, 136, 536–548. [Google Scholar] [CrossRef] [PubMed]
- Amemiya, H.M.; Kundaje, A.; Boyle, A.P. The ENCODE Blacklist: Identification of Problematic Regions of the Genome. Sci. Rep. 2019, 9, 9354. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mancini, C.; Chiriatti, L.; Bruselles, A.; D’ambrosio, P.; Ciolfi, A.; Ferilli, M.; Cappelletti, C.; Carvetta, M.; Radio, F.C.; Cordeddu, V.; et al. The p.Ile202Thr Substitution in TUBB2B Can Be Associated with Syndromic Presentation of Congenital Fibrosis of the Extraocular Muscles. Genes 2025, 16, 1182. https://doi.org/10.3390/genes16101182
Mancini C, Chiriatti L, Bruselles A, D’ambrosio P, Ciolfi A, Ferilli M, Cappelletti C, Carvetta M, Radio FC, Cordeddu V, et al. The p.Ile202Thr Substitution in TUBB2B Can Be Associated with Syndromic Presentation of Congenital Fibrosis of the Extraocular Muscles. Genes. 2025; 16(10):1182. https://doi.org/10.3390/genes16101182
Chicago/Turabian StyleMancini, Cecilia, Luigi Chiriatti, Alessandro Bruselles, Paola D’ambrosio, Andrea Ciolfi, Marco Ferilli, Camilla Cappelletti, Mattia Carvetta, Francesca Clementina Radio, Viviana Cordeddu, and et al. 2025. "The p.Ile202Thr Substitution in TUBB2B Can Be Associated with Syndromic Presentation of Congenital Fibrosis of the Extraocular Muscles" Genes 16, no. 10: 1182. https://doi.org/10.3390/genes16101182
APA StyleMancini, C., Chiriatti, L., Bruselles, A., D’ambrosio, P., Ciolfi, A., Ferilli, M., Cappelletti, C., Carvetta, M., Radio, F. C., Cordeddu, V., Niceta, M., Parrino, M., Capolino, R., Mammì, C., Senese, R., Muto, M., Priolo, M., & Tartaglia, M. (2025). The p.Ile202Thr Substitution in TUBB2B Can Be Associated with Syndromic Presentation of Congenital Fibrosis of the Extraocular Muscles. Genes, 16(10), 1182. https://doi.org/10.3390/genes16101182






