A New Variant in the NALCN Channel Is Responsible for Cerebellar Ataxia and Cognitive Impairment
Abstract
1. Introduction
2. Materials and Methods
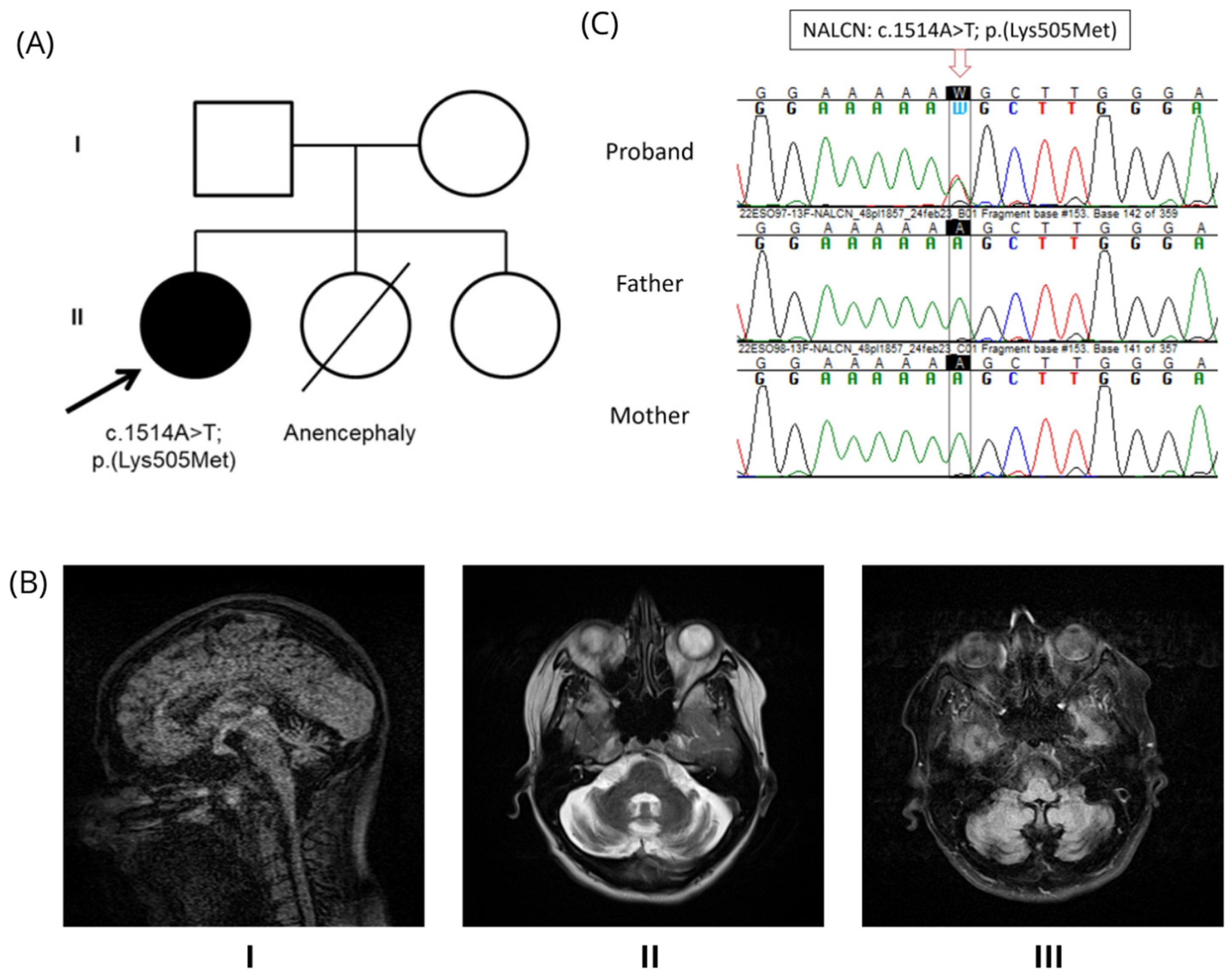
2.1. Family Trio
2.2. Whole Exome Sequencing and Sanger Sequencing
3. Results
3.1. Clinical Description
3.2. Genetic Findings
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CLIFAHDD | Congenital Contractures of the Limbs and Face, Hypotonia, and Developmental Delay |
| WES | Whole Exome Sequencing |
| DA2A | Distal arthrogryposis type 2A |
| HPO | Human Phenotype Ontology |
| CNV | Copy Number Variations |
| IGV | Integrative Genomic Viewer |
| ACMG | American College of Medical Genetics and Genomics |
| SARA | Scale of Assessment and Rating of Ataxia |
| SCA | Spinocerebellar Ataxia |
| FLAIR | Fluid Attenuated Inversion Recovery |
| LOVD | Leiden Open Variation Database |
| MAF | Minor allele Frequency |
| MRI | Magnetic Resonance Imaging |
| GnomAD | Genome Aggregation Database |
| SIFT | Sorting Intolerant From Tolerant |
| PolyPhen-2 | Polymorphism Phenotyping v2 |
| WAIS | Wechsler Adult Intelligence Scale |
References
- Chong, J.X.; McMillin, M.J.; Shively, K.M.; Beck, A.E.; Marvin, C.T.; Armenteros, J.R.; Buckingham, K.J.; Nkinsi, N.T.; Boyle, E.A.; Berry, M.N.; et al. De Novo mutations in NALCN cause a syndrome characterized by congenital contractures of the limbs and face, hypotonia, and developmental delay. Am. J. Hum. Genet. 2015, 96, 462–473. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Abe, C.; Holloway, B.B.; Shu, S.; Kumar, N.N.; Weaver, J.L.; Sen, J.; Perez-Reyes, E.; Stornetta, R.L.; Guyenet, P.G.; et al. Nalcn is a “leak” sodium channel that regulates excitability of brainstem chemosensory neurons and breathing. J. Neurosci. 2016, 36, 8174–8187. [Google Scholar] [CrossRef] [PubMed]
- Cochet-Bissuel, M.; Lory, P.; Monteil, A. The sodium leak channel, NALCN, in health and disease. Front Cell Neurosci. 2014, 8, 132. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Wu, J.X.; Chen, L. Structure of voltage-modulated sodium-selective NALCN-FAM155A channel complex. Nat. Commun. 2020, 11, 6199. [Google Scholar] [CrossRef] [PubMed]
- Bouasse, M.; Impheng, H.; Servant, Z.; Lory, P.; Monteil, A. Functional expression of CLIFAHDD and IHPRF pathogenic variants of the NALCN channel in neuronal cells reveals both gain- and loss-of-function properties. Sci. Rep. 2019, 9, 11791. [Google Scholar] [CrossRef] [PubMed]
- Kschonsak, M.; Chua, H.C.; Noland, C.L.; Weidling, C.; Clairfeuille, T.; Bahlke, O.Ø.; Ameen, A.O.; Li, Z.R.; Arthur, C.P.; Ciferri, C.; et al. Structure of the human sodium leak channel NALCN. Nature 2020, 587, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wei, Y. Role of sodium leak channel (NALCN) in sensation and pain: An overview. Front Pharmacol. 2024, 14, 1349438. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.; FitzPatrick, D.R.; Azam, T.; Gibson, N.A.; Somerville, L.; Joss, S.K.; Deciphering Developmental Disorders Study; Urquhart, D.S. NALCN dysfunction as a cause of disordered respiratory rhythm with central apnea. Pediatrics 2018, 141 (Suppl. 5), S485–S490. [Google Scholar] [CrossRef] [PubMed]
- Harvey, J.R.M.; Plante, A.E.; Meredith, A.L. Ion channels controlling circadian rhythms in suprachiasmatic nucleus excitability. Physiol. Rev. 2020, 100, 1415–1454. [Google Scholar] [CrossRef] [PubMed]
- Matthijs, G.; Souche, E.; Alders, M.; Corveleyn, A.; Eck, S.; Feenstra, I.; Race, V.; Sistermans, E.; Sturm, M.; Weiss, M.; et al. Guidelines for diagnostic next-generation sequencing. Eur. J. Hum. Genet. 2016, 24, 1515–1525. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.C.; Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef] [PubMed]
- Davydov, E.V.; Goode, D.L.; Sirota, M.; Cooper, G.M.; Sidow, A.; Batzoglou, S. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol. 2010, 6, e1001025. [Google Scholar] [CrossRef] [PubMed]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Aguilera, M.A.; Meyer, R.; Massouras, A. VarSome: The human genomic variant search engine. Bioinformatics 2019, 35, 1978–1980. [Google Scholar] [CrossRef] [PubMed]
- Landrum, M.J.; Lee, J.M.; Riley, G.R.; Jang, W.; Rubinstein, W.S.; Church, D.M.; Maglott, D.R. ClinVar: Public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014, 42, D980–D985, (Database issue). [Google Scholar] [CrossRef] [PubMed]
- Schmitz-Hubsch, T.; Du Montcel, S.T.; Baliko, L.; Berciano, J.; Boesch, S.; Depondt, C.; Giunti, P.; Globas, C.; Infante, J.; Kang, J.S.; et al. Scale for the assessment and rating of ataxia: Development of a new clinical scale. Neurology 2006, 66, 1717–1720. [Google Scholar] [CrossRef] [PubMed]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alfoldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Ke, M.; Xu, L.; Lin, S.; Huang, J.; Zhang, J.; Yang, F.; Wu, J.; Yan, Z. Structure of the human sodium leak channel NALCN in complex with FAM155A. Nat. Commun. 2020, 11, 6148. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Koh, K.; Ichinose, Y.; Yasumura, M.; Ohtsuka, T.; Takiyama, Y. A de novo mutation in the NALCN gene in an adult patient with cerebellar ataxia associated with intellectual disability and arthrogryposis. Clin. Genet. 2016, 90, 556–557. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cabrita Pinto, R.L.; Fancellu, R.; Markushi, T.B.; Viaggi, S.; Testa, B.; Conteduca, G.; Fitzsimmons, L.; Coviello, D.; Covone, A.E. A New Variant in the NALCN Channel Is Responsible for Cerebellar Ataxia and Cognitive Impairment. Genes 2025, 16, 1181. https://doi.org/10.3390/genes16101181
Cabrita Pinto RL, Fancellu R, Markushi TB, Viaggi S, Testa B, Conteduca G, Fitzsimmons L, Coviello D, Covone AE. A New Variant in the NALCN Channel Is Responsible for Cerebellar Ataxia and Cognitive Impairment. Genes. 2025; 16(10):1181. https://doi.org/10.3390/genes16101181
Chicago/Turabian StyleCabrita Pinto, Rute Luísa, Roberto Fancellu, Tiziana Benzi Markushi, Silvia Viaggi, Barbara Testa, Giuseppina Conteduca, Lane Fitzsimmons, Domenico Coviello, and Angela Elvira Covone. 2025. "A New Variant in the NALCN Channel Is Responsible for Cerebellar Ataxia and Cognitive Impairment" Genes 16, no. 10: 1181. https://doi.org/10.3390/genes16101181
APA StyleCabrita Pinto, R. L., Fancellu, R., Markushi, T. B., Viaggi, S., Testa, B., Conteduca, G., Fitzsimmons, L., Coviello, D., & Covone, A. E. (2025). A New Variant in the NALCN Channel Is Responsible for Cerebellar Ataxia and Cognitive Impairment. Genes, 16(10), 1181. https://doi.org/10.3390/genes16101181






