Uncovering the PML::RARA Fusion in Cytogenetically Cryptic and FISH-Negative Acute Promyelocytic Leukemia—A Case Report and Comprehensive Literature Review
Abstract
1. Introduction
2. Materials and Methods
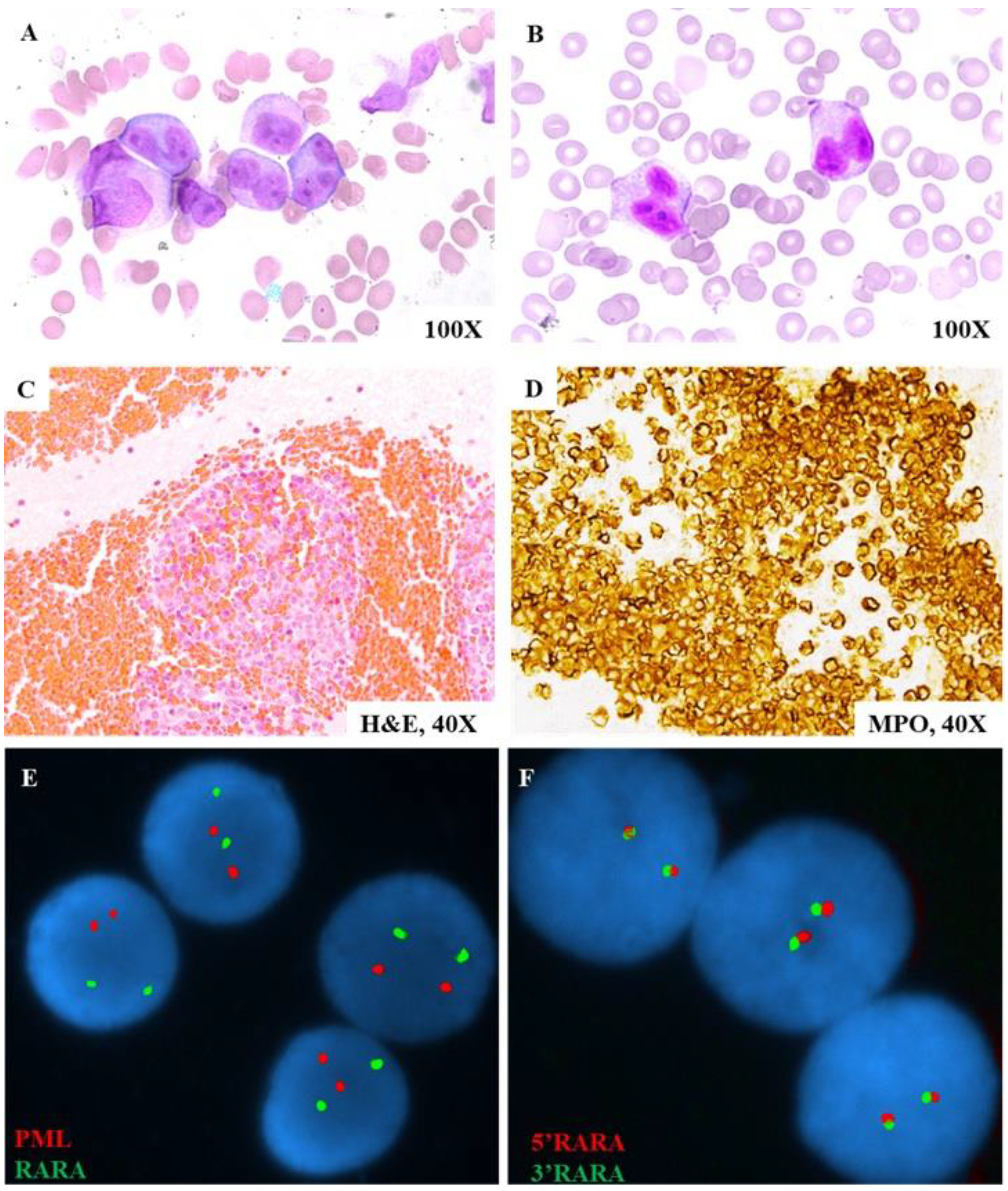
2.1. Clinical and Pathological Examination
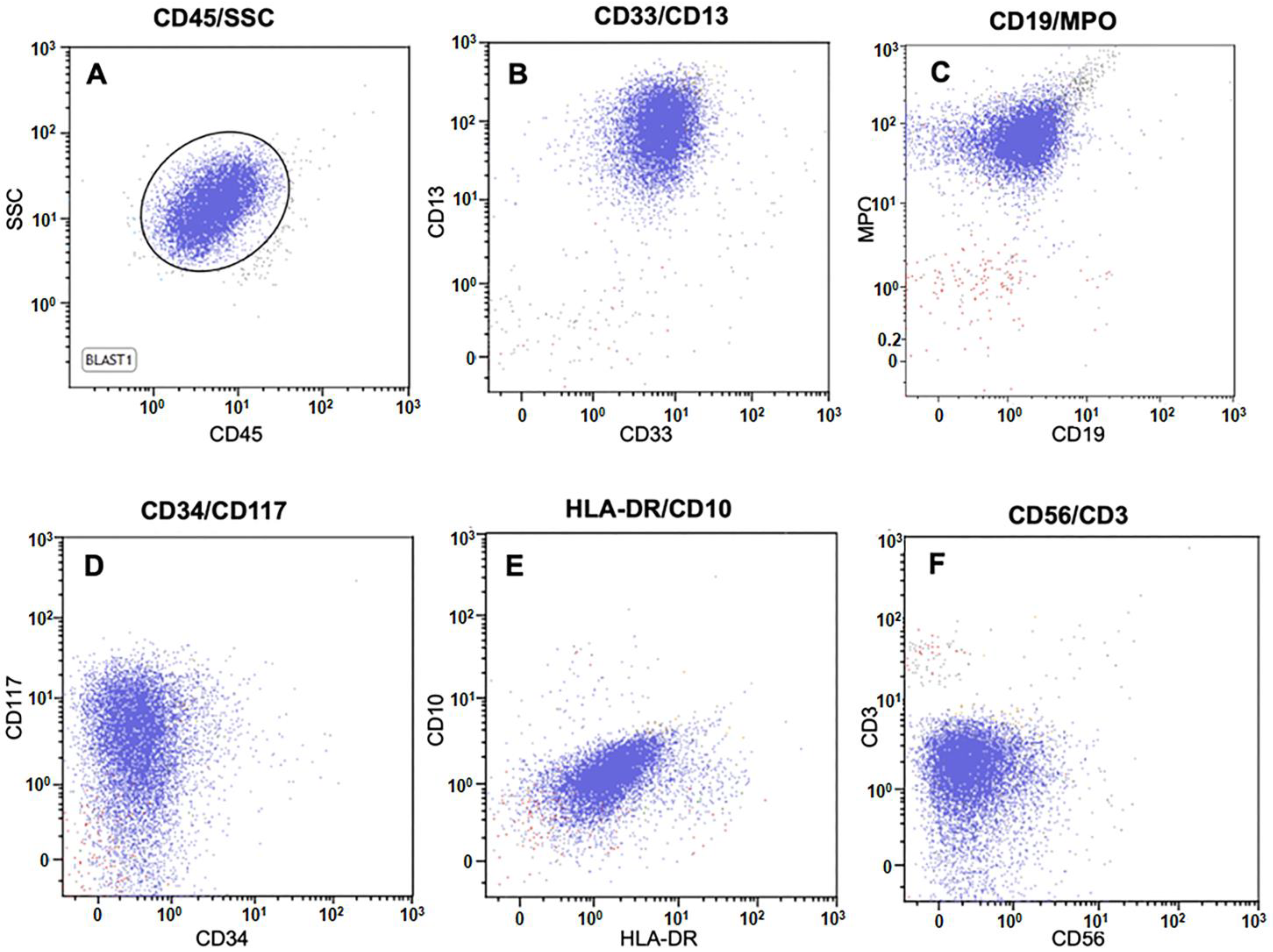
2.2. Flow Cytometry
2.3. Conventional Cytogenetic and Interphase Fluorescence In Situ Hybridization (FISH) Studies
2.4. Literature Review
3. Results
3.1. Case Study
3.2. Literature Review
| No. | Age/ Sex | WBC (109/L) | Hb (g/dL) | Plt (109/L) | Pro-myelocytes | Immuno-phenotype | Coagulation Tests | DIC at Dx | Reason for Presentation | Karyogram | FISH # | RT-PCR * | Treatment | Survival (Months) /Terminal Event | CR | PMID |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 39/F | 242.2 | 8.8 | 20 | Yes 1 | CD13+, CD33+, MPO+, HLA- DR+; CD34− | PT↑, aPTT↑ | Yes | History of CML with persistent fever, malaise, and weight loss | 46,XX,t(9;22)[20] | ND | S (bcr3) | IDA + Ara-C | No (5) /Bilateral mycotic pneumonia | Yes | 7736444 [8] (Emilia, 1995) |
| 2 | 25/F | 2.9 | 8.3 | 9 | Yes | CD33+; HLA-DR− | PT↑, FDP↑, fibrinogen↓, aPTT (nl) | Yes | Purpura and nasal bleeding | 46,XX,del(9)(q22)[20] | ND | L (bcr1) | IDA + ATRA + Ara-C | No (<1) /ICH | No | 10484977 [9] (Yamamoto, 1999) |
| 3 | N/A | N/A | N/A | N/A | Yes | N/A | N/A | N/A | N/A | 45,XY,add(2)(q37),−7,add(9)(p22)/45,idem,add(10)(p14) | ND | L (bcr1) | N/A | N/A | N/A | 10942371 [10] (Grimwade, 2000) |
| 4 | N/A | N/A | N/A | N/A | Yes | N/A | N/A | N/A | N/A | 46,XY[20] | ND | L (bcr1) | N/A | N/A | N/A | 10942371 [10] (Grimwade, 2000) |
| 5 | N/A | N/A | N/A | N/A | Yes | N/A | N/A | N/A | N/A | 46,XX[20] | ND | Positive (unclear variant) | N/A | N/A | N/A | 10942371 [10] (Grimwade, 2000) |
| 6 | N/A | N/A | N/A | N/A | Yes | N/A | N/A | N/A | N/A | 46,XX[20] | ND | S (bcr3) | N/A | N/A | N/A | 10942371 [10] (Grimwade, 2000) |
| 7 | N/A | N/A | N/A | N/A | Yes | N/A | N/A | N/A | N/A | 46,XY[29] | ND | Positive (unclear variant) | N/A | N/A | N/A | 10942371 [10] (Grimwadel, 2000) |
| 8 | N/A | N/A | N/A | N/A | Yes | N/A | N/A | N/A | N/A | 46,XY[20] | ND | V (bcr2) | N/A | N/A | N/A | 10942371 [10] (Grimwade, 2000) |
| 9 | 48/F | 14.93 | 8.8 | 36 | Yes | CD13+, CD33+, MPO+, CD56+; HLA-DR−, CD34− | PT↑, D-dimer↑, fibrinogen↓, AT (nl), aPTT (nl) | No | Vaginal bleeding, history of uterine myoma | 47,XY,+8[14]/46,XX[2] | ND | S (bcr3) | IDA + ATRA | Yes (>1) | Yes | 16797070 [11] (Han, 2007) |
| 10 | 14/M | N/A | N/A | N/A | Yes/hypo-cellular | Confirmed APL, WHO criteria | N/A | N/A | N/A | 46,XY[20] | ND | S (bcr3) | IDA + ATRA | No (10) /GVHD | Yes | 17943164 [12] (Kim, 2008) |
| 11 | 63/F | N/A | N/A | N/A | Yes | Confirmed APL, WHO criteria | N/A | N/A | N/A | 46,XX[20] | ND | S (bcr3) | IDA + Ara-C | No (6) /Pulmonary hemorrhage | Yes | 17943164 [12] (Kim, 2008) |
| 12 | 44/F | 1.5 | 12.6 | 49 | Yes | CD13+, CD33+, MPO+; HLA-DR−, CD34− | N/A | N/A | Immature cells in peripheral blood | 46,XX,i(17)(q10)[12]/46,XX[8] | ND | L (bcr1) | IDA + ATRA + Ara-C | Yes (>1) | Yes | 18294238 [13] (Huh J, 2008) |
| 13 | 50/F | 101.73 | 9.9 | 16 | Yes | CD13+, CD33+, CD117+, CD7+, MPO+; HLA-DR−, CD34− | PT↑, FDP↑, fibrinogen↓, D-dimer↑, aPTT (nl), AT (nl) | No | Cough, fever, and general weakness for a month | 47,XY,+8[19]/46,XX[1] | ND | S (bcr3) | IDA + ATRA | Yes (>5) | Yes | 19893344 [14] (Kim, 2009) |
| 14 | 39/F | 16.9 | 7.6 | 174 | Yes | CD13+, CD33+, MPO+; HLA-DR−, CD34− | fibrinogen↓, FDP↑, D-dimer↑, PT (nl), aPTT (nl) | No | Two weeks of fatigue, five days of high fever and irregular uterine bleeding | 46,XX,7q[7]+/46,XX[8] | ND | L (bcr1) | ATRA + ATO | Yes (>2) | Yes 2 | 19162322 [15] (Wang, 2009) |
| 15 | 26/F | 0.6 | 7.7 | 155 | Yes | CD13+, CD33+, CD117+; HLA-DR−, CD34− | PT↑, INR↑, aPTT (nl) | No | Fever, pallor and bleeding gums | del(5q) | ND | S (bcr 3) | IDA + ATRA | Yes (>8) | Yes | 19224461 [16] (Choughule, 2009) |
| 16 | 33/F | 1.39 | 7.1 | 9.8 | Yes | CD13+, CD33+, CD117+; HLA-DR−, CD34− | Normal | No | Fever and fatigue | Normal | ND | S (bcr 3) | ATO | Yes (>12) | Yes | 19224461 [16] (Choughule, 2009) |
| 17 | 46/M | 63.8 | 6.6 | 146 | Yes | CD13+, CD33+, MPO+; CD117−, HLA-DR−, CD34− | PT↑, aPTT↑, INR↑ | No | Three weeks of fatigue and intermittent fever | del(19p13), del(12q24.1), del(5) | ND | L (bcr1) | IDA + ATRA | Yes (>8) | Yes | 19224461 [16] (Choughule, 2009) |
| 18 | 18/M | 16.5 | 9.5 | 37 | Yes | CD13+, CD33+, CD45+, CD117+; HLA-DR−, CD34− | N/A | No | Hematuria and hematochezia | 46,XY[20] | ND | L (bcr1), V (bcr2) | ATRA | Yes (>1) | N/A | 21156244 [6] (Kim, 2010) |
| 19 | 46/M | 1.7 | 10.6 | 18 | Yes | CD13+, CD33+, MPO+; HLA-DR−, CD34− | PT↑, INR↑, D-dimer↑ | No | Gengival bleeding | 92,XXYY[13]/46,XY[7] | ND | S (bcr3) | IDA + ATRA | Yes (>14) | Yes | 20417966 [17] (Soriani, 2010) |
| 20 | 24/M | 64.3 | N/A | N/A | Microgranular variant | CD13+, CD33+, CD117+; CD45+, CD34+; HLA-DR− | N/A | Yes | Suboccipital headaches, nausea, photophobia, gingival bleeding, debilitating nuchal rigidity, and low-grade fevers | 46,XY[20] | ND | Positive (unclear variant) | ATRA + ATO + Ara-C | Yes (>2) | Yes | 22018276 [18] (Lewis, 2011) |
| 21 | 23/M | 3.94 | 12 | 36 | Yes | CD13+, CD33+; CD34− | N/A | N/A | Ecchymosis and thrombocytopenia | 47,XX,+8[19]/46,XY[1] | ND | L (bcr1) | ATRA + Chemo | n/a | Yes | 22402611 [19] (Yang, 2012) |
| 22 | 57/M | 6.82 | 8.9 | 40 | Yes | CD13+, CD33+, CD117+; HLA-DR−, CD34− | N/A | N/A | Oral ulcer, odynophagia, anemia, and thrombocytopenia | 46,XY[20] | ND | L (bcr1) | ATRA + ATO + Ara-C | Yes (>5) | Yes | 23370423 [20] (Gruver, 2013) |
| 23 | 67/M | 1.9 | 10.8 | 89 | Yes | MPO+; CD34−, HLA-DR− | PTT↓, fibrinogen↓, INR↑, D-dimer↑ | Yes | Easy bruising, fatigue, pancytopenia | 46,XY[20] | ND | Positive (unclear variant) | ATRA | N/A | N/A | 25580502 [21] (Rashidi, 2014) |
| 24 | 17/M | 9.9 | n/a | n/a | Yes | CD13+, CD33+, CD117+; HLA-DR−, CD34−, CD11b− | N/A | Yes | gum bleeding, multiple ecchymoses, abdominal pain, and fever | 46,XY[20] | ND | L (bcr1) | ATRA + ATO + Chemo | Yes (>24) | Yes | 24561214 [22] (Blanco, 2014) |
| 25 | 25/M | 5.2 | 11.6 | 30 | Yes | CD117+, CD64+, CD123+, CD13+, MPO+; CD34−, HLA-DR− | PT↑, INR↑, TT↑ | N/A | Gum bleeding for over 20 days | 46,XY[20] | ND | S (bcr3) | ATRA + ATO + IDA | Yes (>2) | Yes | 27995890 [23] (Wang, 2016) |
| 26 | 66/M | 2.95 | 78 | 7 | No 3 | CD7+, CD13+, CD33+, CD34+, Cd38+, CD117+, CD38, HLA-DR+, MPO+ | PT↑, fibrinogen↑, D-dimer↑, aPTT (nl) | N/A | Petechiae or bruises on the lower limbs that lasted for half a month with intermittent fever and coughing | 46,XY[20] | ND | V (atypical) 4 | IDA + ATRA + Ara-C | Yes (>7) | Yes | 31959056 [24] (Zhang, 2020) |
| 27 | 57/F | n/a | 9.7 | 57 | Microgranular variant | CD13+, CD33+, CD34+, CD117+, MPO+, CD2+; HLA-DR−, CD11b−, | fibrinogen↓ D-dimer↑, PT (nl), PTT (nl), | No | Bruising and gingival bleeding over a two-week period | 46,XX[20] | ND | L (bcr1) | ATRA + ATO | Yes (>1) | Yes | 31809670 [25] (Schultz, 2020) |
| 28 | 17/M | 48.4 | 8.3 | 14 | Yes | CD13+, CD33+, CD38+, CD45+, CD64+, CD117+, HLA-DR+ (small subset), MPO +; CD34− | fibrinogen↓, D-dimer↑, PTT↑, TT (nl) | Yes | Unresponsive during a seizure after two-day history of nausea, blood-tinged vomiting, lethargy, and right-sided weakness | 46,XY[20] | ND | Positive (unclear variant) | ATRA | N/A | Yes | 32366568 [1] (Mai, 2020) |
| 29 | 12/F | 22.5 | 7.4 | 16 | Yes | CD33+, CD34+, MPO+, CD117+, HLA-DR+; CD11b− | fibrinogen↓, PT↑, D-dimer (nl) | Yes | Multiple ecchymoses | 46,XX[20] | ND | S (bcr3) | ATRA + Chemo | Yes (>26) | Yes | 32909480 [26] (Avgerinou, 2020) |
| 30 | 56/F | 0.7 | 8 | 96 | Yes | CD13+, CD33+, CD117+, CD34+, MPO+, CD56+; HLA DR− | N/A | No | Generalized pruritis for 1 month, fever and headache for 1 week | 46,XX[20] | ND | L (bcr1) | ATRA + DNR + Ara-C | No (<1) /Refractory septic shock with encephalopathy | No | 33851647 [27] (Arumugam, 2021) |
| 31 | 54/M | 1.6 | 9.4 | 69 | Yes | MPO+, CD117+; CD34−, HLA-DR− | N/A | N/A | Left leg swelling and a left femoral vein thrombosis, syncopal episode | 46,XY[20] | ND | L (bcr1) | ATRA + ATO | N/A | Yes | 35572917 [28] (Karlin, 2022) |
| 32 | 27/M | 4.67 | 15 | 144 | Yes | CD13+, CD33+, MPO+; CD34−, HLA-DR− | PT↑, D-dimer↓, aPTT (nl), fibrinogen (nl), | no | Imaging findings of marrow heterogeneity in T12 during L4–L5 disk herniation work-up | 46,XY[20] | ND | L (bcr1) | ATRA + ATO | Yes (n/a) | Yes | 37685882 [29] (Mohebnasab, 2023) |
| 33 | 32/F | 1.57 | 8.1 | 68 | Yes | CD2+, CD13+, CD33+, CD117+, MPO+ | PT↑, fibrinogen↓, D-dimer↑, aPTT (nl), | Yes | Several weeks of fatigue and easy bruising | 46,XX[20] | ND | S (bcr3) | ATRA + ATO | Yes (n/a) | Yes | 37685882 [29] (Mohebnasab, 2023) |
| 34 & | 23/M | 80.1 | 4.8 | 22 | Yes | CD13+, CD33+, CD38+, CD64+, CD117+, MPO+; HLA-DR−, D34−, | PT↑, INR↑, PTT (nl) | N/A | Intracranial hemorrhage, 2 weeks post dental cleaning for pain | N/A | ND | L (bcr1) | ATRA | No (<1) /Acute respiratory failure secondary to ICH | No | N/A |
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mai, B.; Liang, C.; Nguyen, A.; Wahed, A.; Chen, L. Cryptic Acute Promyelocytic Leukemia (APL) Presenting as Seizures in an Adolescent. Ann. Clin. Lab. Sci. 2020, 50, 270–273. [Google Scholar]
- Li, G.; Wu, J.; Li, R.; Pan, Y.; Ma, W.; Xu, J.; Nan, M.; Hou, L. Improvement of Early Death in Acute Promyelocytic Leukemia: A Population-Based Analysis. Clin. Lymphoma Myeloma Leuk. 2023, 23, e78–e84. [Google Scholar]
- Zhang, X.; Sun, J.; Yu, W.; Jin, J. Current views on the genetic landscape and management of variant acute promyelocytic leukemia. Biomark. Res. 2021, 9, 33. [Google Scholar] [CrossRef]
- Guarnera, L.; Ottone, T.; Fabiani, E.; Divona, M.; Savi, A.; Travaglini, S.; Falconi, G.; Panetta, P.; Rapanotti, M.C.; Voso, M.T. Atypical Rearrangements in APL-Like Acute Myeloid Leukemias: Molecular Characterization and Prognosis. Front. Oncol. 2022, 12, 871590. [Google Scholar] [CrossRef]
- Gagnon, M.F.; Berg, H.E.; Meyer, R.G.; Sukov, W.R.; Van Dyke, D.L.; Jenkins, R.B.; Greipp, P.T.; Thorland, E.C.; Hoppman, N.L.; Xu, X.; et al. Typical, atypical and cryptic t(15;17)(q24;q21) (PML::RARA) observed in acute promyelocytic leukemia: A retrospective review of 831 patients with concurrent chromosome and PML::RARA dual-color dual-fusion FISH studies. Genes. Chromosomes Cancer 2022, 61, 629–634. [Google Scholar] [CrossRef]
- Kim, M.J.; Cho, S.Y.; Kim, M.H.; Lee, J.J.; Kang, S.Y.; Cho, E.H.; Marschalek, R.; Meyer, C. FISH-negative cryptic PML-RARA rearrangement detected by long-distance polymerase chain reaction and sequencing analyses: A case study and review of the literature. Cancer Genet. Cytogenet. 2010, 203, 278–283. [Google Scholar] [CrossRef]
- Rashidi, A.; Fisher, S.I. FISH-negative, cytogenetically cryptic acute promyelocytic leukemia. Blood Cancer J. 2015, 5, e320. [Google Scholar] [CrossRef]
- Emilia, G.; Marasca, R.; Longo, G.; Ferrari, M.G.; Notohamiprodjo, M.; Temperani, P.; Sacchi, S.; Torelli, G. Detection of PML-RAR alpha fusion transcript in Ph positive leukemia with acute promyelocytic phenotype lacking the t(15;17) cytogenetic abnormality. Cancer Genet. Cytogenet. 1995, 80, 95–99. [Google Scholar] [CrossRef]
- Yamamoto, K.; Hamaguchi, H.; Kobayashi, M.; Tsurukubo, Y.; Nagata, K. Terminal deletion of the long arm of chromosome 9 in acute promyelocytic leukemia with a cryptic PML/RAR alpha rearrangement. Cancer Genet. Cytogenet. 1999, 113, 120–125. [Google Scholar]
- Grimwade, D.; Biondi, A.; Mozziconacci, M.J.; Hagemeijer, A.; Berger, R.; Neat, M.; Howe, K.; Dastugue, N.; Jansen, J.; Radford-Weiss, I.; et al. Characterization of acute promyelocytic leukemia cases lacking the classic t(15;17): Results of the European Working Party. Groupe Francais de Cytogenetique Hematologique, Groupe de Francais d’Hematologie Cellulaire, UK Cancer Cytogenetics Group and BIOMED 1 European Community-Concerted Action “Molecular Cytogenetic Diagnosis in Haematological Malignancies”. Blood 2000, 96, 1297–1308. [Google Scholar]
- Han, J.Y.; Kim, K.E.; Kim, K.H.; Park, J.I.; Kim, J.S. Identification of PML-RARA rearrangement by RT-PCR and sequencing in an acute promyelocytic leukemia without t(15;17) on G-banding and FISH. Leuk. Res. 2007, 31, 239–243. [Google Scholar] [CrossRef]
- Kim, M.; Lim, J.; Kim, Y.; Han, K.; Lee, D.H.; Chung, N.G.; Cho, B.; Kim, H.K.; Eom, K.S.; Min, C.-K.; et al. The genetic characterization of acute promyelocytic leukemia with cryptic t(15;17) including a new recurrent additional cytogenetic abnormality i(17)(q10). Leukemia 2008, 22, 881–883. [Google Scholar] [CrossRef]
- Huh, J.; Moon, H.; Chi, H.; Chung, W. Acute promyelocytic leukemia with i(17)(q10) on G-banding and PML/RARA rearrangement by RT-PCR without evidence of PML/RARA rearrangement on FISH. Int. J. Lab. Hematol. 2009, 31, 372–374. [Google Scholar] [CrossRef]
- Kim, K.E.; Woo, K.S.; Kim, S.H.; Han, J.Y. Detection of PML/RARA rearrangement by reverse transcriptase-PCR and sequencing in a case of microgranular acute promyelocytic leukemia lacking t(15;17) on karyotype and FISH. Korean J. Lab. Med. 2009, 29, 379–383. [Google Scholar]
- Wang, Y.; Fang, M.; Jing, Y.; Li, J.; Jiang, F.; Wang, Y. Derivative (7)t(7;8): The sole karyotype abnormality in acute promyelocytic leukemia with PML/RARA rearrangement identified by RT-PCR and sequence analysis. Leuk. Res. 2009, 33, e55–e58. [Google Scholar]
- Choughule, A.; Polampalli, S.; Amre, P.; Shinde, S.; Banavali, S.; Prabhash, K.; Nair, R.; Subramanian, P.G.; Gujral, S.; Parikh, P.M. Identification of PML/RARalpha fusion gene transcripts that showed no t(15;17) with conventional karyotyping and fluorescent in situ hybridization. Genet. Mol. Res. 2009, 8, 1–7. [Google Scholar]
- Soriani, S.; Cesana, C.; Farioli, R.; Scarpati, B.; Mancini, V.; Nosari, A. PML/RAR-alpha fusion transcript and polyploidy in acute promyelocytic leukemia without t(15;17). Leuk. Res. 2010, 34, e261–e263. [Google Scholar]
- Lewis, C.; Patel, V.; Abhyankar, S.; Zhang, D.; Ketterling, R.P.; McClure, R.F.; Persons, D.L. Microgranular variant of acute promyelocytic leukemia with normal conventional cytogenetics, negative PML/RARA FISH and positive PML/RARA transcripts by RT-PCR. Cancer Genet. 2011, 204, 522–523. [Google Scholar] [CrossRef]
- Yang, J.J.; Park, T.S.; Kim, M.J.; Cho, E.H.; Oh, S.H.; Jeon, B.R.; Oh, D.; Huh, J.Y.; Marschalek, R.; Meyer, C. Acute promyelocytic leukemia with trisomy 8 showing normal PML-RARA FISH signal patterns: Diagnostic application of long-distance polymerase chain reaction in molecularly discrepant leukemia cases. Ann. Hematol. 2012, 91, 1645–1648. [Google Scholar] [CrossRef]
- Gruver, A.M.; Rogers, H.J.; Cook, J.R.; Ballif, B.C.; Schultz, R.A.; Batanian, J.R.; Fesler, M.J.; Tubbs, R.R. Modified array-based comparative genomic hybridization detects cryptic and variant PML-RARA rearrangements in acute promyelocytic leukemia lacking classic translocations. Diagn. Mol. Pathol. 2013, 22, 10–21. [Google Scholar]
- Rashidi, A.; Fisher, S.I. FISH: Negative. Morphology: Positive. Blood 2014, 124, 3501. [Google Scholar] [CrossRef]
- Blanco, E.M.; Curry, C.V.; Lu, X.Y.; Sarabia, S.F.; Redell, M.S.; Lopez-Terrada, D.H.; Roy, A. Cytogenetically cryptic and FISH-negative PML/RARA rearrangement in acute promyelocytic leukemia detected only by PCR: An exceedingly rare phenomenon. Cancer Genet. 2014, 207, 48–49. [Google Scholar] [CrossRef]
- Wang, Y.F.; Liu, J.; Dong, F.; Tian, L.; Wang, J.; Xi, L.Y.; Ke, X.Y. The study of one case of APL with rare cryptic PML-RARalpha fusion gene and the literatures review. Zhonghua Xue Ye Xue Za Zhi 2016, 37, 1001–1002. [Google Scholar]
- Zhang, Z.; Xu, Y.; Jiang, M.; Kong, F.; Chen, Z.; Liu, S.; Li, F. Identification of a new cryptic PML-RARalpha fusion gene without t(15;17) and biallelic CEBPA mutation in a case of acute promyelocytic leukemia: A case detected only by RT-PCR but not cytogenetics and FISH. Cancer Biol. Ther. 2020, 21, 309–314. [Google Scholar] [CrossRef]
- Schultz, M.J.; Blackburn, P.R.; Cogbill, C.H.; Pitel, B.A.; Smadbeck, J.B.; Johnson, S.H.; Vasmatzis, G.; Rech, K.L.; Sukov, W.R.; Greipp, P.T.; et al. Characterization of a cryptic PML-RARA fusion by mate-pair sequencing in a case of acute promyelocytic leukemia with a normal karyotype and negative RARA FISH studies. Leuk. Lymphoma 2020, 61, 975–978. [Google Scholar] [CrossRef]
- Avgerinou, G.; Katsibardi, K.; Filippidou, M.; Tzanoudaki, M.; Papadhimitriou, S.I.; Kattamis, A. Cytogenetically cryptic and fish negative PML/RARA rearrangement in acute promyelocytic leukemia detected by RT-PCR. Leuk. Lymphoma 2020, 61, 3526–3528. [Google Scholar] [CrossRef]
- Arumugam, J.R.; Karthik Bommannan, B.K.; Kalaiyarasi, J.P.; Sundersingh, S. Cytogenetics and FISH negative cryptic acute promyelocytic leukemia with CD56 expression. Indian. J. Pathol. Microbiol. 2021, 64, 406–409. [Google Scholar] [CrossRef]
- Karlin, K.; Bryke, C.; Dias, A.; Michaels, P. Cytogenetically cryptic PML::RARA fusion in acute promyelocytic leukemia: Testing strategies in the modern era. Leuk. Res. Rep. 2022, 17, 100320. [Google Scholar] [CrossRef]
- Mohebnasab, M.; Li, P.; Hong, B.; Dunlap, J.; Traer, E.; Fan, G.; Press, R.D.; Moore, S.R.; Xie, W. Cytogenetically Cryptic Acute Promyelocytic Leukemia: A Diagnostic Challenge. Int. J. Mol. Sci. 2023, 24, 13075. [Google Scholar] [CrossRef]
- George, G.V.; Elsadawi, M.; Evans, A.G.; Ali, S.; Zhang, B.; Iqbal, M.A. Utilization of RT-PCR and Optical Genome Mapping in Acute Promyelocytic Leukemia with Cryptic PML::RARA Rearrangement: A Case Discussion and Systemic Literature Review. Genes. 2024, 16, 7. [Google Scholar] [CrossRef]
- Goldschmidt, N.; Yehuda-Gafni, O.; Abeliovich, D.; Slyusarevsky, E.; Rund, D. Interstitial insertion of RARalpha gene into PML gene in a patient with acute promyelocytic leukemia (APL) lacking the classic t(15;17). Hematology 2010, 15, 332–337. [Google Scholar] [CrossRef]
- Campbell, L.J.; Oei, P.; Brookwell, R.; Shortt, J.; Eaddy, N.; Ng, A.; Chew, E.; Browett, P. FISH detection of PML-RARA fusion in ins(15;17) acute promyelocytic leukaemia depends on probe size. Biomed. Res. Int. 2013, 2013, 164501. [Google Scholar] [CrossRef]
- Carvalho, C.M.; Lupski, J.R. Mechanisms underlying structural variant formation in genomic disorders. Nat. Rev. Genet. 2016, 17, 224–238. [Google Scholar] [CrossRef]
- Weckselblatt, B.; Rudd, M.K. Human Structural Variation: Mechanisms of Chromosome Rearrangements. Trends Genet. 2015, 31, 587–599. [Google Scholar] [CrossRef]
- Brockman, S.R.; Paternoster, S.F.; Ketterling, R.P.; Dewald, G.W. New highly sensitive fluorescence in situ hybridization method to detect PML/RARA fusion in acute promyelocytic leukemia. Cancer Genet. Cytogenet. 2003, 145, 144–151. [Google Scholar] [CrossRef]
- Bennour, A.; Tabka, I.; Youssef, Y.B.; Zaier, M.; Hizem, S.; Khelif, A.; Saad, A.; Sennana, H. A PML/RARA chimeric gene on chromosome 12 in a patient with acute promyelocytic leukemia (M4) associated with a new variant translocation: T(12;15;17)(q24;q24;q11). Med. Oncol. 2013, 30, 409. [Google Scholar] [CrossRef]
- Welch, J.S.; Westervelt, P.; Ding, L.; Larson, D.E.; Klco, J.M.; Kulkarni, S.; Wallis, J.; Chen, K.; Payton, J.E.; Fulton, R.S.; et al. Use of whole-genome sequencing to diagnose a cryptic fusion oncogene. JAMA 2011, 305, 1577–1584. [Google Scholar] [CrossRef]
- Weckselblatt, B.; Hermetz, K.E.; Rudd, M.K. Unbalanced translocations arise from diverse mutational mechanisms including chromothripsis. Genome Res. 2015, 25, 937–947. [Google Scholar] [CrossRef]
- Liquori, A.; Ibanez, M.; Sargas, C.; Sanz, M.A.; Barragan, E.; Cervera, J. Acute Promyelocytic Leukemia: A Constellation of Molecular Events around a Single PML-RARA Fusion Gene. Cancers 2020, 12, 624. [Google Scholar] [CrossRef]
- Fasan, A.; Haferlach, C.; Perglerova, K.; Kern, W.; Haferlach, T. Molecular landscape of acute promyelocytic leukemia at diagnosis and relapse. Haematologica 2017, 102, e222–e224. [Google Scholar] [CrossRef]
- Iaccarino, L.; Ottone, T.; Alfonso, V.; Cicconi, L.; Divona, M.; Lavorgna, S.; Travaglini, S.; Ferrantini, A.; Falconi, G.; Baer, C.; et al. Mutational landscape of patients with acute promyelocytic leukemia at diagnosis and relapse. Am. J. Hematol. 2019, 94, 1091–1097. [Google Scholar] [CrossRef] [PubMed]
| Method | Application | Abnormalities Detected | APL Yield | STAT TAT | Causes of Error | Availability | Cost |
|---|---|---|---|---|---|---|---|
| Karyotyping | Standard of care | t(15;17) and all other chromosomal alterations | >96% | 2 days | Poor metaphase quality, culture failure, submicroscopic cryptic translocations | Widely available | Low |
| FISH–PML/RARA DCDF | Standard of care; Diagnostic confirmation | Typical and atypical PML::RARA fusion | ~98% | 3 h | Hybridization failure, signal overlap, small insertions leading to absent or weak signals | Widely available | Low–Moderate |
| FISH–RARA BAP | Standard of care; Diagnostic confirmation | RARA rearrangements (PML and non-PML partners) | ~100% | 3 h | Hybridization failure, signal overlap, small insertions leading to absent or weak signals | Widely available | Low–Moderate |
| RT-PCR | Diagnostic confirmation | PML::RARA transcript isoforms | >95% | 4 h | Sample contamination, primer errors due to novel breakpoints | Widely available | Low–Moderate |
| qRT-PCR | MRD monitoring | PML::RARA transcript isoforms | >95% | 4 h | Sample contamination, primer errors due to novel breakpoints | Widely available | Low–Moderate |
| RT-PCR + cDNA sequencing | Diagnostic confirmation | PML::RARA transcript isoforms, fusion breakpoints | >95% | 2 days | Sequencing errors, mis-priming, low-quality RNA | Moderate | Moderate |
| tCGH/cCGH/Mpseq | Advanced genomic assessment for rare cases (at low resolution) | PML::RARA transcript isoforms; additional non-diagnostic genomic aberrations | Unknown, very limited studies to date | 3 days | Undetectable balanced rearrangements and unmapped sequences, oversight of low-level mosaicism, inaccurate CNV calls | Limited | Moderate |
| OGM | Advanced genomic assessment for rare cases (at medium resolution) | PML::RARA and non-PML::RARA transcript isoforms; fusion breakpoints; additional non-diagnostic genomic aberrations | Unknown, requires further validation for standard APL diagnostics | 3~5 days | Poor recovery of HMW DNA, complex rearrangement miscalls, undetectable polyploidy, unresolved centromeric/p-arm rearrangements, missed low-level mosaicism | Limited | Moderate–high |
| WGS/WES | Advanced genomic assessment for rare cases (at high resolution) | PML::RARA and non-PML::RARA transcript isoforms; fusion breakpoints; additional non-diagnoatic genomic aberrations | Unknown, primarily used in research or complex cases rather than routine APL diagnosis | 3~5 days | Sequencing artifacts, bioinformatics miscalls, over-sight of low-level mosaicism, variant validation challenges, interpretation challenges, conditional detection of structural rearrangements | Limited | High |
| Tier | Methods | Clinical Relevance |
|---|---|---|
| First line | FISH (DCDF and BAP), Karypotying, RT-PCR | High-yield, fast, easily accessible |
| Advanced | OGM, CMA, targeted sequencing, long read sequencing | Complex cases |
| High-resolution/ research-oriented | WGS/WES, other omics | High-complexity cases, with inconclusive results |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Delikkaya, B.N.; Eberle-Singh, J.; Morton, A.B.; Gong, J.Z.; Liu, J. Uncovering the PML::RARA Fusion in Cytogenetically Cryptic and FISH-Negative Acute Promyelocytic Leukemia—A Case Report and Comprehensive Literature Review. Genes 2025, 16, 1159. https://doi.org/10.3390/genes16101159
Delikkaya BN, Eberle-Singh J, Morton AB, Gong JZ, Liu J. Uncovering the PML::RARA Fusion in Cytogenetically Cryptic and FISH-Negative Acute Promyelocytic Leukemia—A Case Report and Comprehensive Literature Review. Genes. 2025; 16(10):1159. https://doi.org/10.3390/genes16101159
Chicago/Turabian StyleDelikkaya, Busra N., Jaime Eberle-Singh, Arianna B. Morton, Jerald Z. Gong, and Jinglan Liu. 2025. "Uncovering the PML::RARA Fusion in Cytogenetically Cryptic and FISH-Negative Acute Promyelocytic Leukemia—A Case Report and Comprehensive Literature Review" Genes 16, no. 10: 1159. https://doi.org/10.3390/genes16101159
APA StyleDelikkaya, B. N., Eberle-Singh, J., Morton, A. B., Gong, J. Z., & Liu, J. (2025). Uncovering the PML::RARA Fusion in Cytogenetically Cryptic and FISH-Negative Acute Promyelocytic Leukemia—A Case Report and Comprehensive Literature Review. Genes, 16(10), 1159. https://doi.org/10.3390/genes16101159






