Genome-Wide Association Studies of Key Traits in Apis cerana cerana (Hymenoptera: Apidae) from Guizhou Province
Abstract
1. Introduction
2. Materials and Methods
2.1. Source of Experimental A. cerana cerana Workers
2.2. Phenotypic Measurement of A. cerana cerana
2.3. Genomic DNA Extraction and Resequencing Library Construction
2.4. Sequence Alignment and SNP Detection
2.5. Analysis of Kinship Among Individuals
2.6. Genome-Wide Association Study
u~MVN, (0, AT1K), ~MVNn (0, T1)
3. Results
3.1. Phenotypic Trait Measurements
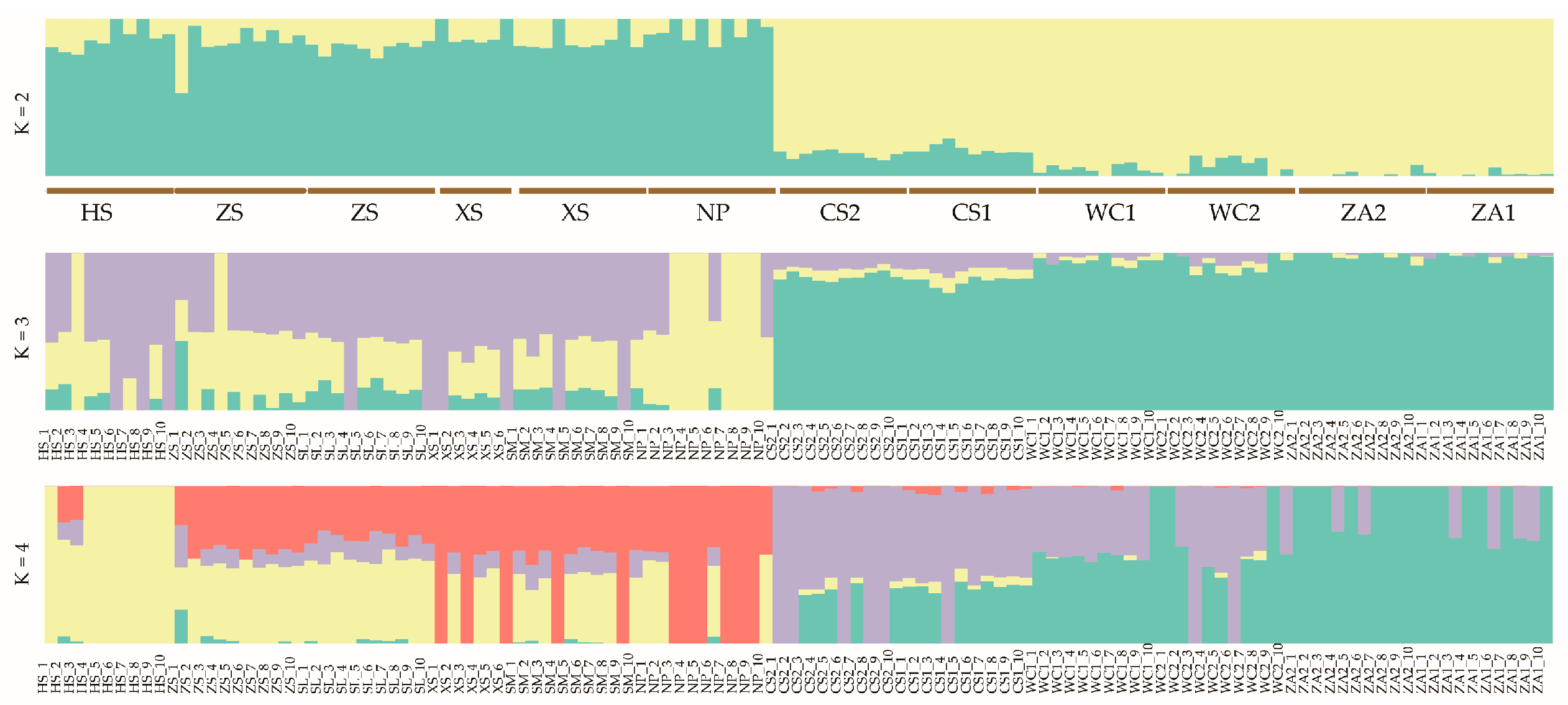
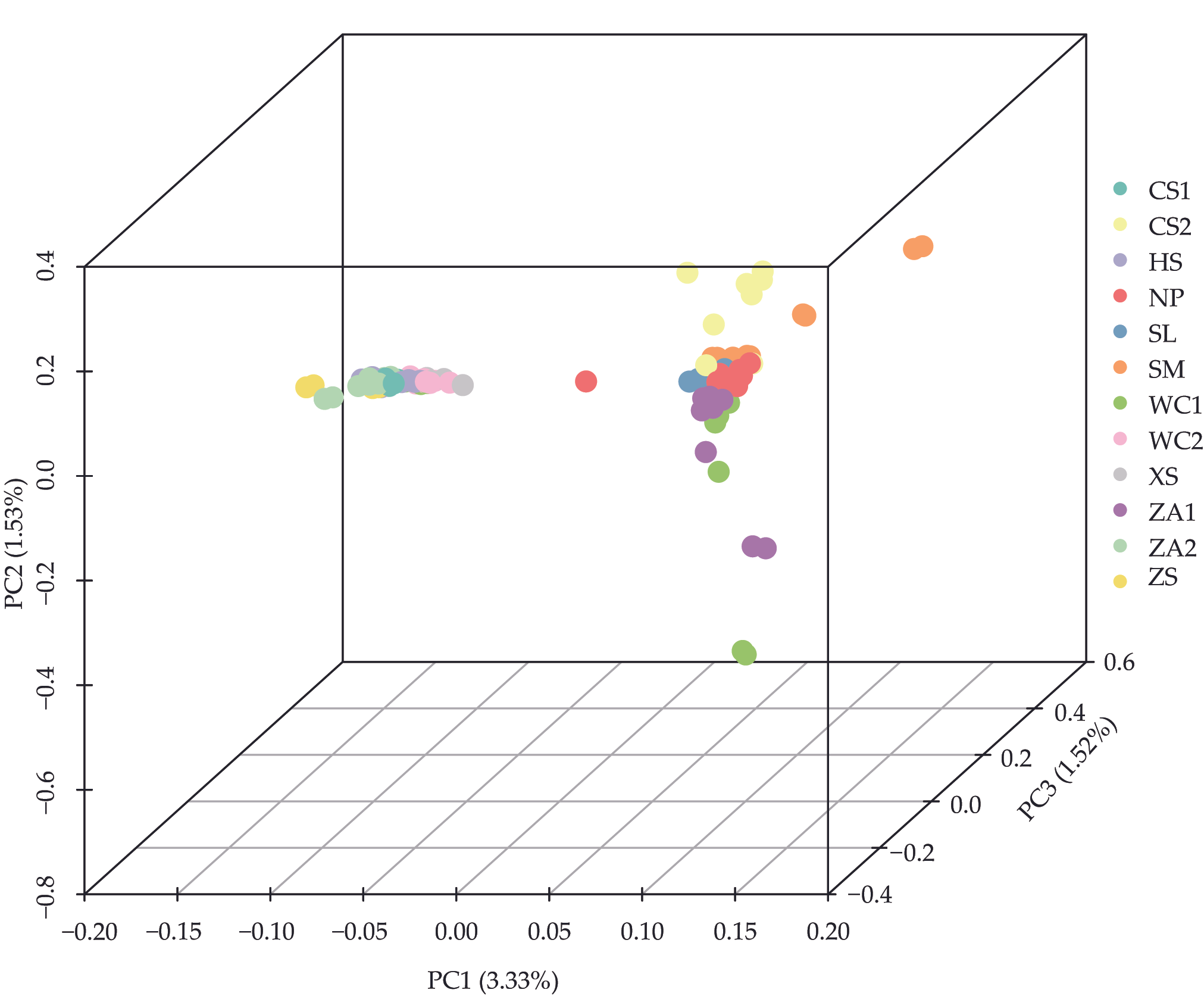
3.2. Genetic Evolution and Population Structure Analysis
3.3. Linkage Disequilibrium Analysis
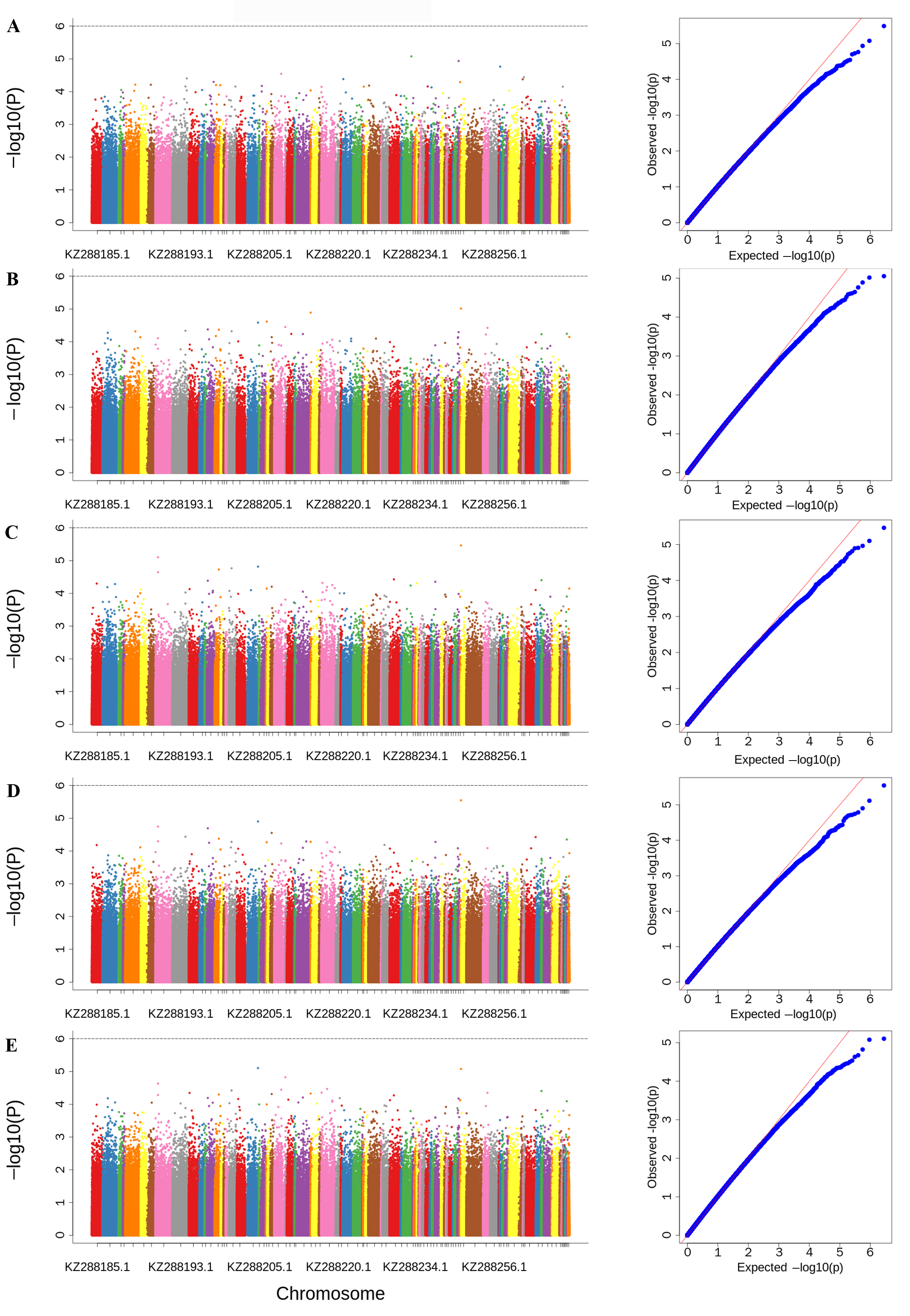
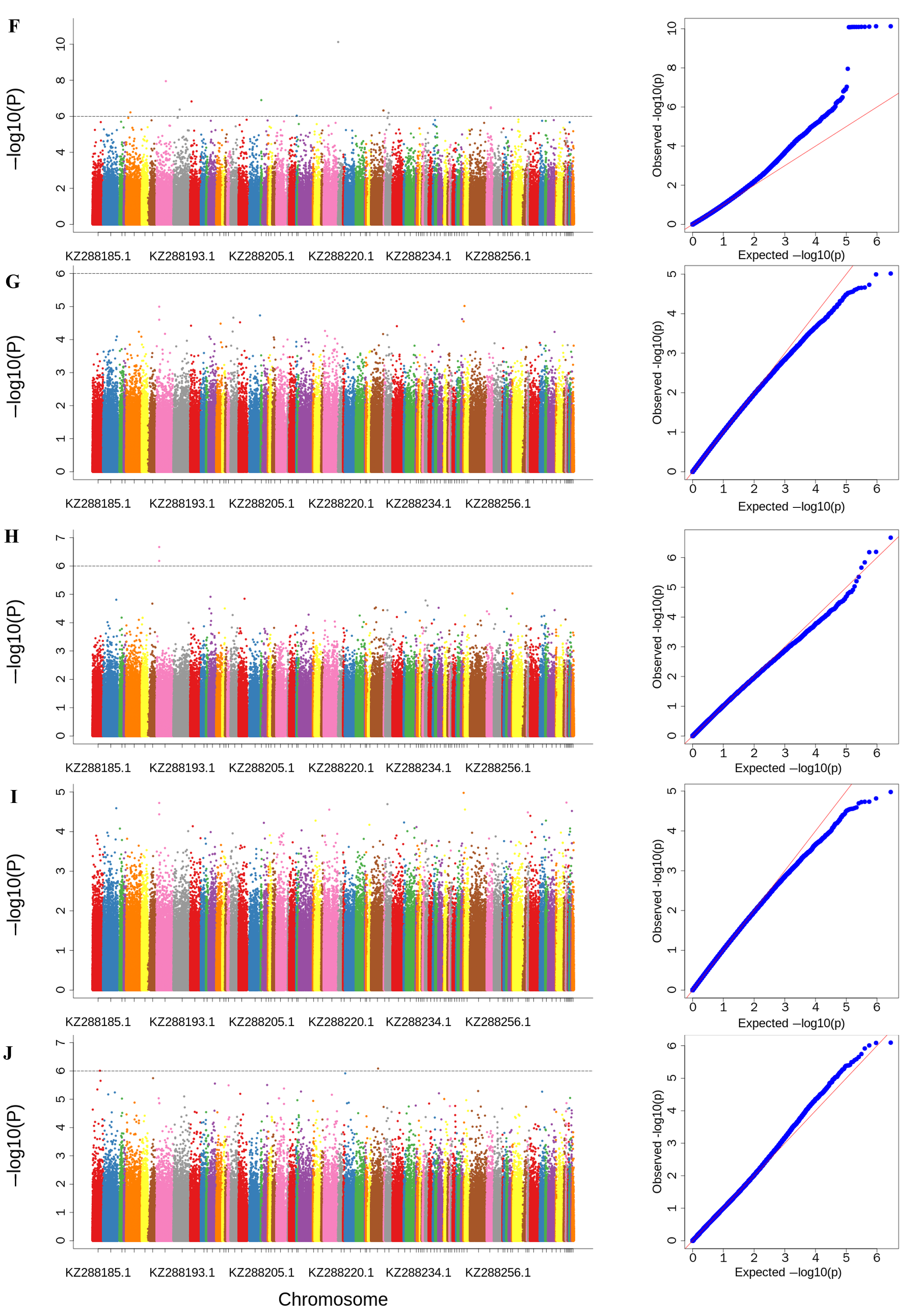
3.4. Genome-Wide Association Studies
3.5. Candidate Gene Screening
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Patel, V.; Pauli, N.; Biggs, E.; Barbour, L.; Boruff, B. Why bees are critical for achieving sustainable development. AMBIO 2020, 50, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.; Zhou, J.; Song, H.L.; Wu, Y.J.; Lan, L.; Tang, X.Y.; Ma, Z.A.; Vossbrinck, C.R.; Vossbrinck, B.; Zhou, Z.Y.; et al. Genomic analysis of Asian honeybee populations in China reveals evolutionary relationships and adaptation to abiotic stress. Ecol. Evol. 2020, 10, 13427–13438. [Google Scholar] [CrossRef]
- Chen, C.; Wang, H.H.; Liu, Z.G.; Chen, X.; Tang, J.; Meng, F.M.; Shi, W. Population genomics provides insights into the evolution and adaptation of the eastern honey bee (Apis cerana). Mol. Biol. Evol. 2018, 35, 2260–2271. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, H.; Wang, Z.; Jie, H.L.; Gao, F.C.; Cai, M.Q.; Wang, K.; Chen, D.F.; Guo, R.; Lin, Z.G.; et al. A key gene for the climatic adaptation of Apis cerana populations in China according to selective sweep analysis. BMC Genom. 2023, 24, 100. [Google Scholar] [CrossRef]
- Lander, E.S. The new genomics: Global views of biology. Science 1996, 274, 536–539. [Google Scholar] [CrossRef]
- Vignal, A.; Milan, D.; SanCristobal, M.; Eggen, A. A review on SNP and other types of molecular markers and their use in animal genetics. Genet. Sel. Evol. 2002, 34, 275–305. [Google Scholar] [CrossRef]
- Würschum, T.; Langer, S.M.; Longin, C.F.; Korzun, V.; Akhunov, E.; Ebmeyer, E.; Schachschneider, R.; Schacht, J.; Kazman, E.; Reif, J.C. Population structure, genetic diversity and linkage disequilibrium in elite winter wheat assessed with SNP and SSR markers. Theor. Appl. Genet. 2013, 126, 1477–1486. [Google Scholar] [CrossRef]
- Xia, X.T.; Zhang, S.J.; Zhang, H.J.; Zhang, Z.J.; Chen, N.B.; Li, Z.G.; Sun, H.X.; Liu, X.; Lyu, S.J.; Wang, X.W.; et al. Assessing genomic diversity and signatures of selection in Jiaxian Red cattle using whole-genome sequencing data. BMC Genom. 2021, 22, 43. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Fan, A.P.; Wang, W.S.; Zhang, J.C.; Jiang, X.J.; Ma, R.J.; Jia, S.Q.; Liu, F.; Lei, C.C.; Huang, Y.Z. Analysis of genetic diversity and genetic structure of Qinchuan cattle conservation population using whole-genome resequencing. Hereditas 2023, 45, 602–616. [Google Scholar] [CrossRef]
- QI, L.N.; Lu, X.L.; Yang, K.X.; Zhou, J.; Li, X.Y.; Liu, K.; Zhang, W.G.; Gu, X.; Zhang, W.J. Analysis of genetic diversity and genetic structure of new Pudong chicken based on SNP chips. Acta Vet. Zootech. Sin. 2023, 12, 4962–4971. [Google Scholar]
- Jalving, R.; van’t Slot, R.; van Oost, B.A. Chicken single nucleotide polymorphism identification and selection for genetic mapping. Poult. Sci. 2004, 83, 1925–1931. [Google Scholar] [CrossRef] [PubMed]
- Tolone, M.; Sardina, M.T.; Criscione, A.; Lasagna, E.; Senczuk, G.; Rizzuto, I.; Riggio, S.; Moscarelli, A.; Macaluso, V.; Di Gerlando, R.; et al. High-density single nucleotide polymorphism markers reveal the population structure of 2 local chicken genetic resources. Poult. Sci. 2023, 102, 102692. [Google Scholar] [CrossRef]
- Harpur, B.A.; Kent, C.F.; Molodtsova, D.; Lebon, J.M.D.; Alqarni, A.S.; Owayss, A.A.; Zayed, A. Population genomics of the honey bee reveals strong signatures of positive selection on worker traits. Proc. Natl. Acad. Sci. USA 2014, 111, 2614–2619. [Google Scholar] [CrossRef]
- Guichard, M.; Dainat, B.; Eynard, S.; Vignal, A.; Servin, B.; Neuditschko, M. Identification of quantitative trait loci associated with calmness and gentleness in honey bees using whole-genome sequences. Anim. Genet. 2021, 52, 472–481. [Google Scholar] [CrossRef]
- Zayed, A.; Whitfield, C.W. A genome-wide signature of positive selection in ancient and recent invasive expansions of the honey bee Apis mellifera. Proc. Natl. Acad. Sci. USA 2008, 105, 3421–3426. [Google Scholar] [CrossRef]
- Spötter, A.; Gupta, P.; Nürnberg, G.; Reinsch, N.; Bienefeld, K. Development of a 44K SNP assay focussing on the analysis of a varroa-specific defence behaviour in honey bees (Apis mellifera carnica). Mol. Ecol. Resour. 2012, 12, 323–332. [Google Scholar] [CrossRef]
- Bessoltane, N.; Toffano-Nioche, C.; Solignac, M.; Mougel, F. Fine scale analysis of crossover and non-crossover and detection of recombination sequence motifs in the honeybee (Apis mellifera). PLoS ONE 2012, 7, e36229. [Google Scholar] [CrossRef]
- Parejo, M.; Wragg, D.; Gauthier, L.; Vignal, A.; Neumann, P.; Neuditschko, M. Using whole-genome sequence information to foster conservation efforts for the European dark honey bee, Apis mellifera mellifera. Front. Ecol. Evol. 2016, 4, 140. [Google Scholar] [CrossRef]
- Hassanyar, A.K.; Nie, H.; Li, Z.; Lin, Y.; Huang, J.; Woldegiorgis, S.T.; Hussain, M.; Feng, W.; Zhang, Z.; Yu, K.; et al. Discovery of SNP molecular markers and candidate genes associated with sacbrood virus resistance in Apis cerana cerana larvae by whole-genome resequencing. Int. J. Mol. Sci. 2023, 24, 6238. [Google Scholar] [CrossRef] [PubMed]
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; de Vries, J.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome-wide association studies. Nat. Rev. Methods Primers 2021, 1, 59. [Google Scholar] [CrossRef]
- Tam, V.; Patel, N.; Turcotte, M.; Bossé, Y.; Paré, G.; Meyre, D. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef]
- Li, C.; Cai, W.T.; Zhou, C.H.; Yin, H.W.; Zhang, Z.Q.; Loor, J.J.; Sun, D.X.; Zhang, Q.; Liu, J.F.; Zhang, S.L. RNA-Seq reveals 10 novel promising candidate genes affecting milk protein concentration in the Chinese Holstein population. Sci. Rep. 2016, 6, 26813. [Google Scholar] [CrossRef]
- Jiang, J.P.; Liu, L.; Gao, Y.H.; Shi, L.J.; Li, Y.H.; Liang, W.J.; Sun, D.X. Determination of genetic associations between indels in 11 candidate genes and milk composition traits in Chinese Holstein population. BMC Genet. 2019, 20, 48. [Google Scholar] [CrossRef] [PubMed]
- Park, D.; Jung, J.W.; Choi, B.S.; Jayakodi, M.; Lee, J.; Lim, J.; Yu, Y.; Choi, Y.S.; Lee, M.L.; Park, Y.; et al. Uncovering the novel characteristics of Asian honey bee, Apis cerana, by whole genome sequencing. BMC Genom. 2015, 16, 1. [Google Scholar] [CrossRef]
- Diao, Q.Y.; Sun, L.X.; Zheng, H.J.; Zeng, Z.J.; Wang, S.Y.; Xu, S.F.; Zheng, H.Q.; Chen, Y.P.; Shi, Y.Y.; Wang, Y.Z.; et al. Genomic and transcriptomic analysis of the Asian honeybee Apis cerana provides novel insights into honeybee biology. Sci. Rep. 2018, 8, 822. [Google Scholar] [CrossRef]
- Wang, Z.L.; Zhu, Y.Q.; Yan, Q.; Yan, W.Y.; Zheng, H.J.; Zeng, Z.J. A chromosome-scale assembly of the Asian honeybee Apis cerana genome. Front. Genet. 2020, 11, 279. [Google Scholar] [CrossRef]
- Radwan, J.; Babik, W. The genomics of adaptation. Proc. R. Soc. B 2012, 279, 5024–5028. [Google Scholar] [CrossRef]
- Darragh, K.; Kay, K.M.; Ramírez, S.R. The convergent evolution of hummingbird pollination results in repeated floral scent loss through gene downregulation. Mol. Biol. Evol. 2025, 42, msaf027. [Google Scholar] [CrossRef]
- Burger, H.; Dötterl, S.; Ayasse, M. Host-plant finding and recognition by visual and olfactory floral cues in an oligolectic bee. Funct. Ecol. 2010, 24, 1234–1240. [Google Scholar] [CrossRef]
- He, Y. GWAS Analysis and Morphological Diversity of Production Traits of Apis cerana cerana in East China. Master’s Thesis, Shandong Agricultural University, Tai’an, China, 2021. [Google Scholar]
- Ruttner, F. Biogeography and Taxonomy of Honeybees; Springer: Heidelberg, Germany, 1987. [Google Scholar]
- Tan, K.; Meixner, M.D.; Fuchs, S.; Zhang, X.; He, S.; Kandemir, I.; Sheppard, W.S.; Koeniger, N. Geographic distribution of the eastern honeybee, Apis cerana (Hymenoptera: Apidae), across ecological zones in China: Morphological and molecular analyses. Syst. Biodivers. 2006, 4, 473–482. [Google Scholar] [CrossRef]
- Wang, Y.C.; Zeng, B.; Deng, M.Q.; Zhao, T.; Liao, Y.; Ren, R.Q.; Wang, H.; Yuan, Y. Whole-genome resequencing reveals genetic diversity and adaptive evolution in Chinese honeybee (Apis cerana cerana) in Guizhou, China. Front. Genet. 2024, 15, 1352455. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Li, L.; Li, Y.F.; Ma, Q.; Liu, S.Q.; Ma, Y.H.; Jiang, L. Analysis of family structure and paternity test of Tan sheep in Yanchi area, China. Animals 2022, 12, 3099. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Dong, S.S.; Xu, J.Y.; He, W.M.; Yang, T.L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef]
- Zondervan, K.T.; Cardon, L.R. The complex interplay among factors that influence allelic association. Nat. Rev. Genet. 2004, 5, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Bellido, A.; Merriam, J.R. Clonal parameters of tergite development in Drosophila. Dev. Biol. 1971, 26, 264–276. [Google Scholar] [CrossRef]
- Li, N.N.; Tong, X.L.; Zeng, J.; Meng, G.; Sun, F.Z.; Hu, H.; Song, J.B.; Lu, C.; Dai, F.Y. Hippo pathway regulates somatic development and cell proliferation of silkworm. Genomics 2019, 111, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.B. Bmdelta phenotype implies involvement of Notch signaling in body segmentation and appendage development of silkworm, Bombyx mori. Arthropod Struct. Dev. 2013, 42, 143–151. [Google Scholar] [CrossRef]
- Wang, G.L.; Gutzwiller, L.; Li-Kroeger, D.; Gebelein, B. A Hox complex activates and potentiates the Epidermal Growth Factor signaling pathway to specify Drosophila oenocytes. PLoS Genet. 2017, 13, e1006910. [Google Scholar] [CrossRef]
- Araujo, H.; Bier, E. sog and dpp exert opposing maternal functions to modify Toll signaling and pattern the dorsoventral axis of the Drosophila embryo. Development 2000, 127, 3631–3644. [Google Scholar] [CrossRef]
- Xie, B.G.; Zhu, W.J. Role of Wnt/β-catenin signaling pathway in embryo implantation and development. Reprod. Contracept. 2013, 33, 328–332. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Z.; He, X.J.; Wang, Z.L.; Zeng, Z.J. H3K4me1 modification functions in caste differentiation in honey bees. Int. J. Mol. Sci. 2023, 24, 6217. [Google Scholar] [CrossRef] [PubMed]
- Mattu, V.K.; Verma, L.R. Comparative morphometric studies on the Indian honeybee of the north-west Himalayas. 3. Hind leg, tergites and sternites. J. Apic. Res. 1984, 23, 117–122. [Google Scholar] [CrossRef]
- Jiang, Y.; Hu, J.S.; Li, Y.H.; Tang, X.Y.; Peng, X.M.; Xie, L.X.; Song, H.L.; Zhou, Z.Y.; Xu, J.S. Comprehensive genomic analysis reveals novel transposable element-derived microRNA regulating caste differentiation in honeybees. Mol. Biol. Evol. 2025, 42, msaf074. [Google Scholar] [CrossRef]
- Li, X.Y. Study on Physiological Response and Molecular Mechanism of Bees to High Temperature and High Humidity Stress. Ph.D. Thesis, Shanxi Agricultural University, Jinzhong, China, 2020. [Google Scholar]
- Martins, J.R.; Bitondi, M.M.G. Nuclear immunolocalization of hexamerins in the fat body of metamorphosing honey bees. Insects 2012, 3, 1039–1055. [Google Scholar] [CrossRef] [PubMed]
- Martins, J.R.; Anhezini, L.; Dallacqua, R.P.; Simões, Z.L.P.; Bitondi, M.M.G. A honey bee hexamerin, HEX 70a, is likely to play an intranuclear role in developing and mature ovarioles and testioles. PLoS ONE 2011, 6, e29006. [Google Scholar] [CrossRef]
- Martins, J.R.; Morais Franco Nunes, F.; Luz Paulino Simões, Z.; Maria Gentile Bitondi, M. A honeybee storage protein gene, hex 70a, expressed in developing gonads and nutritionally regulated in adult fat body. J. Insect Physiol. 2008, 54, 867–877. [Google Scholar] [CrossRef]
- Dogantzis, K.A.; Tiwari, T.; Conflitti, I.M.; Dey, A.; Patch, H.M.; Muli, E.M.; Garnery, L.; Whitfield, C.W.; Stolle, E.; Alqarni, A.S.; et al. Thrice out of Asia and the adaptive radiation of the western honey bee. Sci. Adv. 2021, 7, eabj2151. [Google Scholar] [CrossRef]
- Jackson, J.M.; Pimsler, M.L.; Oyen, K.J.; Strange, J.P.; Dillon, M.E.; Lozier, J.D. Local adaptation across a complex bioclimatic landscape in two montane bumble bee species. Mol. Ecol. 2020, 29, 920–939. [Google Scholar] [CrossRef] [PubMed]
- Vaudo, A.D.; Dyer, L.A.; Leonard, A.S. Pollen nutrition structures bee and plant community interactions. Proc. Natl. Acad. Sci. USA 2024, 121, e2317228120. [Google Scholar] [CrossRef]
- Hoiss, B.; Krauss, J.; Potts, S.G.; Roberts, S.; Steffan-Dewenter, I. Altitude acts as an environmental filter on phylogenetic composition, traits and diversity in bee communities. Proc. R. Soc. B 2012, 279, 4447–4456. [Google Scholar] [CrossRef]
- Harder, L.D. Morphology as a predictor of flower choice by bumble bees. Ecology 1985, 66, 198–210. [Google Scholar] [CrossRef]
- Kudo, G.; Ishii, H.S.; Kawai, Y.; Kohyama, T.I. Key drivers of flowering phenology of alpine plant communities: Exploring the contributions of climatic restriction and flower-visitor composition across geographic regions. Alp. Bot. 2024, 134, 151–169. [Google Scholar] [CrossRef]
- Ogilvie, J.E.; Forrest, J.R.K. Interactions between bee foraging and floral resource phenology shape bee populations and communities. Curr. Opin. Insect Sci. 2017, 21, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Bryś, M.S.; Olszewski, K.; Strachecka, A. The relationship between pollen monodiets and the activities of proteolytic systems in the fat body and hemolymph of honeybee workers. PLoS ONE 2025, 20, e0326175. [Google Scholar] [CrossRef]
- Ren, C.S.; Chang, Z.M.; Han, L.; Chen, X.S.; Long, J.K. Higher essential amino acid and crude protein contents in pollen accelerate the oviposition and colony foundation of Bombus breviceps (Hymenoptera: Apidae). Insects 2023, 14, 203. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.S. Effects of Different Pollens on the Growth and Reproductive Performance of Bombus breviceps (Hymenoptera: Apidae). Master’s Thesis, Guizhou University, Guiyang, China, 2023. [Google Scholar]
- Ricigliano, V.A.; Williams, S.T.; Oliver, R. Effects of different artificial diets on commercial honey bee colony performance, health biomarkers, and gut microbiota. BMC Vet. Res. 2022, 18, 52. [Google Scholar] [CrossRef] [PubMed]



| Abbreviation | Sampling Site and Workers’ Repeated Number | Rearing Method | Longitude (E) | Latitude (N) | Altitude (m) |
|---|---|---|---|---|---|
| Niupeng (NP) | Fahong Village (10) | Movable-frame | 103.77 | 27.12 | 1816.40 |
| Zhongshui (ZS) | Huahongyuan Village (10) | Movable-frame | 103.85 | 27.22 | 2071.00 |
| Chishui 1 (CS1) | Malu Village (10) | Movable-frame | 105.47 | 28.18 | 866.50 |
| Chishui 2 (CS1) | Hongxin Village (10) | Movable-frame | 106.18 | 28.51 | 549.80 |
| Wuchuan 1 (WC1) | Huangyang Village (10) | Movable-frame | 108.02 | 28.66 | 929.00 |
| Wuchuan 2 (WC2) | Tongxin Village (10) | Movable-frame | 107.93 | 28.73 | 745.60 |
| Shimen (SM) | Quanfa Village (10) | Movable-frame | 104.80 | 27.00 | 2247.90 |
| Shilong (SL) | Shilong Village (10) | Movable-frame | 103.93 | 27.40 | 1964.80 |
| Zheng’an 1 (ZA1) | Jianshan Village (10) | Movable-frame | 107.45 | 28.49 | 655.00 |
| Zheng’an 2 (ZA2) | Miaoding Village (10) | Movable-frame | 107.34 | 28.41 | 1138.40 |
| Heishi (HS) | Shuiping Village (10) | Movable-frame | 104.08 | 26.79 | 2482.30 |
| Xueshan (XS) | Baimo Village (6) | Movable-frame | 104.08 | 27.20 | 2370.70 |
| Traits | Mean | Standard Deviation | Max | Min | CV (%) |
|---|---|---|---|---|---|
| PL (mm) | 4.46 | 0.27 | 4.94 | 3.76 | 0.06 |
| FL (mm) | 1.99 | 0.44 | 2.75 | 1.42 | 0.22 |
| TL (mm) | 2.31 | 0.50 | 3.01 | 1.67 | 0.22 |
| TaL (mm) | 1.54 | 0.35 | 2.05 | 1.11 | 0.23 |
| TaW (mm) | 0.96 | 0.21 | 1.31 | 0.66 | 0.22 |
| T3&4L (mm) | 2.46 | 0.14 | 2.81 | 1.49 | 0.06 |
| S3L (mm) | 1.89 | 0.37 | 2.44 | 1.33 | 0.20 |
| WML3 (mm) | 0.76 | 0.14 | 1.06 | 0.54 | 0.18 |
| WMSL3 (mm) | 1.28 | 0.26 | 1.80 | 0.69 | 0.20 |
| WMI3 (mm) | 0.31 | 0.16 | 0.83 | 0.13 | 0.51 |
| S6L (mm) | 1.85 | 0.38 | 2.36 | 1.37 | 0.21 |
| S6W (mm) | 2.29 | 0.47 | 2.95 | 1.66 | 0.21 |
| FWL (mm) | 6.91 | 1.58 | 9.29 | 3.94 | 0.23 |
| FWW (mm) | 2.45 | 0.50 | 3.13 | 1.48 | 0.21 |
| CI | 2.38 | 0.53 | 3.80 | 0.88 | 0.22 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Ren, C.; Yuan, Y.; Yang, X.; Deng, M.; Zhao, T.; Ren, R.; Liao, Y.; Wang, H.; Jiang, Z.; et al. Genome-Wide Association Studies of Key Traits in Apis cerana cerana (Hymenoptera: Apidae) from Guizhou Province. Genes 2025, 16, 1148. https://doi.org/10.3390/genes16101148
Wang Y, Ren C, Yuan Y, Yang X, Deng M, Zhao T, Ren R, Liao Y, Wang H, Jiang Z, et al. Genome-Wide Association Studies of Key Traits in Apis cerana cerana (Hymenoptera: Apidae) from Guizhou Province. Genes. 2025; 16(10):1148. https://doi.org/10.3390/genes16101148
Chicago/Turabian StyleWang, Yinchen, Changshi Ren, Yang Yuan, Xu Yang, Mengqing Deng, Tian Zhao, Rongqing Ren, Yan Liao, Hua Wang, Ziwei Jiang, and et al. 2025. "Genome-Wide Association Studies of Key Traits in Apis cerana cerana (Hymenoptera: Apidae) from Guizhou Province" Genes 16, no. 10: 1148. https://doi.org/10.3390/genes16101148
APA StyleWang, Y., Ren, C., Yuan, Y., Yang, X., Deng, M., Zhao, T., Ren, R., Liao, Y., Wang, H., Jiang, Z., Xue, X., & Fang, X. (2025). Genome-Wide Association Studies of Key Traits in Apis cerana cerana (Hymenoptera: Apidae) from Guizhou Province. Genes, 16(10), 1148. https://doi.org/10.3390/genes16101148






