GhTGA2, a Potential Key Regulator of Salt Stress Response: Insights from Genome-Wide Identification of TGA Family Genes Across Ten Cotton Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of TGA Gene Families
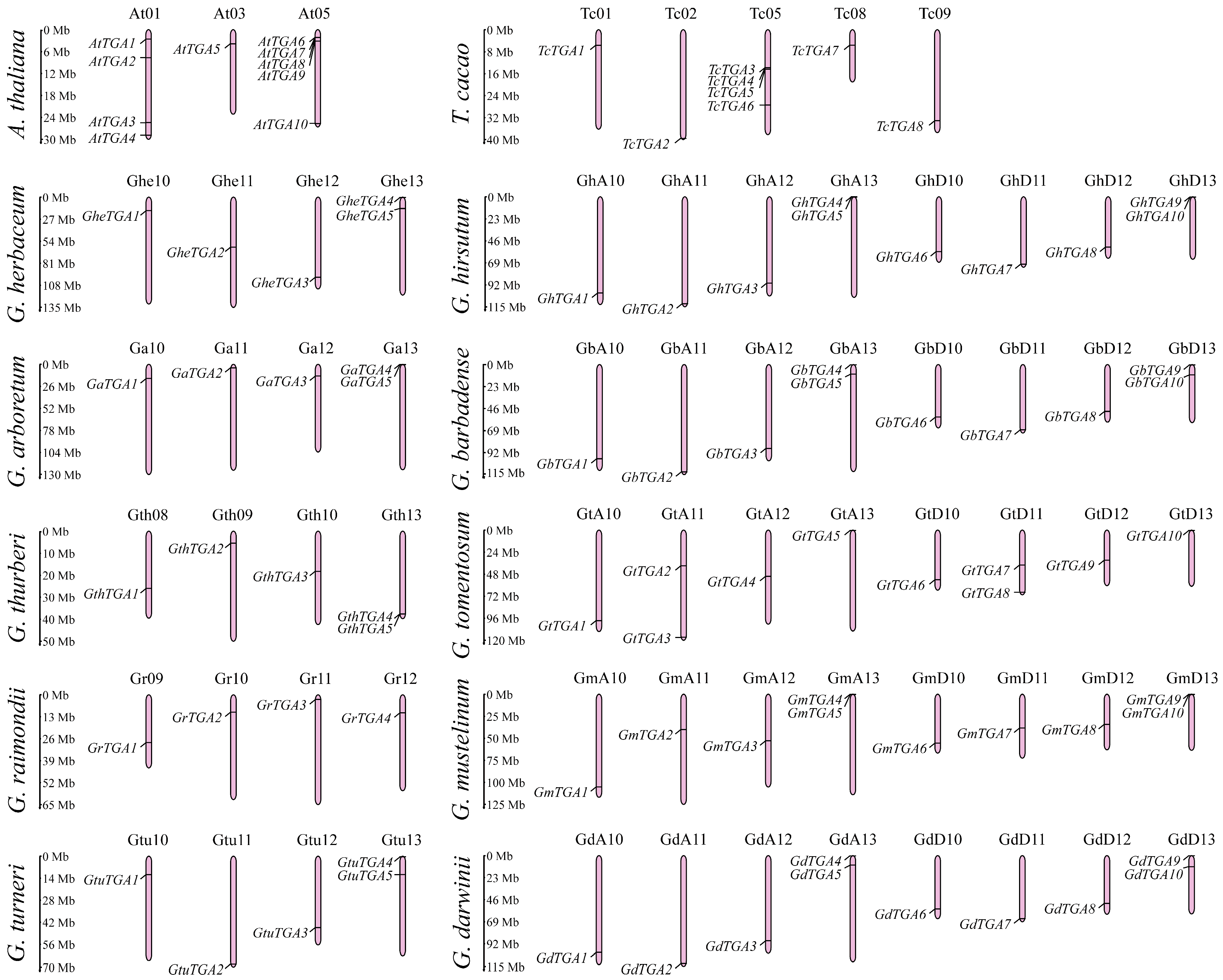
2.2. Chromosomal Localization and Properties of Proteins of the TGA Family
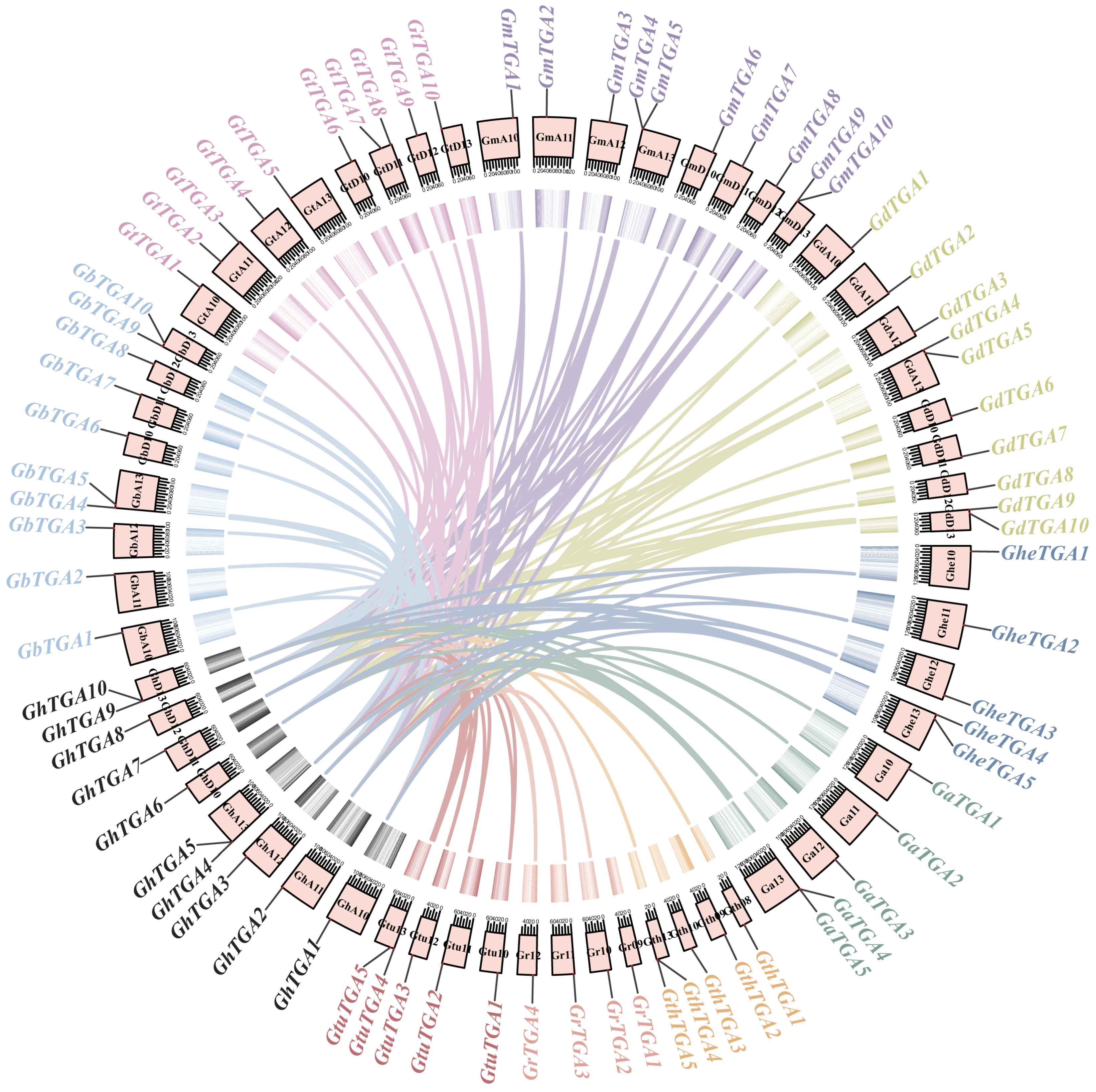
2.3. Phylogenetic and Collinearity Analysis of TGA Gene Families
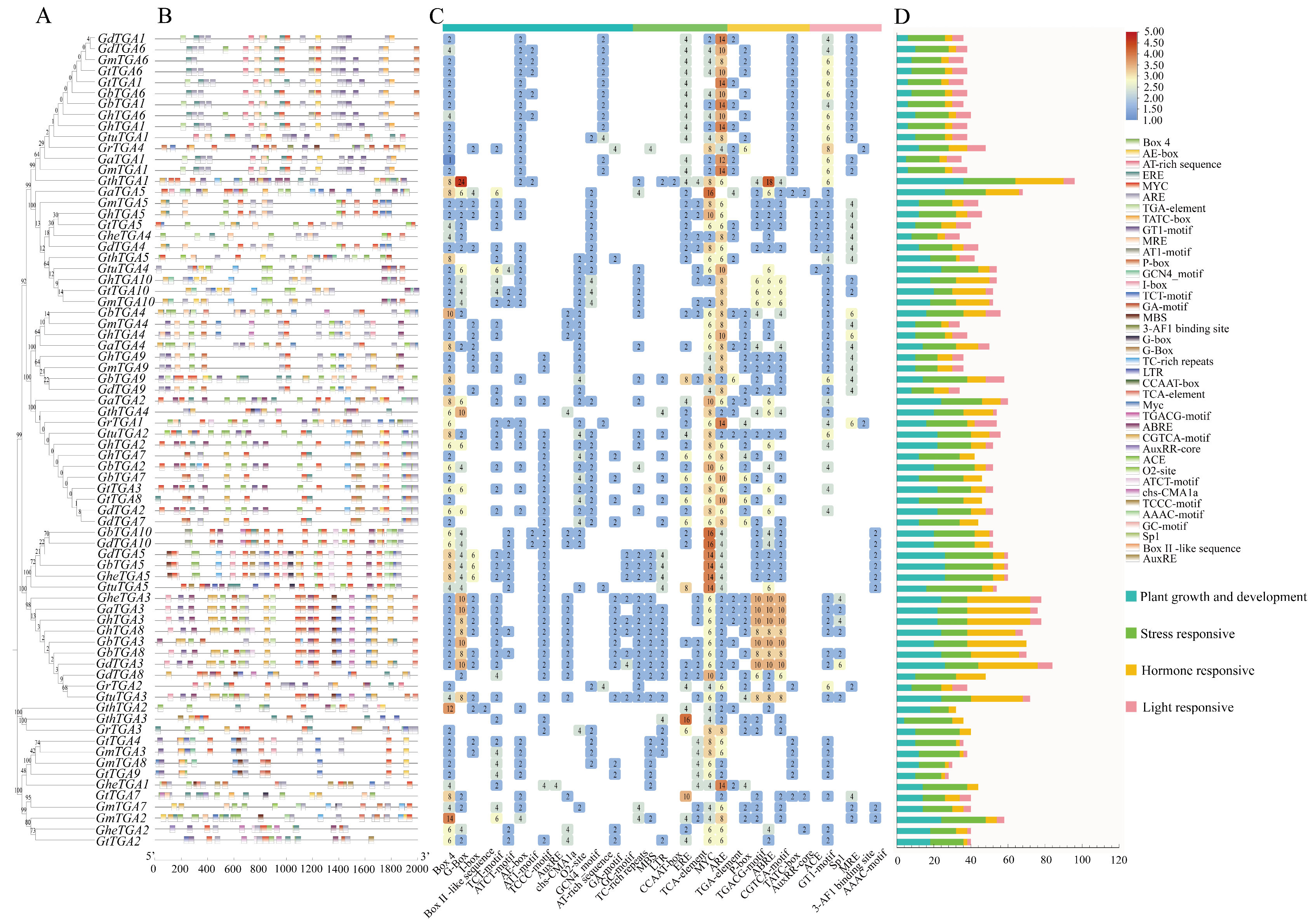
2.4. Genomic Architecture, Evolutionarily Conserved Motifs, and Structurally Preserved Domains in Cotton TGA Family
2.5. Identification of cis-Regulatory Motifs in Gossypium TGA Transcription Factor Promoters
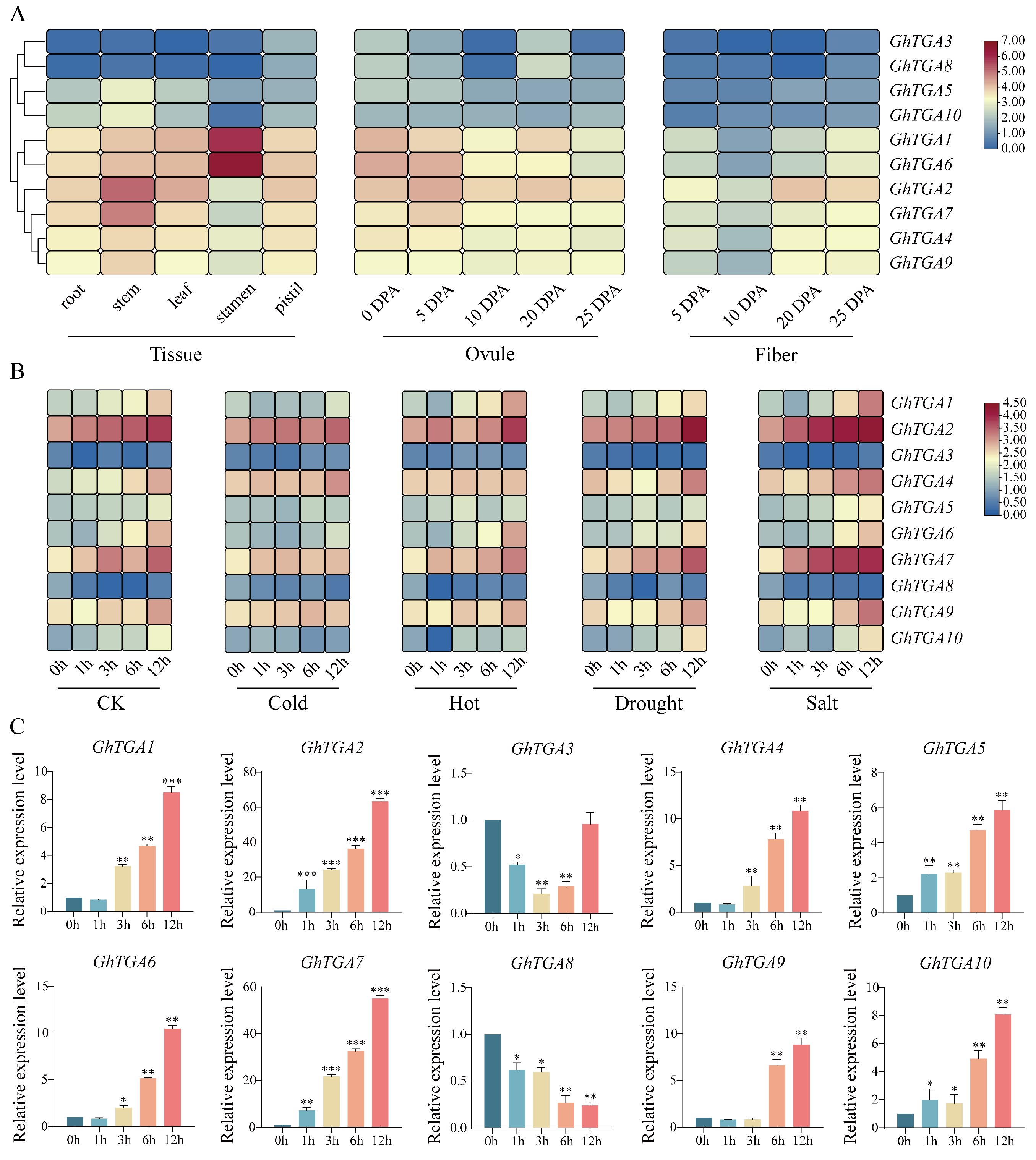
2.6. Expression Patterns of the TGA Gene Family in G. hirsutum and Their Responses to Stress
2.7. Subcellular Localization of the Candidate Gene GhTGA2
2.8. Functional Validation of GhTGA2
2.9. Statistical Analysis
3. Results
3.1. Identification and Chromosomal Localization of the TGA Gene Family
3.2. Evolutionary Relationship Analysis of TGA Gene Family
3.3. Chromosomal Collinearity Analysis of the TGA Gene Family in Gossypium
3.4. Analysis of the Gene Structure, Conserved Motifs, and Conserved Domains of the TGA Gene Family in Gossypium
3.5. Prediction of cis-Acting Elements in the Promoter Region of the TGA Gene Family of Cotton
3.6. Expression Analysis of TGA Gene Family in G. hirsutum
3.7. Subcellular Localization of GhTGA2
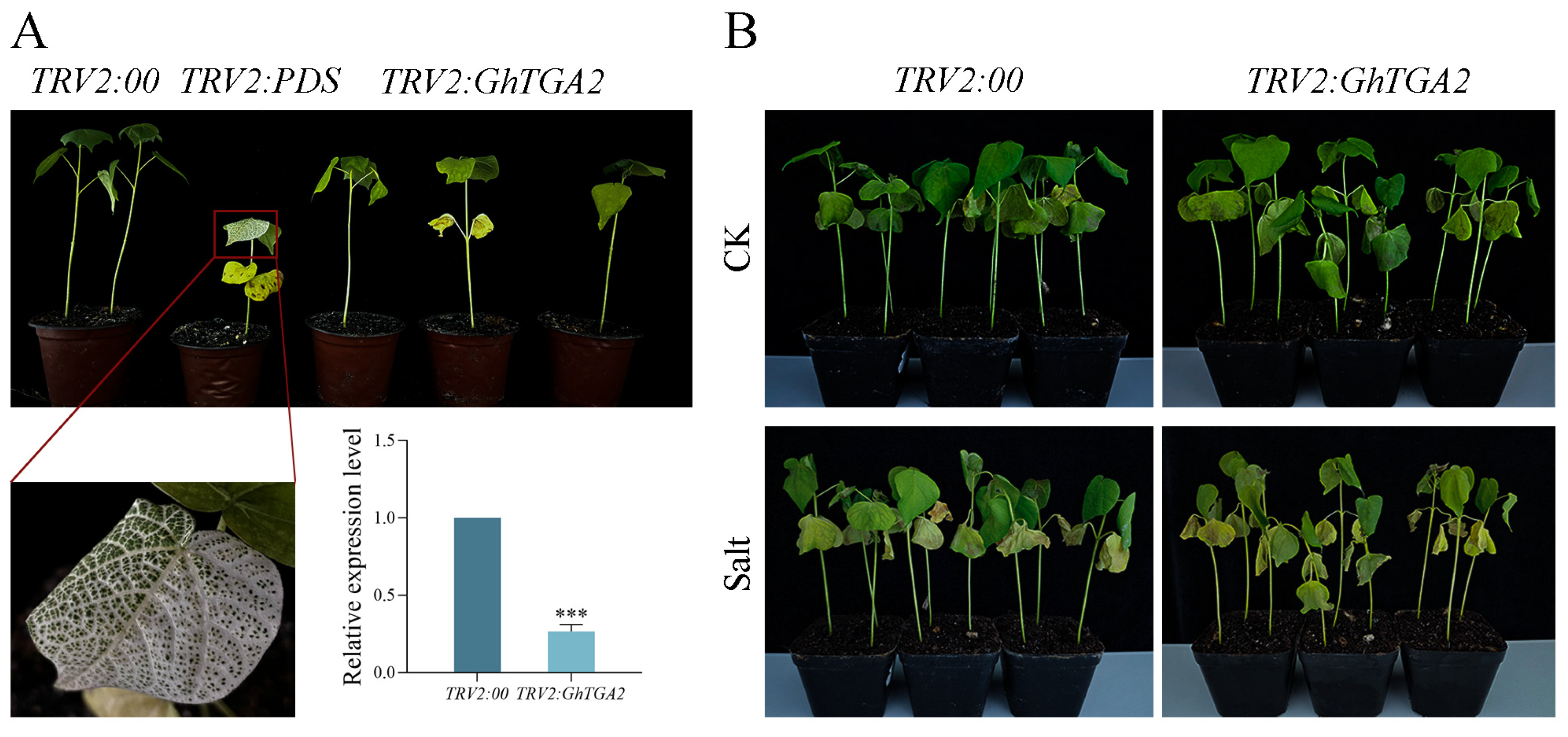
3.8. GhTGA2-Silenced Cotton Plants Showed High Sensitivity to Salt Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ahmad, I.; Zhu, G.; Zhou, G.; Song, X.; Hussein Ibrahim, M.E.; Ibrahim Salih, E.G.; Hussain, S.; Younas, M.U. Pivotal Role of Phytohormones and Their Responsive Genes in Plant Growth and Their Signaling and Transduction Pathway under Salt Stress in Cotton. Int. J. Mol. Sci. 2022, 23, 7339. [Google Scholar] [CrossRef]
- Dong, Y.; Hu, G.; Grover, C.E.; Miller, E.R.; Zhu, S.; Wendel, J.F. Parental legacy versus regulatory innovation in salt stress responsiveness of allopolyploid cotton (Gossypium) species. Plant J. 2022, 111, 872–887. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, Y.; Wan, H.; Tang, J.; Ni, Z. The sea-island cotton GbTCP4 transcription factor positively regulates drought and salt stress responses. Plant Sci. 2022, 322, 111329. [Google Scholar] [CrossRef]
- Xu, P.; Guo, Q.; Meng, S.; Zhang, X.; Xu, Z.; Guo, W.; Shen, X. Genome-wide association analysis reveals genetic variations and candidate genes associated with salt tolerance related traits in Gossypium hirsutum. BMC Genom. 2021, 22, 26. [Google Scholar] [CrossRef]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How Plant Hormones Mediate Salt Stress Responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of Plant Responses to Salt Stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef] [PubMed]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Alves, M.S.; Dadalto, S.P.; Gonçalves, A.B.; De Souza, G.B.; Barros, V.A.; Fietto, L.G. Plant bZIP transcription factors responsive to pathogens: A review. Int. J. Mol. Sci. 2013, 14, 7815–7828. [Google Scholar] [CrossRef] [PubMed]
- Foster, R.; Izawa, T.; Chua, N.H. Plant bZIP proteins gather at ACGT elements. FASEB J. 1994, 8, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Manzoor, M.A.; Manzoor, M.M.; Li, G.; Abdullah, M.; Han, W.; Wenlong, H.; Shakoor, A.; Riaz, M.W.; Rehman, S.; Cai, Y. Genome-wide identification and characterization of bZIP transcription factors and their expression profile under abiotic stresses in Chinese pear (Pyrus bretschneideri). BMC Plant Biol. 2021, 21, 413. [Google Scholar] [CrossRef]
- Liu, H.; Tang, X.; Zhang, N.; Li, S.; Si, H. Role of bZIP Transcription Factors in Plant Salt Stress. Int. J. Mol. Sci. 2023, 24, 7893. [Google Scholar] [CrossRef]
- Sun, T.; Busta, L.; Zhang, Q.; Ding, P.; Jetter, R.; Zhang, Y. TGACG-BINDING FACTOR 1 (TGA1) and TGA4 regulate salicylic acid and pipecolic acid biosynthesis by modulating the expression of SYSTEMIC ACQUIRED RESISTANCE DEFICIENT 1 (SARD1) and CALMODULIN-BINDING PROTEIN 60g (CBP60g). New Phytol. 2018, 217, 344–354. [Google Scholar] [CrossRef]
- Pontier, D.; Privat, I.; Trifa, Y.; Zhou, J.-M.; Klessig, D.F.; Lam, E. Differential regulation of TGA transcription factors by post-transcriptional control. Plant J. 2002, 32, 641–653. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, F.; Lam, E.; Chua, N.H. Two tobacco DNA-binding proteins with homology to the nuclear factor CREB. Nature 1989, 340, 727–730. [Google Scholar] [CrossRef] [PubMed]
- Bergman, M.E.; Evans, S.E.; Kuai, X.; Franks, A.E.; Despres, C.; Phillips, M.A. Arabidopsis TGA256 Transcription Factors Suppress Salicylic-Acid-Induced Sucrose Starvation. Plants 2023, 12, 3284. [Google Scholar] [CrossRef] [PubMed]
- Schiermeyer, A.; Thurow, C.; Gatz, C. Tobacco bZIP factor TGA10 is a novel member of the TGA family of transcription factors. Plant Mol. Biol. 2003, 51, 817–829. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.C.; Yin, H.M.; Zhang, X.Z.; Wang, L.L.; Cao, K.; Piao, J.P.; Liu, J.F. Genome-wide identification of the TGA transcription factor family in maize and expression analysis related to salt stress. J. Jiangxi Agric. Univ. 2023, 45, 631–642. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, W.; Wang, C.; Cai, B.; Feng, J.; Zhou, D.; Chen, Y.; Zhang, M.; Qi, D.; Wang, Z.; et al. Genome-Wide Identification and Expression Analysis of the TGA Gene Family in Banana (Musa nana Lour.) Under Various Nitrogen Conditions. Int. J. Mol. Sci. 2025, 26, 2168. [Google Scholar] [CrossRef]
- Tian, M.; Dai, Y.; Noman, M.; Li, R.; Li, X.; Wu, X.; Wang, H.; Song, F.; Li, D. Genome-wide characterization and functional analysis of the melon TGA gene family in disease resistance through ectopic overexpression in Arabidopsis thaliana. Plant Physiol. Biochem. 2024, 212, 108784. [Google Scholar] [CrossRef]
- Zeng, Q.; Gu, J.; Cai, M.; Wang, Y.; Xie, Q.; Han, Y.; Zhang, S.; Lu, L.; Chen, Y.; Zeng, Y.; et al. Genome-Wide Identification and Expression Analysis of TGA Family Genes Associated with Abiotic Stress in Sunflowers (Helianthus annuus L.). Int. J. Mol. Sci. 2024, 25, 4097. [Google Scholar] [CrossRef]
- Wang, M.; Ma, Y.; Qiu, Y.-X.; Long, S.-S.; Dai, W.-S. Genome-wide characterization and expression profiling of the TGA gene family in sweet orange (Citrus sinensis) reveal CsTGA7 responses to multiple phytohormones and abiotic stresses. Front. Plant Sci. 2025, 16, 1530242. [Google Scholar] [CrossRef]
- Duan, Y.; Xu, Z.; Liu, H.; Wang, Y.; Zou, X.; Zhang, Z.; Xu, L.; Xu, M. Genome-Wide Identification of the TGA Gene Family and Expression Analysis under Drought Stress in Brassica napus L. Int. J. Mol. Sci. 2024, 25, 6355. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Liu, Y.; Li, Z.; Wang, X.; Jiang, C.; Zhao, X.; Kang, S.; Liu, X.; Zhao, S.; Wang, J.; et al. Genome-wide analysis reveals regulatory mechanisms and expression patterns of TGA genes in peanut under abiotic stress and hormone treatments. Front. Plant Sci. 2023, 14, 1269200. [Google Scholar] [CrossRef] [PubMed]
- Idrovo Espín, F.M.; Peraza-Echeverria, S.; Fuentes, G.; Santamaría, J.M. In silico cloning and characterization of the TGA (TGACG MOTIF-BINDING FACTOR) transcription factors subfamily in Carica papaya. Plant Physiol. Biochem. 2012, 54, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Tomaž, Š.; Gruden, K.; Coll, A. TGA transcription factors-Structural characteristics as basis for functional variability. Front. Plant Sci. 2022, 13, 935819. [Google Scholar] [CrossRef]
- Lu, C.; Liu, X.; Tang, Y.; Fu, Y.; Zhang, J.; Yang, L.; Li, P.; Zhu, Z.; Dong, P. A comprehensive review of TGA transcription factors in plant growth, stress responses, and beyond. Int. J. Biol. Macromol. 2024, 258, 128880. [Google Scholar] [CrossRef]
- Fan, W.; Dong, X. In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell 2002, 14, 1377–1389. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gutsche, N.; Zachgo, S. The N-Terminus of the Floral Arabidopsis TGA Transcription Factor PERIANTHIA Mediates Redox-Sensitive DNA-Binding. PLoS ONE 2016, 11, e0153810. [Google Scholar] [CrossRef]
- Chern, M.; Bai, W.; Ruan, D.; Oh, T.; Chen, X.; Ronald, P.C. Interaction specificity and coexpression of rice NPR1 homologs 1 and 3 (NH1 and NH3), TGA transcription factors and Negative Regulator of Resistance (NRR) proteins. BMC Genom. 2014, 15, 461. [Google Scholar] [CrossRef]
- Gatz, C. From pioneers to team players: TGA transcription factors provide a molecular link between different stress pathways. Mol. Plant Microbe Interact. 2013, 26, 151–159. [Google Scholar] [CrossRef]
- Ke, D.X.; Huo, Y.Y.; Liu, Y.; Li, J.Y.; Liu, X.X. Functional analysis of GmTGA26 gene under salt stress in soybean. Acta Agron. Sin. 2022, 48, 1697–1708. [Google Scholar] [CrossRef]
- Li, X.; Zhang, X.; Guo, W.; Zhang, P.; Zhang, W. Cloning and Expression of Transcription Factor Genes Related to Fusarium Wilt Resistance in Cotton. Acta Bot. Boreali-Sin. 2017, 37, 67–73. [Google Scholar] [CrossRef]
- Yu, J.; Jung, S.; Cheng, C.-H.; Ficklin, S.P.; Lee, T.; Zheng, P.; Jones, D.; Percy, R.G.; Main, D. CottonGen: A genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 2014, 42, D1229–D1236. [Google Scholar] [CrossRef]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.; Li, Z.; Sun, Y.; Aluko, O.O.; Wu, X.; Wang, Q.; Liu, G. MG2C: A user-friendly online tool for drawing genetic maps. Mol. Hortic. 2021, 1, 16. [Google Scholar] [CrossRef]
- Zhao, D.; Gao, F.; Guan, P.; Gao, J.; Guo, Z.; Guo, J.; Cui, H.; Li, Y.; Zhang, G.; Li, Z.; et al. Identification and analysis of differentially expressed trihelix genes in maize (Zea mays) under abiotic stresses. PeerJ 2023, 11, e15312. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Duan, L.; Wang, F.; Shen, H.; Xie, S.; Chen, X.; Xie, Q.; Li, R.; Cao, A.; Li, H. Identification, evolution, and expression of GDSL-type Esterase/Lipase (GELP) gene family in three cotton species: A bioinformatic analysis. BMC Genom. 2023, 24, 795. [Google Scholar] [CrossRef]
- Zhou, X.; Su, L.; Tang, R.; Dong, Y.; Wang, F.; Li, R.; Xie, Q.; Zhang, X.; Xiao, G.; Li, H. Genome-wide analysis of Hsp40 and Hsp70 gene family in four cotton species provides insights into their involvement in response to Verticillium dahliae and abiotic stress. Front. Genet. 2023, 14, 1120861. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef]
- Su, Y.; Guo, A.; Hua, J. Discussion on Methods for Salt Tolerance Evaluation of Cotton. J. China Agric. Univ. 2021, 26, 11–19. [Google Scholar] [CrossRef]
- He, H.; Wang, J.; Meng, Z.; Dijkwel, P.P.; Du, P.; Shi, S.; Dong, Y.; Li, H.; Xie, Q. Genome-Wide Analysis of the SRPP/REF Gene Family in Taraxacum kok-saghyz Provides Insights into Its Expression Patterns in Response to Ethylene and Methyl Jasmonate Treatments. Int. J. Mol. Sci. 2024, 25, 6864. [Google Scholar] [CrossRef]
- Shimizu, K.; Suzuki, H.; Uemura, T.; Nozawa, A.; Desaki, Y.; Hoshino, R.; Yoshida, A.; Abe, H.; Nishiyama, M.; Nishiyama, C.; et al. Immune gene activation by NPR and TGA transcriptional regulators in the model monocot Brachypodium distachyon. Plant J. 2022, 110, 470–481. [Google Scholar] [CrossRef]
- Thurow, C.; Schiermeyer, A.; Krawczyk, S.; Butterbrodt, T.; Nickolov, K.; Gatz, C. Tobacco bZIP transcription factor TGA2.2 and related factor TGA2.1 have distinct roles in plant defense responses and plant development. Plant J. 2005, 44, 100–113. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Xu, X.; Xu, J.; Yuan, C.; Hu, Y.; Liu, Q.; Chen, Q.; Zhang, P.; Shi, N.; Qin, C. Characterization of genes associated with TGA7 during the floral transition. BMC Plant Biol. 2021, 21, 367. [Google Scholar] [CrossRef]
- Zhang, H.; Mao, L.; Xin, M.; Xing, H.; Zhang, Y.; Wu, J.; Xu, D.; Wang, Y.; Shang, Y.; Wei, L.; et al. Overexpression of GhABF3 increases cotton(Gossypium hirsutum L.) tolerance to salt and drought. BMC Plant Biol. 2022, 22, 313. [Google Scholar] [CrossRef]
- Jin, H.; Choi, S.-M.; Kang, M.-J.; Yun, S.-H.; Kwon, D.-J.; Noh, Y.-S.; Noh, B. Salicylic acid-induced transcriptional reprogramming by the HAC-NPR1-TGA histone acetyltransferase complex in Arabidopsis. Nucleic Acids Res. 2018, 46, 11712–11725. [Google Scholar] [CrossRef] [PubMed]
- Butterbrodt, T.; Thurow, C.; Gatz, C. Chromatin immunoprecipitation analysis of the tobacco PR-1a- and the truncated CaMV 35S promoter reveals differences in salicylic acid-dependent TGA factor binding and histone acetylation. Plant Mol. Biol. 2006, 61, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Ndamukong, I.; Abdallat, A.A.; Thurow, C.; Fode, B.; Zander, M.; Weigel, R.; Gatz, C. SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 2007, 50, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Zander, M.; Chen, S.; Imkampe, J.; Thurow, C.; Gatz, C. Repression of the Arabidopsis thaliana jasmonic acid/ethylene-induced defense pathway by TGA-interacting glutaredoxins depends on their C-terminal ALWL motif. Mol. Plant 2012, 5, 831–840. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Guo, Z.; Zhang, L.; Zhang, F.; Jiang, W.; Shen, Q.; Fu, X.; Yan, T.; Shi, P.; Hao, X.; et al. Interaction of bZIP transcription factor TGA6 with salicylic acid signaling modulates artemisinin biosynthesis in Artemisia annua. J. Exp. Bot. 2019, 70, 3969–3979. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, L.; Fan, J.; Shi, S.; Zhu, F.; Zhang, G.; Wang, J.; Li, Z.; Wang, F.; Li, H. GhTGA2, a Potential Key Regulator of Salt Stress Response: Insights from Genome-Wide Identification of TGA Family Genes Across Ten Cotton Species. Genes 2025, 16, 1143. https://doi.org/10.3390/genes16101143
Meng L, Fan J, Shi S, Zhu F, Zhang G, Wang J, Li Z, Wang F, Li H. GhTGA2, a Potential Key Regulator of Salt Stress Response: Insights from Genome-Wide Identification of TGA Family Genes Across Ten Cotton Species. Genes. 2025; 16(10):1143. https://doi.org/10.3390/genes16101143
Chicago/Turabian StyleMeng, Lu, Jiliang Fan, Shandang Shi, Faren Zhu, Ganggang Zhang, Junwei Wang, Zihan Li, Fei Wang, and Hongbin Li. 2025. "GhTGA2, a Potential Key Regulator of Salt Stress Response: Insights from Genome-Wide Identification of TGA Family Genes Across Ten Cotton Species" Genes 16, no. 10: 1143. https://doi.org/10.3390/genes16101143
APA StyleMeng, L., Fan, J., Shi, S., Zhu, F., Zhang, G., Wang, J., Li, Z., Wang, F., & Li, H. (2025). GhTGA2, a Potential Key Regulator of Salt Stress Response: Insights from Genome-Wide Identification of TGA Family Genes Across Ten Cotton Species. Genes, 16(10), 1143. https://doi.org/10.3390/genes16101143





